Abstract
Topical application of nucleic acids offers many potential therapeutic advantages for suppressing genes in the skin, and potentially for systemic gene delivery. However, the epidermal barrier typically precludes entry of gene-suppressing therapy unless the barrier is disrupted. We now show that spherical nucleic acid nanoparticle conjugates (SNA-NCs), gold cores surrounded by a dense shell of highly oriented, covalently immobilized siRNA, freely penetrate almost 100% of keratinocytes in vitro, mouse skin, and human epidermis within hours after application. Significantly, these structures can be delivered in a commercial moisturizer or phosphate-buffered saline, and do not require barrier disruption or transfection agents, such as liposomes, peptides, or viruses. SNA-NCs targeting epidermal growth factor receptor (EGFR), an important gene for epidermal homeostasis, are > 100-fold more potent and suppress longer than siRNA delivered with commercial lipid agents in cultured keratinocytes. Topical delivery of 1.5 uM EGFR siRNA (50 nM SNA-NCs) for 3 wk to hairless mouse skin almost completely abolishes EGFR expression, suppresses downstream ERK phosphorylation, and reduces epidermal thickness by almost 40%. Similarly, EGFR mRNA in human skin equivalents is reduced by 52% after 60 h of treatment with 25 nM EGFR SNA-NCs. Treated skin shows no clinical or histological evidence of toxicity. No cytokine activation in mouse blood or tissue samples is observed, and after 3 wk of topical skin treatment, the SNA structures are virtually undetectable in internal organs. SNA conjugates may be promising agents for personalized, topically delivered gene therapy of cutaneous tumors, skin inflammation, and dominant negative genetic skin disorders.
Keywords: transdermal delivery, nanocluster, antisense, nanotechnology, dermatology
The recent development of small molecule inhibitors and antibodies that target components of signaling pathways has revolutionized the treatment of cancers, inflammatory diseases, and genetic disorders, including those that largely manifest in skin (1–3). These protein-based therapeutics, however, are costly, have limited targeting ability, and, when delivered through traditional intravenous or gastrointestinal routes, can lead to systemic toxicity (4, 5). Topical delivery is particularly attractive for the therapy of skin disorders. However, proteins larger than a few hundred daltons cannot easily enter the skin, and high concentrations of proteins must be applied for a cutaneous effect (6). An alternative to protein-based pathway inhibition involves the blocking and/or degradation of precursor mRNA before translation into protein. Gene silencing leads to down-regulation of protein expression and functions with greater specificity than inhibitors of protein function (7). In fact, targeted gene suppression by antisense DNA and siRNA has shown promising preclinical results, and/or is currently in clinical trials for a variety of diseases, including many forms of cancer (e.g., melanoma, neuroblastoma, and pancreatic adenocarcinoma), genetic disorders, and macular degeneration (8). For gene suppression in skin, a topical delivery route is optimal, given the easy accessibility of skin and the reduced risk of systemic side effects. However, penetration of oligonucleotides through the epidermal barrier has remained a major technological challenge that must be overcome before the therapeutic potential of oligonucleotides in skin can be realized (9–11).
Recently, we introduced spherical nucleic acid nanoparticle conjugates (SNA-NCs, inorganic gold nanoparticles densely coated with highly oriented oligonucleotides) (Scheme S1) as agents capable of simultaneous transfection and gene regulation (12–15). In contrast with conventional approaches to gene regulation (16–20), these nanoparticle conjugates require neither cationic transfection materials nor additional modifications to drive cellular entry. SNA-NCs enter almost 100% of cells in the more than 50 cell lines and primary cells tested to date, as well as cultured tissues and whole organs (21, 22). These unusual constructs exhibit remarkable stability, nuclease-degradation resistance, and decreased immunogenicity as a result of their high-density oligonucleotide shells (23, 24). To date, they have shown no toxicity at the cellular level at concentrations required for effective gene knockdown. Therefore, we hypothesized that siRNA-based SNA gold nanoparticle conjugates might be ideal topical delivery agents to treat diseases of the skin.
Results
To evaluate the potential for targeted gene suppression by siRNA-based SNA-NCs through noninvasive topical delivery, we first investigated their uptake in primary normal human keratinocytes (hKCs), which are typically difficult to transfect (25). hKCs were incubated with Cy3 fluorophore-labeled SNA-NCs (Table S1), and entry was studied by confocal fluorescence microscopy. Within 2 h, 100% of the observed hKCs showed strong cytoplasmic fluorescence (Fig. 1A). The number of gold nanoparticles in cells (and tissues) can be quantified by inductively coupled plasma mass spectrometry (ICP-MS). Incubation of 0.025–0.1 nM SNA-NCs (0.75–3.0 nM siRNA) for 48 h with hKCs leads to uptake of approximately 1.4·103 SNA-NCs per cell, which is approximately five times greater than that in either HaCaT (spontaneously immortalized hKCs) or HeLa cells (Fig. 1B). Although the reason for this high KC uptake is still under investigation, these observations suggested SNA-NCs would be good candidates for topical delivery experiments.
Fig. 1.
