Abstract
Proofreading mechanisms increase specificity in biochemical reactions by allowing for the dissociation of intermediate complexes. These mechanisms disrupt and reset the reaction to undo errors at the cost of increased time of reaction and free energy expenditure. Here, we draw an analogy between proofreading and microtubule growth which share some of the features described above. Our analogy relates the statistics of growth and shrinkage of microtubules in physical space to the cycling of intermediate complexes in the space of chemical states in proofreading mechanisms. Using this analogy, we find a new kinetic regime of proofreading in which an exponential speed-up of the process can be achieved at the cost of a somewhat larger error rate. This regime is analogous to the transition region between two known growth regimes of microtubules (bounded and unbounded) and is sharply defined in the limit of large proofreading networks. We find that this advantageous regime of speed-error tradeoff might be present in proofreading schemes studied earlier in the charging of tRNA by tRNA synthetases, in RecA filament assembly on ssDNA, and in protein synthesis by ribosomes.
Keywords: enzyme-substrate reaction, nonequilibrium thermodynamics, speed-error trade-off, dynamic instability
Kinetic proofreading is a mechanism for error correction in biochemical processes, introduced in 1974 by John Hopfield (1) and independently by Jacques Ninio (2). Proofreading mechanisms enhance the effect of small differences in binding energy on reaction rates at the cost of consuming extra free energy. Such mechanisms involve a network of parallel pathways leading to product formation, in contrast to linear Michaelis-Menten-like schemes. When undesirable reactants participate in a reaction, free energy is used to drive the molecular system in cycles through these pathways, revisiting chemical intermediates many times before completing the reaction (1, 2).
Such cycling is central to the error correcting properties of such schemes. On the other hand, the cycling has a cost in the form of increased time and energy to form products. While keeping error rates low is important in biological processes like protein synthesis, DNA replication and other enzyme-substrate reactions, slowing down these processes can directly impact fitness of organisms by slowing down their growth and reproduction. The resulting speed vs accuracy tradeoff is central to many biological and evolutionary questions.
We explore an analogy between the nonequilibrium dynamics of proofreading schemes and the nonequilibrium growth of microtubules. Despite very different biological and chemical underpinnings of these two processes, there is much in common in the statistics of their fluctuations. Through this analogy, we discover that proofreading schemes naturally have two distinct kinetic regimes of operation. In the traditional regime, all reactants are cycled, minimizing errors at a large cost in time and energy. However, we identify another regime where undesirable reactants are caught up in cycling while desirable reactants can complete the reaction with little wasted time. There is a sharp transition between the two regimes in the limit of large biochemical networks but the effect persists qualitatively down to the simplest of proofreading schemes.
We conclude by connecting our work to experimental results on proofreading in tRNA charging, in the sequence-dependent assembly of RecA filaments on single stranded DNA, and in protein synthesis by ribosomes. These experiments measured differences in reaction kinetics between correct and incorrect substrates undergoing the same reaction through techniques ranging from indirect stoichiometric methods to single molecule manipulation. In light of our present study, the measurements reveal that the proofreading mechanisms might operate in a regime with shorter reaction times at only a small cost in error rate. Further experiments on reaction network kinetics for different substrates should be able to reveal precisely where such systems are positioned with respect to time-energy-error tradeoffs.
Model
Hopfield’s Kinetic Proofreading Scheme.
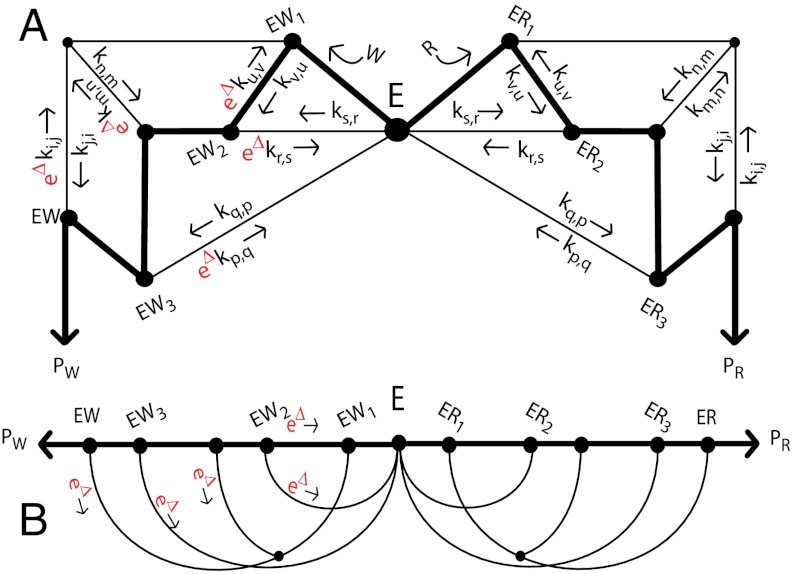
Consider the case of an enzyme E with two competing substrates: a correct or “right” one, R, and an incorrect or “wrong” one, W. (We will use S to denote either of the substrates R, W). Enzyme E can bind to R forming two intermediate complexes, ER∗ and ER, before leading to the final product PR, as shown in Fig. 1. E can also undergo a similar set of reactions with the wrong substrate W leading to product PW.
Fig. 1.
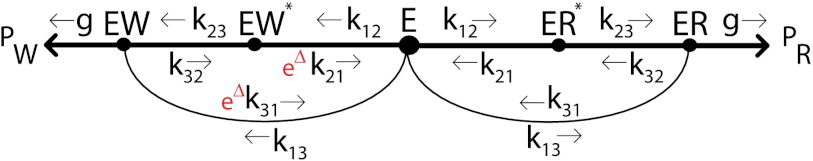
Proofreading mechanism proposed by Hopfield (1). Substrates R, W compete to react with E, and form either the right product PR or the wrong one PW. The ES↔ES∗ reactions (S = R, W) are coupled to external out-of-equilibrium ATP- hydrolysis reactions.