Uptake, safety, and gene suppression efficacy of SNA-NCs in hKCs. (A) Uptake of Cy3-labeled nonsense SNA-NCs (red) was noted in the cytoplasm of approximately 100% of the primary hKCs after 24 h incubation. Blue, Hoechst 33343-stained nuclei. Scale bar, 20 μm. (B) KCs exhibit dose-dependent SNA uptake that is higher than the uptake in the HeLa and HaCaT cell lines. (C) Cellular proliferation during 96 h of treatment with nonsense SNA-NCs is similar to that of control treatment with PBS only. (D) Heat map of cellular gene array study (Dataset S1). (E) Western blot showing EGFR protein levels. hKCs were treated for 48 h with SNA-NCs or DharmaFECT1-delivered siRNA, or with controls of PBS and/or nonsense SNA-NCs. After treatment, growth medium was replaced with fresh medium, and the cells were incubated for an additional 12 h. Cell lysates were then harvested for blotting. Note the greater suppression by 0.01 nM EGFR SNA-NCs (equivalent to 0.3 nM siRNA) as compared with the 30 nM EGFR siRNA delivered with DharmaFECT1 at 60 h. (F) EGFR mRNA suppression after 48 h treatment with EGFR SNA-NCs or DhamaFECT1-delivered EGFR siRNA (treatment terminated after 48 h). The decrease and eventual loss of gene suppression during the subsequent 48 h after termination of treatment was evaluated by RT-qPCR. By 48 h after removal of SNA-NCs from the medium, 50% of the maximum suppression is still observed in EGFR SNA-NC—treated KCs, whereas suppression by DharmaFECT1-delivered EGFR siRNA is no longer evident. All data are expressed as mean ± SD.
With any gene regulation therapy or experiment, cell toxicity is a major concern. hKCs incubated with SNA-NCs are morphologically indistinguishable from untreated cells by routine microscopic evaluation. Proliferation assays also show no evidence of cytotoxicity after treatment of hKCs for 48 h with 0.05–0.1 nM SNA-NCs with nonsense siRNA sequences (Fig. 1C, Table S1). Finally, genome-wide expression profiling showed that only seven genes are up-regulated and none are down-regulated in hKCs treated with nonsense SNA-NCs, although the same cells treated with an equivalent amount of siRNA delivered through a cationic lipid-based transfection agent (DharmaFECT1®) exhibit up- or down-regulation of 427 genes (Fig. 1D and Dataset S1). These observations are consistent with previous studies in the HeLa cell line, which show that SNA-NCs enter cells in a stealth-like fashion and exhibit limited interference with normal cellular behavior (24, 26, 27).
Given the importance of EGFR signaling in epidermal cell proliferation and its recognized overexpression in many malignancies, EGFR was chosen as an initial target for gene suppression. EGFR knockdown in hKCs with the SNA-NCs is extremely effective. As little as 0.01 nM of SNA-NCs (0.3 nM of total siRNA, Table S1) in 1 mL of cell culture medium down-regulates EGFR protein expression by more than 90% as compared to control experiments involving PBS-treated cells and cells treated with nonsense SNA-NCs (Fig. 1E, group 5, lane 1 vs. groups 1–3). Total depletion of protein expression, within the sensitivity of Western blots, can be effected with 0.05 nM EGFR SNA-NCs in cell culture medium (1.5 nM of total siRNA) (Fig. 1E, group 5, lane 2). Significantly, knockdown of EGFR with SNA-NCs in hKCs can be achieved with 100-fold less total siRNA than that required to observe an effect with DharmaFECT1® (Fig. 1E, group 5, lane 1 vs. group 4, lane 2). Interestingly, target knockdown with SNA-NCs is also more persistent than with DharmaFECT1-delivered siRNAs (Fig. 1F). Here, at 48 h, greater than 90% mRNA knockdown is seen in both groups; however, suppression is completely lost at 96 h with DharmaFECT1-delivered siRNA, whereas SNA-NC—treated cells still exhibit 50% knockdown. This persistence is likely caused by the known resistance of SNA constructs to nuclease degradation, and therefore increased intracellular stability of the nucleic acid sequences on the SNA construct surface (23).
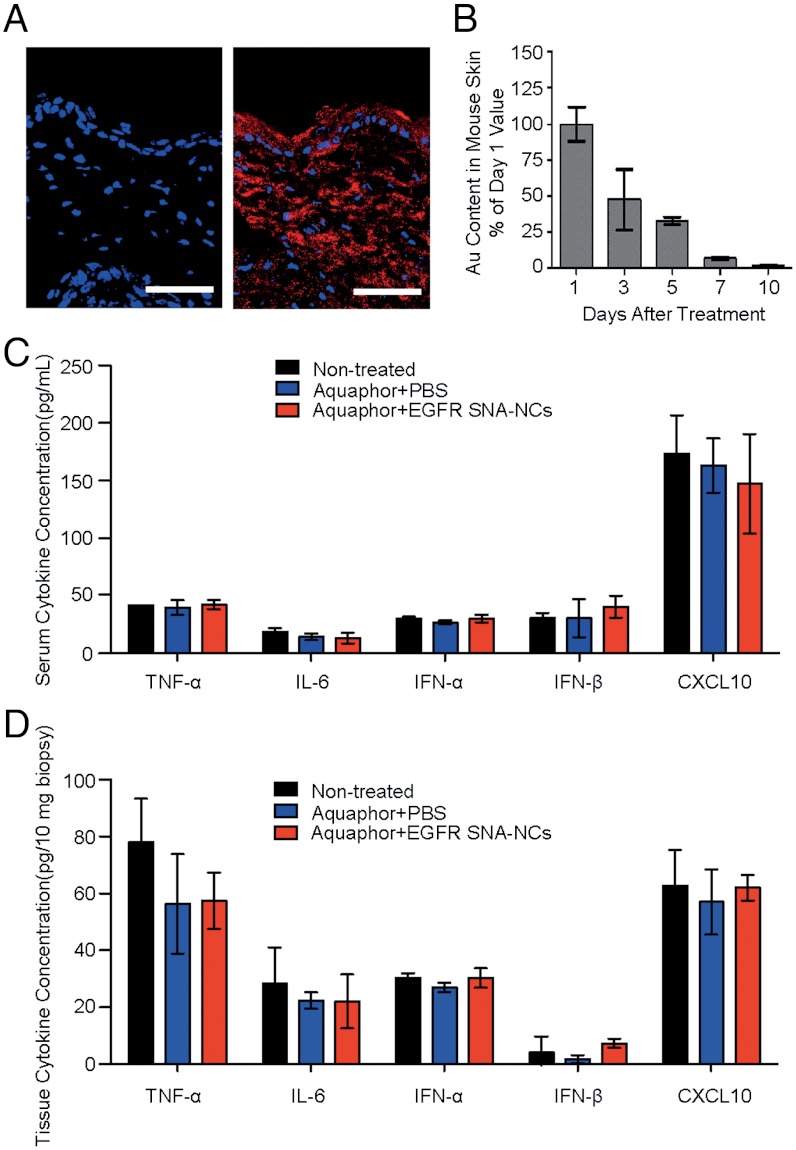
Because the SNA-NCs easily enter hKCs in in vitro experiments, we explored their ability to penetrate both mouse skin and human skin models. The SKH1-E hairless strain was chosen for studies because of its ease in use (eliminating the need for depilation), immunocompetency, and increased normal epidermal thickness (to better simulate human skin) in comparison with most mouse strains (28). Studies were also conducted in hair-bearing C57BL/6J mice (shaved 24 h before application to minimize epidermal barrier disruption). Cy5 fluorophore-labeled SNA-NCs in PBS were first mixed with a commonly used petrolatum-based moisturizer, Aquaphor® (1∶1, vol/vol), and then topically applied to mouse dorsal skin. In both strains, SNA-NCs penetrated through the stratum corneum and into the epidermis and dermis as early as 3 h after a single application (Fig. 2A). To evaluate the clearance of SNA-NCs from the skin after topical application, SKH1-E hairless mice were treated daily for 3 d with 50 nM nonsense SNA-NCs, and the gold content was quantified by ICP-MS in mouse skin during the subsequent 10 d. At the end of the experiment, only 2% of the initial gold was present (Fig. 2B), demonstrating the clearance of the SNA constructs from skin.
Fig. 2.
Penetration and clearance of SNA-NCs in hairless mouse skin. (A) Mouse (SKH1-E) skin treated topically with 1∶1 Aquaphor only (Left) or with 50 nM Cy5-labeled (red) SNA-NCs dispersed in the 1∶1 Aquaphor (Right); 3 h after application, the SNA-NCs are seen in the cytoplasm of epidermal cells and the dermis as well. Blue, DAPI-stained nuclei. Scale bars, 100 μm. (B) Mouse skin was treated daily for 3 d with 50 nM nonsense SNA-NCs; samples were harvested at 24 h to 10 d after treatment discontinuation and analyzed by ICP-MS for gold content. The gold content in mouse skin progressively decreases after cessation of topical treatment; 10 d after the final treatment, only 2% of the original gold content remains (n = 3 at each time point). (C) ELISA cytokine assays for TNF-α, IFN-α, IFN-β, IL-6, and CXCL10 in serum show no stimulation of innate immune responses after 3 wk of thrice-weekly treatment (every other day but Sunday; n = 3 per treatment group). (D) EGFR SNA-NC treatment does not induce the cytokine expression locally in the treated skin site after 3 wk of treatment. Results are presented in pg per 10 mg biopsy.
To take an initial step toward determining the potential toxicity of the SNA-NCs in vivo, we topically applied nonsense SNA-NCs in Aquaphor to the skin of shaved C57BL/6J mice. SNA-NCs were delivered daily for 10 d at four different concentrations (500 nM, 250 nM, 100 nM, and 50 nM; n = 5 per concentration; these values refer to the final SNA concentrations in Aquaphor). Animals were killed and skin and internal organs were harvested and studied histologically. Analogous experiments were done with hairless mice (n = 8) for 4 wk using 50 nM SNA-NCs, the effective knockdown dose (see below and Fig. 3). Significantly, no cutaneous inflammation, ulceration, scaling, or color alteration was ever observed in all experiments. In addition, blinded histological assessment showed no abnormalities in the liver, lungs, kidneys, or skin of mice treated for the 10 d or 4 wk. ICP-MS showed low gold accumulation in the internal organs of hairless mice that were treated for 3 wk with 50 nM SNA in Aquaphor; indeed, gold content was undetectable in the kidneys and lungs, and it was very low in the liver (0.0003%) and spleen (0.00015%) when normalized to the total amount of applied gold. ELISA assays showed no significant increase of tumor necrosis factor-α (TNF-α), interleukin-6 (IL-6), interferon-α (IFN-α), interferon-β (IFN-β), and chemokine (C-X-C motif) ligand 10 (CXCL10; IP-10) in the serum of hairless mice treated three times weekly for 3 wk with 50 nM EGFR SNA-NCs (Fig. 2C). Similarly, no immune response was observed locally in the treated skin tissue as evidenced by lack of cytokine expression and activation of protein kinase R (PKR) pathway (Fig. 2D, and Figs. S1 and S2). Finally, these skin samples were also subjected to a microarray assay. At 6 h after application, only one off-target gene (cystatin A1) was found to be differentially expressed by > 1.5-fold in skin treated with EGFR SNA-NCs in comparison with Aquaphor, and at 3 wk after initiation of treatment, only two off-target genes were differentially expressed (lactotransferrin and cathelicidin antimicrobial peptide). Off-target gene expression between the nonsense SNA-NCs and EGFR SNA-NCs was not significantly different after 3 wk.