The enzyme E can be viewed as exploring this reaction network when the enzyme-substrate complex is undergoing stochastic transitions between different states. A small difference in binding energy Δ (measured in units of kBT, where kB is Boltzmann’s constant and T is the absolute temperature) between the complexes ER and EW will ultimately bias the enzyme towards producing PR over PW. The rate at which PW is produced relative to PR defines the error rate η. The exploration of the network shown in Fig. 1 can happen with different statistics of returns to the free enzyme state E. The error rate η depends of course on such statistics. If the enzyme executes all reactions at equilibrium, the error rate is given simply by the Boltzmann factor, η ∼ e-Δ. The equilibrium error rate e-Δ can range from 10-4 in the case of DNA nucleotide base pairing to 10-2 in protein synthesis. In the proofreading mechanisms proposed by Hopfield and Ninio, the enzyme-substrate complexes are driven out-of-equilibrium by ATP hydrolysis and much lower error rates η can be achieved.
The kinetics of the original Hopfield’s kinetic proofreading scheme, depicted in Fig. 1, can be characterized by four ratios of kinetic constants, 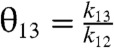 ,
, 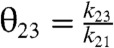 ,
, 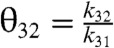 ,
, 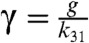 . In this scheme, the proofreading is most effective when
. In this scheme, the proofreading is most effective when
| [1] |
In these limits, the reaction E + S↔ES∗ rapidly equilibrates compared to the rest of the network and the enzyme-substrate complex reaches the state ES primarily through ES∗ and not directly from E. Typical dynamics for the enzyme-substrate complex obeying the inequalities in [1] are schematically shown in Fig. 2A.
Fig. 2.
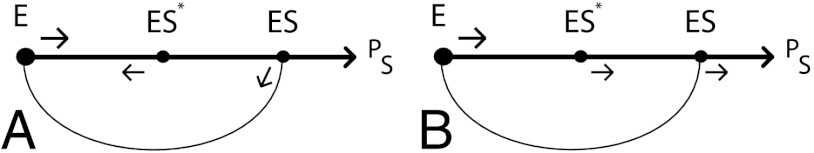
Dominant transitions for Hopfield’s proofreading scheme in different kinetic limits. (A) In the limit of [1], both substrates S = W, R cycle many times around the loop in the reaction network which helps achieve the lowest possible error rate of η ∼ e-2Δ. (B) In the limit of [11], substrate S = R completes the reaction without much backtracking as shown here while W still backtracks as shown in (A).
The reaction path involves multiple back and forths between E + S and ES∗ until equilibration. Occasional further progress takes the complex past ES∗ to ES. Typically, the ES complex disintegrates back into E + S and only occasionally does it proceed to form the final product PS. This type of kinetics leads, therefore, to many cyclic trajectories around E - ES∗ - ES loops, driven by ATP hydrolysis. These stochastic cycling trajectories magnify the small bias towards the correct R substrate, leading to enhanced error correction but at a price—stochastic trajectories typically take a long time due to many cycles. In the limits of [1], the error rate η and time TR taken to produce a molecule of the correct product PR are given by (1) ,
| [2] |
The error rate η is well below the equilibrium bound of e-Δ but TR is very large compared to typical reaction time scales in the network, because γ ≪ 1 and θ23 ≪ 1. In the following, we will address the issue of this slowing down and the related expenditure of energy. We begin by generalizing the results of Hopfield’s kinetic proofreading scheme to more complex biochemical networks.
Generalized Kinetic Proofreading Schemes.
A typical proofreading scheme can be represented by a large network of biochemical reactions with two isomorphic subnetworks, leading to correct PR and incorrect PW products (Fig. 3A). In analogy with Hopfield’s original mechanism, we will assume that the only difference between the two subnetworks is the existence of a set of “discriminatory” reactions whose R and W kinetics differ by a factor eΔ. These factors distinguishing between two substrates are distributed in such a way that any individual pathway leading to a final product carries one net factor of eΔ that suppresses the production of PW relative to PR.
Fig. 3.
A generalized proofreading network. (A) The enzyme E forms a complex with one with one of two substrates R, W and explores the network shown by executing a random walk on it. PR is the desired product (or final state) while PW is an incorrect product (an error). For good proofreading, one of the paths (bold line) must dominate the walk. (B) Schematic redrawing of the generalized network in (A) emphasizing the central reaction path from the initial free enzyme state E to final product molecules PR, PW. A typical trajectory involves moving a certain distance along the dominant path towards PR or PW before randomly taking one of the sideways paths back, undoing part or all of the reaction. The probability of taking a sideways path is higher for W than for R and hence the final product is reached exponentially less often for the incorrect substrate W.
Through an appropriate choice of kinetic constants associated with such a biochemical network, one can achieve enhanced proofreading with very low error rates. If driven with sufficient chemical potential by ATP hydrolysis, such a mechanism can achieve its lowest error rate η given by
| [3] |
when all discriminatory reactions in the network work in concert. Here n + 1 is the number of independent pathways leading to final product formation, each of which is restricted to carry only one (net) discriminatory factor of eΔ. Equivalently, n is the number of independent loops in the network. The example shown in Fig. 3A has n = 4 loops and five pathways from E to ER (or to EW.) This result can be viewed as a generalization of the original n = 1 loop kinetic scheme of Hopfield (1).