Fig. 3.
Targeted gene knockdown by SNA-NCs correlates with biological effects in vivo. (A) Studies were performed to assess EGFR knockdown after 3 wk of treatment with EGFR SNA-NCs (three times per week). RT-qPCR analysis showed a 65% reduction in mRNA expression. Hairless mice, untreated or similarly treated with PBS:Aquaphor, nonsense SNA-NCs, or free siRNAs, showed no significant EGFR mRNA knockdown (n = 5 mice in each group at each time point). (B) The protein expression of EGFR was nearly eliminated in the EGFR SNA-NC—treated group, whereas the downstream phosphorylation of ERK was inhibited by 74%; total ERK expression remained constant. (C) The mean thickness of EGFR SNA-NC—treated skin was 40% less than that of control-treated skin (P < 0.001), as measured by computerized morphometry. Epidermal thickness was measured from the top of the stratum granulosum to the basement membrane (arrows) at three equidistant sites. (D) EGFR SNA-NC—treated skin samples show approximately 40% reduction in the percentage of basal layer KCs that stain positively for Ki-67 antigen, a marker of cellular proliferation, in comparison with controls. Scale bars, 100 μm. Data in all figures are expressed as mean ± SD.
To test SNA-NC efficacy in suppressing EGFR in mouse skin, 50 nM SNA-NCs in Aquaphor were applied three times per week Table S1. After 3 wk of application, EGFR mRNA expression was reduced significantly (by 65%) (Fig. 3A), and protein levels were almost completely suppressed (Fig. 3B). EGFR expression in comparison with untreated skin was not diminished by Aquaphor alone, free EGFR siRNAs, or nonsense SNA-NCs. The specificity of EGFR knockdown was further confirmed by the concomitant 74% decrease in phosphorylation of downstream ERK 1/2, although total ERK 1/2 expression remained the same (Fig. 3B). The knockdown effect of EGFR SNA-NCs was also confirmed phenotypically with computerized morphometric analyses of histological sections of mouse skin treated with EGFR SNA-NCs, which showed an almost 40% reduction in thickness as compared to skin treated with either nonsense SNA-NCs or the Aquaphor control. Representative examples of mouse skin from these studies are seen in Fig. 3C. Moreover, the number of basal epidermal nuclei, which were immunostained for Ki-67 (a proliferation marker), was reduced, demonstrating the ability of EGFR SNA-NCs to inhibit cellular proliferation in the epidermis (Fig. 3D). Skin samples from hairless mice treated with Aquaphor, nonsense SNA-NCs in Aquaphor, and EGFR SNA-NCs in Aquaphor were also subjected to microarray analysis. Taken together, our studies provide evidence that the SNA-NCs are able to knock down a specific gene target in vivo and lead to a biological effect with minimal off-target effects.
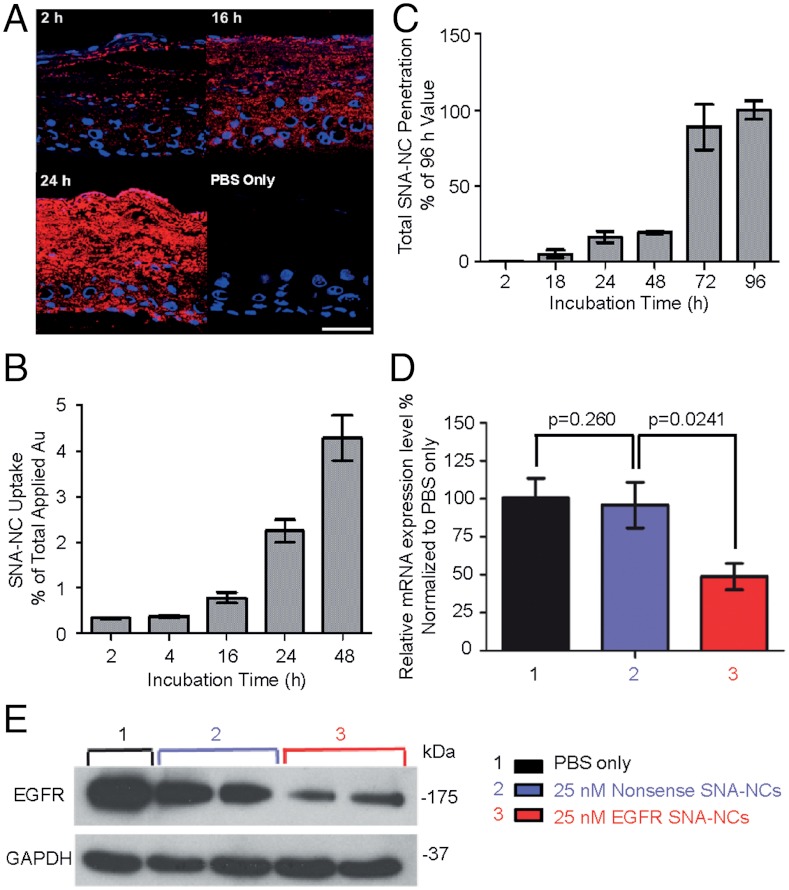
We then evaluated SNA-NC penetration through human skin using Franz cell chambers (28). Flux of nonsense SNA-NCs through human skin in Franz cell chambers was maximal between 8 and 24 h after topical application of nonsense SNA-NCs, regardless of solvent (SNA-NCs in PBS, glycerol, or Aquaphor) (Fig. S3). We also assessed penetration and knockdown efficacy of SNA-NCs in human skin equivalents, given that human skin is known to be thicker and more difficult to penetrate than mouse skin. Human skin equivalents (3D organotypic raft cultures) simulate in vivo human epidermis, including an intact lipid and well-differentiated protein epidermal barrier (29). Given the excellent penetration and knockdown in mouse skin with 50 nM EGFR SNA-NCs, the applied concentration was reduced to 25 nM for testing in human skin equivalents. In as little as 2 h after topical treatment, SNA-NCs were found in the basal layer of the epidermis (Fig. 4A), and the uptake and penetration through human skin equivalents increased in a time-dependent manner (Fig. 4 A–C). EGFR SNA-NCs (25 nM) knocked down EGFR mRNA expression by 52% (Fig. 4D) and EGFR protein expression by 75% (Fig. 4E) compared to nonsense SNA-NC—treated skin equivalents, even though only 4.7% of the applied SNA-NCs entered the epidermis based on ICP-MS quantification. Light microscopic images of skin equivalents treated with EGFR SNA-NCs appeared similar to skin equivalents treated with nonsense SNA-NCs in PBS or control nontreated skin equivalents, as expected after 60 h of treatment.