To get a better understanding of this result, it is useful to redraw the biochemical network as in Fig. 3B. The biochemical kinetics that achieve lowest error leads to a dominant path through the network (bold line in Fig. 3B) through which the reaction is actually completed. However, most attempts to follow the dominant path result in the enzyme being ejected off this path through one of the n sideways paths leading back to an earlier intermediate. Such ejections are accompanied by a free energy releasing reaction such as ATP hydrolysis. Each of these sideways paths contains a discriminatory reaction carrying a factor of eΔ (for substrate W) along it. Hence the probability of not taking a given sideways path is greater for R than for W by eΔ. The probability of reaching the final state is a product of n (small) probabilities of not taking sideways paths, giving exponential purification ([3].
Proofreading models with n-stages were studied in past work (3–6) that obtained detailed results on minimal error rates ([3] above) and the relation between time, energy, chemical potential, and error. We discuss the relationship between these earlier models and our networks represented by Fig. 3 later and in the SI Appendix.
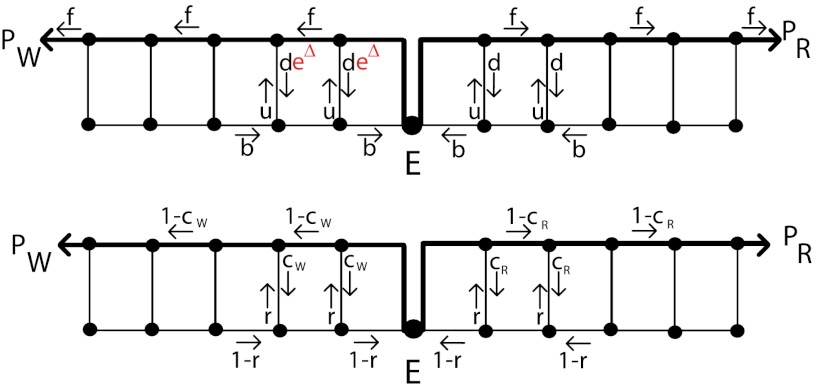
Kinetic Proofreading on a Ladder Network.
One of the difficulties of analyzing the behavior of a general biochemical network, such as Fig. 3, is its complexity. In particular, it is difficult to establish whether there are different proofreading regimes, each corresponding to different scaling of the error rate η and the completion time TR. To explore the possible existence of new kinetic regimes, we will first consider a particular ladder-like topology of the biochemical network, depicted in Fig. 4. In this network the dominant reaction path leading to final product is the upper rail of the ladder, while the sideways paths follow the rungs of the ladder to the lower rail. Due to these sideways paths, at every step, substrate R has a probability
 |
[4] |
of switching from the top to the bottom rail. We also assume that on the bottom rail the reactions can proceed only in the backward direction (i.e., away from the final products). Thus typically the enzyme-substrate complex proceeds along the top rail towards completion of the reaction (states PR, PW in Fig. 4), but it may switch randomly to the lower rail and undo some of that progress. The complex can then be randomly moved back to the upper rail to make another attempt to complete the reaction. The probability of switching from the backward mode, on the lower rail, to the forward mode, on the upper rail, is given by 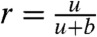 . The kinetic constant r is independent of cR and cW and is assumed to be the same for W and R. On the other hand, although both W and R undergo stochastic switching from the upper to lower rail, the corresponding kinetic constant is higher for W (deΔ instead of d), due to its lower binding energy with the enzyme E. As a result, the probability of switching to the backward mode for W,
. The kinetic constant r is independent of cR and cW and is assumed to be the same for W and R. On the other hand, although both W and R undergo stochastic switching from the upper to lower rail, the corresponding kinetic constant is higher for W (deΔ instead of d), due to its lower binding energy with the enzyme E. As a result, the probability of switching to the backward mode for W,
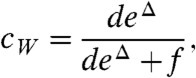 |
[5] |
is higher than cR. It is this difference between cR and cW that is amplified in the present proofreading mechanism leading to strong discrimination between the two substrates. (Note, however, that the conclusions of this paper do not qualitatively depend on all the details of which kinetic parameters in the network of Fig. 4 carry the discriminating factor eΔ. Explicit calculations with alternative assumptions but similar results are shown in the SI Appendix). The irreversible kinetic constants shown in Fig. 4 break detailed balance and result from the coupling to free energy releasing reactions like ATP hydrolysis.
Fig. 4.
Ladder network for kinetic proofreading. For both substrates, the reaction can proceed only in the forward direction along the top “rails” of their subgraphs and only backwards along the lower rail. This network can be defined using kinetic rate constants f, d, u and b, or equivalently using probabilities cR, cW and r.
Results
Proofreading and Dynamical Instability of Microtubules.
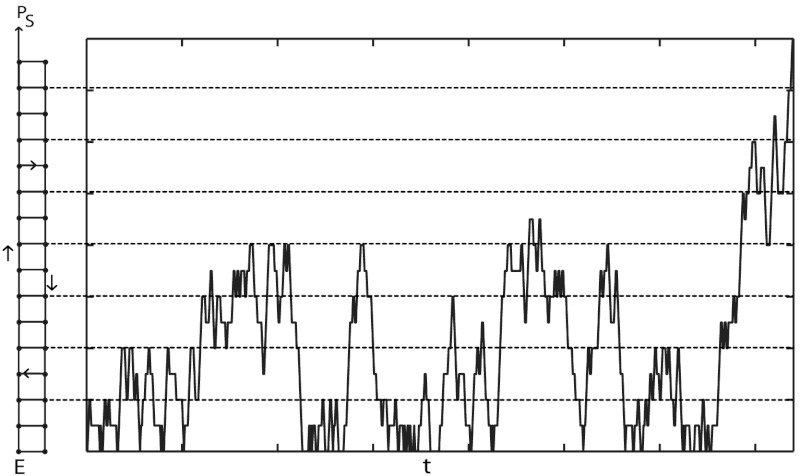
A typical trajectory on the ladder network that results in a low error rate η makes many forward and backward moves before reaching the final product PS. As measured along the dominant path (here the upper rail of the ladder), the variance of excursions scales linearly with the mean excursion length L, rather than as 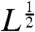 as we would expect for random walks. One such trajectory is schematically depicted in Fig. 5. Here kinetic parameters were specially chosen for visualization purposes: typical proofreading trajectories will include more short, failed excursions before reaching the final state PS.
as we would expect for random walks. One such trajectory is schematically depicted in Fig. 5. Here kinetic parameters were specially chosen for visualization purposes: typical proofreading trajectories will include more short, failed excursions before reaching the final state PS.
Fig. 5.
Stochastic trajectory of bounded enzyme kinetics on a ladder network. The trajectory consists of forward moves on one rail, alternating with backward moves on the other rail. After many such stochastic cycles, the reaction ultimately reaches the final product S. For better visualization, enzyme kinetics were chosen to be close to the bounded-unbounded transition (but on the bounded side).
This behavior is reminiscent of the so-called dynamical instability of microtubules (7), growing and collapsing, for example within a “microtubule aster” (8). We will now use this analogy to gain insight into proofreading mechanisms.
Microtubules are polymers made of tubulin monomers (or more precisely dimers) that can be subject to dynamical instability—an out-of equilibrium assembly driven by the hydrolysis of GTP associated with tubulin. Assembly and disassembly of tubulin monomers into/from rigid and long microtubules is a complex molecular process involving many structural transitions (e.g., nucleation involving γ-tubulin, lateral interactions between many “protofilaments” forming tubes, hydrolysis-induced strain within the tubes, etc). For our purposes, however, we will simplify these assembly phenomena and assume that microscopic molecular mechanisms result in the following growth dynamics: the polymer typically grows at some rapid rate vg. However, there is a constant probability per unit time, fcat, of a “catastrophe”, after which the polymer starts to shrink at a rate vs. The shrinking polymer might then be “rescued” and brought back to the growing state at a rate fres (9). Alternatively it might reach zero length and start growing again after a new nucleation (which we assume here to be instantaneous). In reality, there are many higher-order effects not captured by this model, including variation in the rates fcat, fres as a function of the length of the polymer. See refs. 10, 11 for recent work and summary of the rich phenomena constituting microtubule growth.
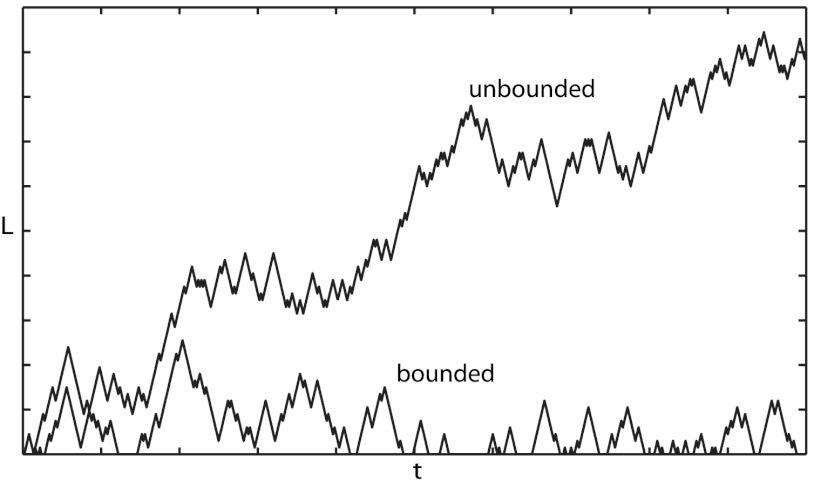
The four phenomenological parameters vg, vs, fcat, and fres are functions of kinetic constants characterizing transitions between different states of monomers. Depending on the values of these parameters there are two qualitatively different regimes of microtubule growth. If 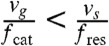 , catastrophes dominate over rescue events and the polymer’s length repeatedly returns to zero. Thus the polymer’s length stays bounded with time, and the standard deviation of its fluctuations is of same order as its average length. The polymer is said to be in the bounded growth regime . It is analogous to stochastic kinetics of proofreading on the ladder network—indeed the time variations of the microtubule length, L, (Fig. 6) closely resemble the stochastic kinetics depicted in Fig. 5. We call the biochemical kinetics in this regime “bounded walk kinetics”. There is a direct analogy between the phenomenological parameters describing the dynamical instability and the probabilities of the ladder network scheme: for instance cR and cW are analogous to the catastrophe rate fcat, while r plays the role of the rescue rate fres.
, catastrophes dominate over rescue events and the polymer’s length repeatedly returns to zero. Thus the polymer’s length stays bounded with time, and the standard deviation of its fluctuations is of same order as its average length. The polymer is said to be in the bounded growth regime . It is analogous to stochastic kinetics of proofreading on the ladder network—indeed the time variations of the microtubule length, L, (Fig. 6) closely resemble the stochastic kinetics depicted in Fig. 5. We call the biochemical kinetics in this regime “bounded walk kinetics”. There is a direct analogy between the phenomenological parameters describing the dynamical instability and the probabilities of the ladder network scheme: for instance cR and cW are analogous to the catastrophe rate fcat, while r plays the role of the rescue rate fres.
Fig. 6.
Examples of bounded and unbounded growth of microtubules. The fluctuations in the bounded walk are of size comparable to the mean displacement (σ ∼ μ). The unbounded walk amounts to a random walk and the fluctuations scale as the square root of the mean (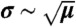 ).