Fig. 4.
Penetration and gene knockdown in human skin equivalents. (A) Skin equivalents (EpiDerm; MatTek) treated with a single application of 25 nM Cy5-labeled (red) SNA-NCs or PBS for up to 48 h. Blue, Hoechst 33343-stained nuclei. Note the presence of SNA-NCs throughout the stratum corneum and nucleated epidermis. Scale bar, 50 μm. (B) ICP-MS measurements of gold show time-dependent uptake of the SNA-NCs in the epidermis. (C) The amount of gold in the cell culture medium, representing the number of particles passing through skin equivalents as measured by ICP-MS, increases with time. EGFR mRNA expression measured by RT-qPCR (D) and EGFR protein expression measured by immunoblotting (E) in skin equivalents treated with a single application of 25 nM EGFR SNA-NCs for 60 h demonstrate effective gene knockdown in human skin. Each study was performed at least three times in triplicate. Data are expressed as mean ± SD.
Discussion
In this work we have discovered that SNA-NC constructs have the unusual ability to suppress gene expression. Historically, researchers have relied on physicochemical methods to facilitate epidermal penetration and cellular delivery of negatively charged nucleic acids (11, 30–33). However, these methods require high concentrations of free siRNAs, can have unacceptable toxicity in in vivo models, and are limited by the instability of conventional siRNAs. In contrast, SNA-NCs exhibit long-lasting knockdown ability at picomolar to nanomolar concentrations with no apparent toxicity. We attribute this high efficacy to the dense monolayer of siRNAs on the nanoparticle surface. Our mechanistic studies with SNA-NCs show that the ion cloud associated with the high-density oligonucleotide shell, combined with steric inhibition at the surface of the particles, inhibits enzymatic nucleic acid degradation and results in an increased stability of siRNAs that comprise the SNA-NC (13, 14, 23).
Conventional RNA and DNA are also limited in their ability to cross the cellular membrane, even in monolayer cultures, without the use of viral vectors, lipid and polymer agents, or techniques such as electroporation. In contrast, SNA-NCs in PBS are taken up by approximately 100% of KCs without ancillary agents. Although the increased uptake in serum-free media suggests that a factor in serum suppresses uptake, the mechanism for better uptake in a primary cell (the hKCs) compared to immortalized cell lines (HaCaT and HeLa cells) remains unclear. Previous studies in HeLa and C166 endothelial cells have shown that the high-density SNA-NC shell recruits scavenger receptors from cells to facilitate endocytosis and is responsible for the superior cell entry of SNA-NCs in comparison to free siRNA of an identical sequence (22). How oligonucleotide density and charge and nanoparticle core size affect uptake, and how nanoparticles exit cells, are currently being investigated.
Proof that siRNA could successfully suppress gene expression in vivo was recently achieved in a mouse model (32) and a patient (11) with a genetic condition of thickened soles (pachyonychia congenita) through siRNA injection, which bypassed the epidermal barrier. Although the small treated areas showed a marked reduction in skin thickening, the pain and the impracticality associated with these injections made the approach untenable. Other mechanical approaches, such as ultrasound and laser, have been used to facilitate penetration through the mouse stratum corneum and drive siRNA into skin, but require specialized equipment (30, 31, 33), limit the area of delivery, and can potentially harm the skin. Recent investigations have demonstrated uptake of siRNAs and DNAs into the epidermis in mice with no stratum corneum (e.g., open wounds or mucosae) or with atopic dermatitis, an inflammatory skin disorder with an ineffective epidermal barrier (33, 34). Indeed, traversing the intact multilayered epidermis in vivo for intracellular delivery is particularly challenging. Stratum corneum-penetrating peptides have been some of the most promising topical constructs. Topical administration of hybrid MITF siRNA—peptide conjugates reduced MITF mRNA by 50% in mouse skin and reduced pigmentation in Chinese human subjects with facial melisma when the constructs were applied for 12 wk to their skin (35). Most recently, a skin-permeating and cell-entering peptide was shown to enhance siRNA delivery, leading to 43% knockdown of GAPDH protein in shaved BALB/c mouse skin at the maximal concentration tested, 10 μM (36). Our studies show that SNA-NCs, in contrast, virtually abolish EGFR protein expression in hairless mouse skin with its intact stratum corneum at siRNA concentrations of 1.5 μM (50 nM SNA-NCs), emphasizing the promise of this delivery system. We speculate that the delay in observation of significant EGFR knockdown beyond 2 wk likely reflects the predominant expression of EGFR in proliferating basal KCs and the 2 wk turnover of mouse epidermis, but could also suggest that accumulation of siRNA through continued delivery is required.