).
On the other hand, if 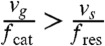 , the microtubules tend to grow more than they shrink and the total length of the polymer, L, grows linearly with time t, with a standard deviation increasing as
, the microtubules tend to grow more than they shrink and the total length of the polymer, L, grows linearly with time t, with a standard deviation increasing as 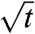 . The microtubule is said to be in the unbounded growth regime (Fig. 6). As we will see below, an analogue of this second growth regime also exists for the stochastic kinetics of kinetic proofreading and it has quite unexpected implications for the possible speeding up of proofreading reaction schemes. By analogy with microtubules, we call biochemical kinetics in this regime “unbounded walk kinetics”.
. The microtubule is said to be in the unbounded growth regime (Fig. 6). As we will see below, an analogue of this second growth regime also exists for the stochastic kinetics of kinetic proofreading and it has quite unexpected implications for the possible speeding up of proofreading reaction schemes. By analogy with microtubules, we call biochemical kinetics in this regime “unbounded walk kinetics”.
Speeding Up Proofreading on the Ladder Network.
The behavior of the ladder network can be understood in terms of two independent parameters: 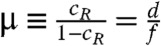 related to catastrophes and
related to catastrophes and  related to rescues. (Parameter cW can be determined in terms of μ using Eqn. 5. The binding energy difference Δ is determined by the chemical structure of the final products and not a variable parameter in this work.). For convenience, we also define
related to rescues. (Parameter cW can be determined in terms of μ using Eqn. 5. The binding energy difference Δ is determined by the chemical structure of the final products and not a variable parameter in this work.). For convenience, we also define 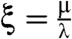 .
.
For large λ (i.e., probability of rescues r close to 1) with fixed ξ, we can derive simple expressions for the error rate η, and the average time TR taken to produce the right product in terms of 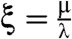 ,
,
 |
[6] |
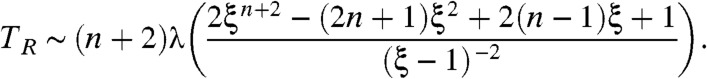 |
[7] |
We find three distinct regimes of behavior for time TR and error η as a function of μ and λ (or equivalently ξ), depicted in Fig. 7. These regimes, defined precisely below, correspond to different probabilities of catastrophes cR and cW relative to the probability of rescue r:
Fig. 7.
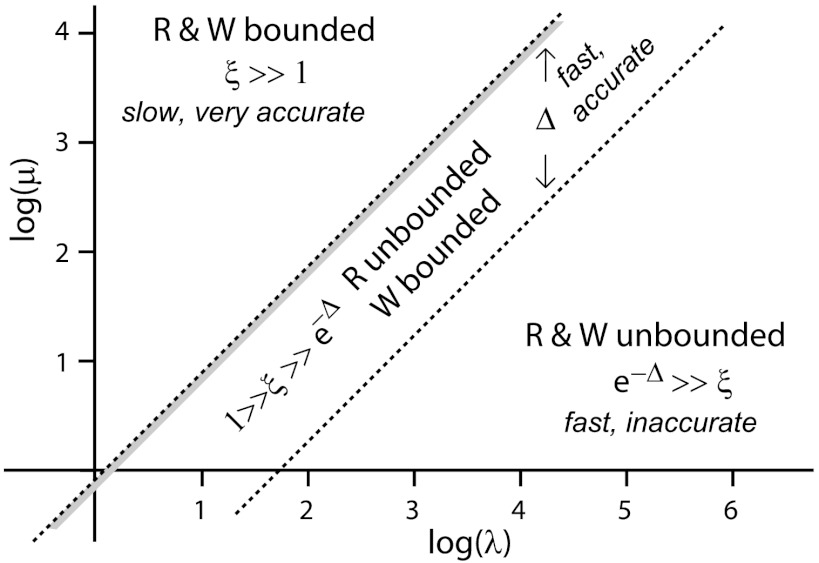
Behavior in different regions of parameter space (log(μ), log(λ)), where  and
and  . λ is related to events of “rescues” (r is the probability of rescue for both R and W), while μ is related to the probability of “catastrophes” for R, cR. We distinguish three regions in this parameter space—in the upper region, both R and W undergo bounded walks kinetics, giving very low error rate at the cost of slow product formation. In the diagonal central strip, R exhibits unbounded walk kinetics, while kinetics of W is that of bounded walks. Such kinetics save much time for proofreading, at a small expense in error rate. The shaded subregion minimizes the small expense in error rate and hence is particularly advantageous. In the third, lower region, both R and W exhibit unbounded walks. The width of the central strip is proportional to Δ.
. λ is related to events of “rescues” (r is the probability of rescue for both R and W), while μ is related to the probability of “catastrophes” for R, cR. We distinguish three regions in this parameter space—in the upper region, both R and W undergo bounded walks kinetics, giving very low error rate at the cost of slow product formation. In the diagonal central strip, R exhibits unbounded walk kinetics, while kinetics of W is that of bounded walks. Such kinetics save much time for proofreading, at a small expense in error rate. The shaded subregion minimizes the small expense in error rate and hence is particularly advantageous. In the third, lower region, both R and W exhibit unbounded walks. The width of the central strip is proportional to Δ.
Regime 1:
ξ≫1 (which implies r < cR < cW). Both R and W substrates have the kinetics of bounded walks, returning repeatedly to the origin. The error rate approaches its absolute minimum while time TR is exponentially large,
| [8] |
There is also a large free energy cost to the repeated cycling.
Regime 2:
1≫ξ≫e-Δ (which implies cR < r < cW). Incorrect substrate W undergoes bounded walks (r < cW) and is still subject to frequent backtracking. The kinetics for R correspond to unbounded walks (cR < r) and R proceeds towards reaction completion without much backtracking. This limit greatly speeds up of the proofreading process but has a cost in the form of a higher error rate,
| [9] |
where α < 1 is defined through e-αΔ ≡ ξ. Time TR, linear in the size of the system, is much smaller than the exponentially growing time TR ∼ λξn in the limit 1 ≪ ξ above. (Time TW for the incorrect product remains large in the present limit.). One can minimize α, the cost in error rate of having a linear completion time, by tuning R to be unbounded but very close to the bounded-unbounded transition; i.e., cR = r - ϵ for some small ϵ > 0. This advantageous regime is shown as the shaded subregion of the central region in Fig. 7.
Regime 3:
e-Δ≫ξ (which implies cR < cW < r). In this case, both correct and incorrect substrates, R and W, undergo unbounded walk kinetics. The error rate, which is large in this limit and time TR are given by,
| [10] |
We can summarize these three regimes using an abstract axis which represents the spectrum of bounded and unbounded walks, as shown in Fig. 8. The substrates W and R are positioned on this axis by the values of their kinetic constants and are separated by a fixed distance determined by Δ. The overall position of the substrates, however, can be moved along the axis from the bounded regime (cR < cW < r) to the bounded/unbounded regime (cR < cW < r) and finally to the unbounded regime (r < cR < cW). The middle regime, with R and W on either side of this transition is particularly advantageous in terms of time and free energy consumed at only a small cost in error rate. We later discuss one possible mechanism for changing cR, cW and moving the substrates along the bounded-unbounded axis using magnesium [Mg2+] in the context of protein synthesis, based on recent work (12).
Fig. 8.

An abstract one-dimensional representation of the space of possible kinetics for a proofreading model. The kinetics of R and W can differ only by a fixed amount, determined by their enzyme binding energy difference Δ, but can be shifted together within this space. Placing both R, W kinetics well within the bounded walk region is the traditional choice, leading to the minimal error rate η at the cost of slow product formation. However, placing them at transition area between the bounded and unbounded walks regimes offers a great speed-up of the reaction at a relatively small cost in the error rate η.
Speeding Up the Hopfield-Ninio Proofreading Scheme.
The transition between bounded and unbounded walks kinetics on the ladder network of Fig. 4 is sharp for n≫1 as shown schematically in Fig. 8. For simpler proofreading networks, with small values of n, the transition is no longer sharply defined. However, one can still distinguish between the traditional kinetic regime, in which both R and W kinetics are similar to bounded walks and a faster regime, in which W undergoes bounded walks, while R does not.
We illustrate the existence of this new regime for Hopfield’s original proofreading mechanism shown in Fig. 1 by modifying the traditional kinetic limit defined in [1] to,
| [11] |
In this new limit, the inequalities γ, θ23, θ32 ≪ eΔ, and eΔθ13 ≪ θ23 cause W to still waste time in stochastic cycles like those described by Hopfield. The other inequalities in [11] ensure that R typically proceeds directly to final product formation, through E + R → ER∗ → ER → PR, as shown in Fig. 2B. Such a trajectory resembles the unbounded growth of microtubules. For example, the inequalities 1 ≪ θ23, γ ensure that ER∗ proceeds to ER without dissociation and that ER leads to product formation without much back flow. This limit saves much time compared to the bounded walks limit [2] while keeping the error rate below e-Δ,
| [12] |
One might wonder if the regimes and transition we have identified continue to exist for general disordered networks of the kind shown in Fig. 3. We investigated this question in two models of disordered networks using numerical simulations. We found that a bounded-unbounded transition does exist for these models with weak disorder as long as the network structure does not heavily disfavor rescues. Details and results are presented in the SI Appendix. General disorder could alter the nature of the transition we have found here but such an investigation is beyond the scope of this work.
Networks whose structure does not allow rescues do not exhibit the transition we have found here by balancing rescue and catastrophe rates. For such networks, all discard pathways take the system back to the initial state E + S unlike the case in Figs. 3 and 4. The time-energy-error tradeoff for these networks, which always operate in the bounded regime, was studied in (3–6). The above works identified the conditions necessary to minimize dissipation for a fixed error rate within the bounded regime. These works also identified kinetic limits in which such networks are fast and accurate over a finite interval of n. We elaborate further on the relationship between the time-error tradeoff found in our paper and that in (3–6) in the SI Appendix.
Experimental Evidence for Kinetic Regimes
Charging of tRNA.
Early evidence for proofreading came from experiments on the charging of tRNA with amino acids by tRNA synthetases (13, 14). In these experiments, a given species of tRNA, say isoleucine-tRNA, was charged by isoleucyl-tRNA synthetase with a supply of only the correct amino acids (isoleucine). In a separate run, the same synthetases and isoleucine-tRNA were allowed to react with a supply of incorrect amino acids alone (say valine). In both cases, the amount of ATP hydrolyzed per tRNA charged (or mischarged) was measured. The experiments found that only 1.5 molecules of ATP were hydrolyzed on average per correctly charged tRNA molecule while each incorrect charging of tRNA with valine required approximately 270 molecules of ATP.