As immune systems in biological organisms are capable of mounting responses against foreign nucleic acids, a growing concern for RNA interference (RNAi) technology is the potential induction of inflammatory cytokines by synthetic siRNAs both in vitro and in vivo. KCs and skin dendritic cells are immune cells, capable of the production of inflammatory cytokines such as TNF-α, IL-6, and type I interferons, as well as chemokines (such as CXCL10), further increasing the immune risks of topically applied siRNAs. Remarkably, our data from blood serum and mouse skin show that siRNAs, when densely conjugated to the nanoparticles in the form of SNA-NCs, are minimally stimulatory and have far fewer off-target effects than the free siRNAs of the same sequence introduced by traditional methods (24). Furthermore, our studies show rapid clearance from skin, minimal accumulation in viscera, and no evidence of histological changes in internal organs after topical delivery. The low immunogenicity and few off-target effects, coupled with low toxicity and high efficacy, point to a significant advantage for using SNA-NC technology to introduce siRNAs.
Taken together, the data presented here provide compelling evidence of RNAi-mediated gene suppression in vivo after topical application to intact skin of siRNAs delivered without the need for additional physical measures, keratolytics, or peptides to enhance penetration. The ability to penetrate into the epidermis in sufficient concentrations to cause morphological change through gene knockdown suggests significant therapeutic potential for treating skin diseases. In fact, the generation of siRNAs is much faster and more cost-effective than the discovery of new small molecule or antibody therapies, and may eventually allow for personalized, genotype-based therapy. The SNA-NC structure is highly tailorable, and the gold core can serve as a scaffold for multicomponent functionalization, not just with siRNAs, but with targeting antibodies or peptides, and small molecule drugs as well (22). As such, these SNA conjugates are part of the growing arsenal of nanomaterials that are showing promise as alternatives to conventional molecular formats for the development of novel and effective therapies for a wide variety of diseases (22, 37–40).
Materials and Methods
Cell Culture, Human Skin Equivalents, and Mouse Skin.
Human epidermal keratinocytes were isolated from neonatal foreskin, as previously described, and cultured in supplemented keratinocyte serum-free medium (M154CF; Cascade Biologics) with 0.07 mM CaCl2 (41). Human cervical cancer HeLa cells and human keratinocyte line HaCaT cells were maintained in DMEM (Gibco®; Invitrogen) with 10% heat-inactivated fetal bovine serum. Human skin equivalents were generated by the Skin Disease Research Center (SDRC) at Northwestern University (NU) and used when they were fully differentiated as intact skin (42). Studies were also performed with EpiDerm skin model samples (MatTek), grown in medium provided by MatTek. All cultures were maintained at 37 °C in 5% CO2. SKH1-E hairless (Charles River Labs) and C57BL/6J (Jackson Laboratory) immunocompetent mice were used for penetration, toxicity, and efficacy studies at 6 wk with institutional Animal Care and Use Committee approval.
Visualization of Nanoparticle Uptake in Vitro and in Vivo.
For in vitro uptake analysis, hKCs grown on chamber coverglass (Nalge Nunc International) were incubated with 50 pM Cy3-labeled SNA-NCs for 2–24 h and the uptake of nanoparticles was visualized by a LSM 510 confocal microscope (Zeiss). Hoechst 33342 (Invitrogen) was used to stain the nucleus of hKCs. For penetration studies, 50 nM Cy5-labeled SNA-NCs in PBS or PBS only were added to the top of the EpiDerm 3D skin samples at different time points. EpiDerm samples were then washed and harvested for hematoxylin and eosin staining or DAPI nuclear stain (Sigma) for fluorescent imaging after OCT embedding. In in vivo studies, the 50 nM Cy5-labeled SNA-NCs in PBS:Aquaphor (Beiersdorf) mixture (1∶1) was applied to the skin of SKH-1E and C57BL/6J mice for up to 24 h before skin samples were harvested and sectioned in OCT for fluorescent imaging.
Quantification of Nanoparticle Uptake, Penetration, and Clearance.
Samples from cells, EpiDerm, and mouse skin were harvested for ICP-MS to measure gold content (SI Text), which quantitatively represents nanoparticle uptake (43). Final values were normalized to the total amount of applied SNA-NCs.
Immune Response Studies.
To evaluate immune responses to nanoparticle treatment, SKH1-E hairless mice were treated for 3 wk as described earlier. Blood was collected in HEMATO-CLAD Hematocrit Tubes (1-000-7500-HC/5; Drummond Scientific). Isolated serum samples were used for ELISA cytokine testing for TNF-α, IL-6, IFN-α, IFN-β, and CXCL10 (ELISA Kit KMC3011, KMC0061, and KMC4041; Invitrogen; Elisa Kit BMS6018 and BMS6027; eBioscience). RNA samples from treated mouse skin were analyzed by RT-qPCR to measure mRNA expression.
Cell Proliferation Assay and Gene Microarray Analysis.
hKCs (3·103 cells/well) in 48-well plates were treated with 0.05 or 0.1 nM nonsense SNA-NCs or PBS for 48 h. Proliferation was assessed daily using a Guava EasyCyte flow cytometer (Millipore). Studies were performed four times in triplicate. Microarray expression analyses were performed at the NU Genomics Core Facility and analyzed at the Bioinformatics Core Facility (44) (SI Text).
mRNA Analysis by RT-qPCR.