One can explain this data within a minimal 1-loop proofreading scheme shown in Fig. 1 with ATP-hydrolysis coupled to the ES∗ → ES reaction. The data implies that with the wrong substrate, the enzyme-substrate complex proceeds from EW∗ to EW consuming ATP but then most often (i.e., 269 out of 270 times), dissociates back into E + W without forming the final product. In contrast, with the right substrate, formation of an ER complex results in the final product most of the time and dissociates only a fraction of the time (on average 1 out of 3 times). These numbers imply that incorrect charging of tRNA with valine proceeds in the bounded manner shown in Fig. 2A, with a lot of cycles before the final product is produced. On the other hand, correct charging with isoleucine proceeds with minimal wasted ATP and corresponds either to a fully unbound scheme like that shown in Fig. 2B or a mixed scheme where the reactions at ER∗ behave bounded as in Fig. 2A but the final reactions at ER are unbounded as in Fig. 2B. Such a figure is shown in the SI Appendix. Data on the total error rate in refs. 13, 14 suggests that a mixed scheme as shown in the SI Appendix better describes the experiment because the fully unbounded trajectory in Fig. 2B would have too high an error rate.
Translation by Ribosomes.
Experiments on ribosomes during protein synthesis have revealed similar proofreading characteristics, consistent with the 1-loop network of Fig. 1. In refs. 15, 16, single-molecule FRET measurements were used to probe different internal states of the complex formed by the ribosome, tRNA and the elongation factor EF-Tu. Measurements were performed on ribosomes (loaded with mRNA) incorporating amino acids from tRNA molecules into a growing polypeptide. Such studies reveal an intricate series of steps by which a charged tRNA molecule is at first tentatively accepted into the A-site of a ribosome. The complexes formed up to this stage can be modeled by state ES∗ in Fig. 1. Following this state, GTP attached to EF-Tu undergoes irreversible hydrolysis, modeled by ES∗ → ES in Fig. 1. The hydrolyzed complex can either complete the reaction by incorporating the amino acid from the tRNA into the growing polypeptide chain or dissociate and release the charged tRNA molecule from the ribosome.
FRET time traces were used in ref. 15 to infer rates of making the above transitions, both for cognate and near-cognate tRNAs. For near-cognate tRNAs (modeled by W in Fig. 1), it was found that only 22% proceed from ES∗ to ES without dissociating back into E + S, compared to 80% for cognate tRNAs (modeled by R). Furthermore, measurement of overall error-rates suggests that when the complex does arrives at state ES after GTP hydrolysis, near-cognate tRNAs are again very likely to dissociate from the ribosome while cognate tRNAs proceed forward, first losing EF-Tu and then adding their amino acid to the growing polypeptide. Hence, this measurement suggests that proofreading in ribosomes is likely in the bounded regime for near-cognate tRNAs (Fig. 2A) and in the unbounded regime for cognate tRNAs (Fig. 2B). Such operation would confer a benefit in speed at a small expense in error as discussed earlier. See refs. 17–20 for similar but independent work on ribosomal proofreading.
Recent work (12) found that increasing the amount of [Mg2+] in the buffer increases the rate of tRNA incorporation for both cognate and noncognate tRNAs which in turn increases the overall reaction rate but reduces accuracy. Detailed kinetic measurements (12) show that the increase of [Mg2+] can be viewed as moving the kinetics closer to being unbounded.
Assembly of RecA.
The assembly of filaments of the protein RecA on single-stranded DNA provides an example of multistage proofreading with close parallels to microtubule growth (21, 22). As part of the homology search during recombination, filaments of RecA grow on and cover single-stranded DNA. Experiments in ref. 21 found the extent of coverage by RecA filaments to be highly sensitive to the underlying DNA sequence. Ref. 21 explained this enhanced discrimination between DNA sequences by showing that the growth of RecA filaments experiences large out-of-equilibrium fluctuations. In fact, the growth of RecA was shown to be the “mirror image” of dynamic instability of microtubules. RecA filaments polymerized to the full length of the single-stranded DNA and then experienced occasional periods of shortening. These shortening periods were often arrested and reversed by rapid growth back to the full length. This back and forth motion along the DNA strand was shown to act like a multistage proofreading process, where the DNA sequence was repeatedly “examined” by RecA, enhancing discrimination between sequences.
With disfavored DNA sequences, RecA filaments experienced large length fluctuations—periods of shrinkage and growth caused the growing tip of the RecA filament to repeatedly traverse the length of the DNA sequence. On the other hand, with preferred DNA sequences, RecA filaments had smaller shrinking events than rescue events and covered the entire length of the DNA strand most of the time. RecA-covered DNA strands are biologically active and in this way, the favored DNA strands are kept active for longer periods of time. At the same time, the system benefits from the enhanced discrimination of disfavored sequences through repeated depolymerization of RecA filaments and “examination” of such sequences. In light of the discussion in this paper, the RecA system provides a striking example of a multistage proofreading process tuned to operate at a favorable point in the time-error tradeoff.
Conclusions
We have made a connection between two phenomena seemingly very different on the surface—proofreading in enzymatic reactions and microtubule growth—by focusing on the statistics of exploration of their respective biochemical spaces. We found that the common class of “exploratory statistics” called bounded is most effective for reducing error in proofreading. Exploiting this analogy allowed us to go further. We were able to identify another regime of exploratory statistics—a mix of bounded and unbounded exploration—that is greatly advantageous for proofreading when time and dissipation are also concerns in addition to the error rate. In the SI Appendix, we verified that our qualitative conclusions hold in two models of weak disorder present in the network structure.
In ref. 8, the time taken by microtubules to locate a target (like a centromere) in the bounded phase was compared to a hypothetical model of microtubules without dynamical instability. In the hypothetical model, polymerization takes place at equilibrium and the statistics of polymer growth are simply that of a biased random walk. Locating the centromere target was dramatically faster in the bounded phase with dynamical instability. It might be interesting to speculate if known gradients (23) from chromosomes might shift the dynamics of microtubules growing in the right direction to the unbounded regime, leaving tubules growing in incorrect directions bounded. Such a shift of dynamics would further speed up discovery of the chromosome, in complete analogy with proofreading models discussed here.