Total RNA was extracted and 1 μg of RNA was reverse transcribed using qScript cDNA SuperMix (Quanta BioSciences). Real-time reverse-transcription PCR was performed on the cDNA with LightCycler®480 SYBR Green I Master on a LightCycler®480 system (Roche). The relative abundance of each mRNA transcript was normalized to GAPDH expression, and compared to untreated cells. Primers were purchased from Integrated DNA Technologies (Table S2). Each study was performed three times in triplicate.
Immunoblotting.
Immunoblotting was performed as previously described (45) (SI Text).
Histology and Immunohistologic Analyses.
Human skin equivalents and treated mouse skin, distant skin, and internal organs (liver, lung, and kidney) were harvested, embedded in paraffin, prepared for routine histology, and stained with hematoxylin and eosin. The thickness of mouse epidermis treated with EGFR SNA-NCs, nonsense SNA-NCs, or PBS, each mixed with Aquaphor, was measured (three equidistant sites per specimen) from the basement membrane to the top of the stratum granulosum using the Zeiss AxioPlan2 microscope at 20× magnification and AxioVision software. Samples from treated mouse skin were also stained for Ki-67 using a rabbit monoclonal antibody (SP6; Cell Marque) as a marker for proliferation.
Statistical Analysis.
Multifactor ANOVA was used to assess in vivo study results. Posthoc comparisons after ANOVA and significance testing for in vitro studies were performed by Student’s t test. With all analyses, P < 0.05 was considered significant.
Supplementary Material
ACKNOWLEDGMENTS.
The authors acknowledge the National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant R01 AR060810 (to A.P. and C.M.), National Center for Research Resources Grant UL1 RR025741 (to A.P.), Center for Cancer Nanotechnology Excellence Grant U54CA151880 (to C.M.) from National Cancer Institute, and Army Research Office (C.M.). Core resources and a pilot grant were provided by the Northwestern Skin Disease Research Center (P30AR057216) with support from National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases. Expression analyses were performed at Northwestern University Genomics and Bioinformatics Consulting Cores, and supported by a Cancer Center Support Grant (NCI CA060553) and the National Center for Research Resources (UL1 RR025741). Imaging work and metal analysis were performed at the Northwestern University Quantitative Bioelemental Imaging Center, generously supported by National Science Foundation Grant CHE-9810378/005 and National Aernoautics and Space Administration Ames Research Center Grant NNA04CC36G.
Footnotes
Conflict of interest statement: This technology has been licensed from Northwestern University by AuraSense Therapeutics, LLC. C.A.M., A.S.P., and D.A.G. have financial interests in AuraSense Therapeutics, LLC.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118425109/-/DCSupplemental.
References
- 1.Flaherty KT, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. New Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Von Hoff DD, et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. New Engl J Med. 2009;361:1164–1172. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- 3.Griffiths CEM, et al. Comparison of ustekinumab and etanercept for moderate-to-severe psoriasis. New Engl J Med. 2010;362:118–128. doi: 10.1056/NEJMoa0810652. [DOI] [PubMed] [Google Scholar]
- 4.Mountzios G, Syrigos KN. A benefit-risk assessment of erlotinib in non-small-cell lung cancer and pancreatic cancer. Drug Safety. 2011;34:175–186. doi: 10.2165/11586540-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 5.Adams GP, Weiner LM. Monoclonal antibody therapy of cancer. Nat Biotechnol. 2005;23:1147–1157. doi: 10.1038/nbt1137. [DOI] [PubMed] [Google Scholar]
- 6.Prausnitz MR, Langer R. Transdermal drug delivery. Nat Biotechnol. 2008;26:1261–1268. doi: 10.1038/nbt.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aigner A. Gene silencing through RNA interference (RNAi) in vivo: Strategies based on the direct application of siRNAs. J Biotechnol. 2006;124:12–25. doi: 10.1016/j.jbiotec.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Rayburn ER, Zhang RW. Antisense, RNAi, and gene silencing strategies for therapy: Mission possible or impossible? Drug Discov Today. 2008;13:513–521. doi: 10.1016/j.drudis.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Proksch E, Brandner JM, Jensen J. The skin: An indispensable barrier. Exp Dermatol. 2008;17:1063–1072. doi: 10.1111/j.1600-0625.2008.00786.x. [DOI] [PubMed] [Google Scholar]
- 10.Geusens B, Sanders N, Prow T, Van Gele M, Lambert J. Cutaneous short-interfering RNA therapy. Expert Opinion Drug Deliv. 2009;6:1333–1349. doi: 10.1517/17425240903304032. [DOI] [PubMed] [Google Scholar]
- 11.Leachman SA, et al. First in-human mutation-targeted siRNA phase Ib trial of an inherited skin disorder. Mol Ther. 2010;18:442–446. doi: 10.1038/mt.2009.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosi NL, et al. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 13.Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. Gene regulation with polyvalent siRNA-nanoparticle conjugates. J Am Chem Soc. 2009;131:2072–2073. doi: 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cutler JI, Auyeung E, Mirkin CA. Spherical nucleic acids. J Am Chem Soc. 2012;134:1376–1391. doi: 10.1021/ja209351u. [DOI] [PubMed] [Google Scholar]