Thus microtubule growth and proofreading are both characterized by exploratory statistics beneficial to the location (or reaction with) of a preferred target. For these two biochemical processes, such statistics are made possible by their nonequilibrium coupling to an ATP or GTP driving force. We hope that by focusing only on exploratory statistics and not on the detailed mechanism that make such behavior possible, our analogy might help connect these two processes to other biological exploratory phenomena such as foraging, growth of vascular and nervous systems, or other searches that happen on very different physical scales (24).
Supplementary Material
Acknowledgments.
We benefited greatly from discussions with B. Greenbaum, Z. Frentz, D. Hekstra, J. Hopfield, J. Chuang, D. Jordan, S. Keuhn, A. Libchaber and T. Tlusty. We thank J. Hopfield, T. Tlusty and M. Ehrenberg for providing comments on a draft of this paper. The work of A.M. has been supported by the Institute for Advanced Study through the Hattie and Arnold Broitman fellowship.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119911109/-/DCSupplemental.
References
- 1.Hopfield J. Kinetic proofreading—new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci USA. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ninio J. Kinetic amplification of enzyme discrimination. Biochimie. 1975;57:587–595. doi: 10.1016/s0300-9084(75)80139-8. [DOI] [PubMed] [Google Scholar]
- 3.Ehrenberg M, Blomberg C. Thermodynamic constraints on kinetic proofreading in biosynthetic pathways. Biophys J. 1980;31:333–358. doi: 10.1016/S0006-3495(80)85063-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blomberg C, Ehrenberg M. Energy considerations for kinetic proofreading in biosynthesis. J Theor Biol. 1980;88:631–670. doi: 10.1016/0022-5193(81)90242-3. [DOI] [PubMed] [Google Scholar]
- 5.Freter R, Savageau MA. Proofreading systems of multiple stages for improved accuracy of biological discrimination. J Theor Biol. 1980;85:99–123. doi: 10.1016/0022-5193(80)90284-2. [DOI] [PubMed] [Google Scholar]
- 6.Savageau MA, Lapointe D. Optimization of kinetic proofreading. J Theor Bio. 1981;93:157–177. doi: 10.1016/0022-5193(81)90062-x. [DOI] [PubMed] [Google Scholar]
- 7.Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 8.Holy T, Leibler S. Dynamic instability of microtubules as an efficient way to search in-space. Proc Natl Acad Sci USA. 1994;91:5682–5685. doi: 10.1073/pnas.91.12.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dogterom M, Leibler S. Physical aspects of the growth and regulation of microtubule structures. Phys Rev Lett. 1993;70:1347–1350. doi: 10.1103/PhysRevLett.70.1347. [DOI] [PubMed] [Google Scholar]
- 10.Gardner MK, et al. Rapid microtubule self-assembly kinetics. Cell. 2011;146:582–592. doi: 10.1016/j.cell.2011.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howard J, Hyman AA. Growth, fluctuation and switching at microtubule plus ends. Nat Rev Mol Cell Bio. 2009;10:569–574. doi: 10.1038/nrm2713. [DOI] [PubMed] [Google Scholar]
- 12.Johansson M, Zhang J, Ehrenberg M. Genetic code translation displays a linear trade-off between efficiency and accuracy of tRNA Selection. Proc Natl Acad Sci USA. 2012;109:131–136. doi: 10.1073/pnas.1116480109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hopfield J, Yamane T, Yue V, Coutts S. Direct experimental-evidence for kinetic proofreading in amino acylation of transfer-rnaile. Proc Natl Acad Sci USA. 1976;73:1164–1168. doi: 10.1073/pnas.73.4.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamane T, Hopfield J. Experimental-evidence for kinetic proofreading in aminoacylation of transfer-RNA by synthetase. Proc Natl Acad Sci USA. 1977;74:2246–2250. doi: 10.1073/pnas.74.6.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blanchard S, Gonzalez R, Kim H, Chu S, Puglisi J. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 16.Lee T-H, Blanchard SC, Kim HD, Puglisi JD, Chu S. The role of fluctuations in tRNA selection by the ribosome. Proc Natl Acad Sci USA. 2007;104:13661–13665. doi: 10.1073/pnas.0705988104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen J-D, et al. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gromadski K, Rodnina M. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol Cell. 2004;13:191–200. doi: 10.1016/s1097-2765(04)00005-x. [DOI] [PubMed] [Google Scholar]
- 19.Johansson M, Lovmar M, Ehrenberg M. Rate and accuracy of bacterial protein synthesis revisited. Curr Opin Microbiol. 2008;11:141–147. doi: 10.1016/j.mib.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 20.Lovmar M, Ehrenberg M. Rate, accuracy andcost ofribosomes inbacterial cells. Biochimie. 2006;88:951–961. doi: 10.1016/j.biochi.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 21.Bar-Ziv R, Tlusty T, Libchaber A. Protein-DNA computation by stochastic assembly cascade. Proc Natl Acad Sci USA. 2002;99:11589–11592. doi: 10.1073/pnas.162369099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tlusty T, Bar-Ziv R, Libchaber A. High-fidelity dna sensing by protein binding fluctuations. Phys Rev Lett. 2004;93:258103. doi: 10.1103/PhysRevLett.93.258103. [DOI] [PubMed] [Google Scholar]
- 23.Athale CA, et al. Regulation of microtubule dynamics by reaction cascades around chromosomes. Science. 2008;322:1243–1247. doi: 10.1126/science.1161820. [DOI] [PubMed] [Google Scholar]
- 24.Kirschner MW, Gerhart JC. The Plausibility of Life: Resolving Darwin’s Dilemma. CT: Yale University Press; 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.