- 15.Patel PC, et al. Scavenger receptors mediate cellular uptake of polyvalent oligonucleotide-functionalized gold nanoparticles. Bioconjugate Chem. 2010;21:2250–2256. doi: 10.1021/bc1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: Advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malone RW, Felgner PL, Verma IM. Cationic liposome-mediated RNA transfection. Proc Natl Acad Sci USA. 1989;86:6077–6081. doi: 10.1073/pnas.86.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akinc A, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNamara JO, II, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 20.Song E, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 21.Giljohann DA, et al. Oligonucleotide loading determines cellular uptake of DNA-modified gold nanoparticles. Nano Lett. 2007;7:3818–3821. doi: 10.1021/nl072471q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giljohann DA, et al. Gold nanoparticles for biology and medicine. Angew Chem Int Edit. 2010;49:3280–3294. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seferos DS, Prigodich AE, Giljohann DA, Patel PC, Mirkin CA. Polyvalent DNA nanoparticle conjugates stabilize nucleic acids. Nano Lett. 2009;9:308–311. doi: 10.1021/nl802958f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massich MD, Giljohann DA, Schmucker AL, Patel PC, Mirkin CA. Cellular response of polyvalent oligonucleotide-gold nanoparticle conjugates. ACS Nano. 2010;4:5641–5646. doi: 10.1021/nn102228s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dickens S, et al. Nonviral transfection strategies for keratinocytes, fibroblasts, and endothelial progenitor cells for ex vivo gene transfer to skin wounds. Tissue Eng Part C Methods. 2010;16:1601–1608. doi: 10.1089/ten.TEC.2009.0648. [DOI] [PubMed] [Google Scholar]
- 26.Massich MD, et al. Regulating immune response using polyvalent nucleic acid-gold nanoparticle conjugates. Mol Pharm. 2009;6:1934–1940. doi: 10.1021/mp900172m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cutler JI, et al. Polyvalent nucleic acid nanostructures. J Am Chem Soc. 2011;133:9254–9257. doi: 10.1021/ja203375n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benavides F, Oberyszyn TM, VanBuskirk AM, Reeve VE, Kusewitt DF. The hairless mouse in skin research. J Dermatol Sci. 2009;53:10–18. doi: 10.1016/j.jdermsci.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Batheja P, Song YF, Wertz P, Michniak-Kohn B. Effects of growth conditions on the barrier properties of a human skin equivalent. Pharm Res. 2009;26:1689–1700. doi: 10.1007/s11095-009-9879-1. [DOI] [PubMed] [Google Scholar]
- 30.Tran MA, et al. Targeting V600EB-Raf and Akt3 using nanoliposomal-small interfering RNA inhibits cutaneous melanocytic lesion development. Cancer Res. 2008;68:7638–7649. doi: 10.1158/0008-5472.CAN-07-6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee WR, Shen SC, Zhuo RZ, Wang KC, Fang JY. Enhancement of topical small iinterfering RNA delivery and expression by low-fluence erbium: YAG laser pretreatment of skin. Hum Gene Ther. 2009;20:580–588. doi: 10.1089/hum.2008.156. [DOI] [PubMed] [Google Scholar]
- 32.Garcia M, et al. Development of skin-humanized mouse models of pachyonychia congenita. J Invest Dermatol. 2011;131:1053–1060. doi: 10.1038/jid.2010.353. [DOI] [PubMed] [Google Scholar]
- 33.Ritprajak P, Hashiguchi M, Azuma M. Topical application of cream-emulsified CD86 siRNA ameliorates allergic skin disease by targeting cutaneous dendritic cells. Mol Ther. 2008;16:1323–1330. doi: 10.1038/mt.2008.91. [DOI] [PubMed] [Google Scholar]
- 34.Uchida T, Kanazawa T, Kawai M, Takashima Y, Okada H. Therapeutic effects on atopic dermatitis by anti-RelA short interfering RNA combined with functional peptides Tat and AT1002. J Pharmacol Exp Ther. 2011;338:443–450. doi: 10.1124/jpet.111.180042. [DOI] [PubMed] [Google Scholar]
- 35.Yi XA, et al. MITF-siRNA formulation is a safe and effective therapy for human melasma. Mol Ther. 2011;19:362–371. doi: 10.1038/mt.2010.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsu T, Mitragotri S. Delivery of siRNA and other macromolecules into skin and cells using a peptide enhancer. Proc Natl Acad Sci USA. 2011;108:15816–15821. doi: 10.1073/pnas.1016152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolishetti N, et al. Engineering of self-assembled nanoparticle platform for precisely controlled combination drug therapy. Proc Natl Acad Sci USA. 2010;107:17939–17944. doi: 10.1073/pnas.1011368107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Enlow EM, Luft JC, Napier ME, DeSimone JM. Potent engineered PLGA nanoparticles by virtue of exceptionally high chemotherapeutic loadings. Nano Lett. 2011;11:808–813. doi: 10.1021/nl104117p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis ME, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowman MC, et al. Inhibition of HIV fusion with multivalent gold nanoparticles. J Am Chem Soc. 2008;130:6896–6897. doi: 10.1021/ja710321g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyce ST, Ham RG. Calcium-regulated differentiation of normal human epidermal-keratinocytes in chemically defined clonal culture and serum-free serial culture. J Invest Dermatol. 1983;81:S33–S40. doi: 10.1111/1523-1747.ep12540422. [DOI] [PubMed] [Google Scholar]
- 42.Getsios S, et al. Desmoglein 1-dependent suppression of EGFR signaling promotes epidermal differentiation and morphogenesis. J Cell Biol. 2009;185:1243–1258. doi: 10.1083/jcb.200809044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng D, Seferos DS, Giljohann DA, Patel PC, Mirkin CA. Aptamer nano-flares for molecular detection in living cells. Nano Lett. 2009;9:3258–3261. doi: 10.1021/nl901517b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brazma A, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 45.Wang XQ, et al. Suppression of epidermal growth factor receptor signaling by protein kinase C-alpha activation requires CD82, Caveolin-1, and ganglioside. Cancer Res. 2007;67:9986–9995. doi: 10.1158/0008-5472.CAN-07-1300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.