Abstract
The heterohexameric minichromosome maintenance (MCM2–7) complex is an ATPase that serves as the central replicative helicase in eukaryotes. During initiation, the ring-shaped MCM2–7 particle is thought to open to facilitate loading onto DNA. The conformational state accessed during ring opening, the interplay between ATP binding and MCM2–7 architecture, and the use of these events in the regulation of DNA unwinding are poorly understood. To address these issues in isolation from the regulatory complexity of existing eukaryotic model systems, we investigated the structure/function relationships of a naturally minimized MCM2–7 complex from the microsporidian parasite Encephalitozoon cuniculi. Electron microscopy and small-angle X-ray scattering studies show that, in the absence of ATP, MCM2–7 spontaneously adopts a left-handed, open-ring structure. Nucleotide binding does not promote ring closure but does cause the particle to constrict in a two-step process that correlates with the filling of high- and low-affinity ATPase sites. Our findings support the idea that an open ring forms the default conformational state of the isolated MCM2–7 complex, and they provide a structural framework for understanding the multiphasic ATPase kinetics observed in different MCM2–7 systems.
Keywords: DNA replication, structural dynamics, ATP hydrolysis, helicase loading, helicase activation
The replisome is a large macromolecular machine that coordinates the activity of multiple functional components to ensure the faithful and timely duplication of DNA. Replisome progression is powered, in part, by hexameric helicases, which unwind parental duplexes to provide template strands for DNA synthesis. Because of their ring-shaped structure, the encirclement of single- or double-stranded DNA by replicative helicases is topologically restricted. This restriction can be overcome by either the direct assembly of the motor onto DNA (1–3) or dedicated loading factors that assist ring opening (1, 4, 5). How and why different hexameric helicases use a particular strategy are long-standing questions.
The eukaryotic replicative helicase is composed of six distinct but homologous ATPases associated with various cellular activities (AAA+) subunits, collectively termed the minichromosome maintenance (MCM2–7) complex (6, 7). During the G1-phase of the cell cycle, MCM2–7 heterohexamers are loaded onto DNA by the origin recognition complex together with the Cdc6 and Cdt1 proteins (8, 9). As cells enter S-phase, MCM2–7 is activated through phosphorylation of Mcm2, Mcm4, and Mcm6 by the Dbf4-dependent Cdc7 kinase (10, 11) and the chaperoned association of two accessory factors, Cdc45 and GINS (11). The resultant Cdc45--MCM--GINS (CMG) complex then translocates in the 3′→5′ direction along the leading strand, displacing the lagging strand to facilitate DNA unwinding (12, 13).
Studies of both Saccharomyces cerevisiae (Sce) and Drosophila melanogaster (Dme) MCM2–7 indicated that the helicase ring spontaneously opens between the Mcm2 and Mcm5 subunits (14–16). This breach has been proposed to act as a gate by which DNA enters into the motor interior. What triggers the subsequent closure of the MCM2–7 ring has not been established. In origin recognition complex-dependent loading reactions, MCM2–7 particles become topologically linked to duplex DNA as double hexamers (8, 9), suggesting that the initiation machinery, inter-MCM interactions, and/or nucleic acid binding may seal the Mcm2/5 gate. Biochemical studies of SceMCM2–7 have also indicated that nucleotide alone could promote ring closure (14, 15); however, EM analyses of the DmeMCM2–7 complex have not detected this effect (16). It is currently unknown if the differences between yeast and fly MCMs correspond to species-specific effects or represent distinct intermediates within a general helicase-loading mechanism.
How ATP affects the physical state of the hexamer beyond the Mcm2/5 gate is also unclear. Based on existing structures of homologous archaeal MCMs and AAA+ proteins in general, ATP is thought to bind at the interface between the C-terminal domains of adjacent subunits, generating interprotomer rocking motions that drive processive movement as nucleotide is turned over (17). Interestingly, site-directed mutagenesis studies have indicated that individual MCM2–7 subunits contribute unequally to ATPase and helicase activities (12, 15, 18). ATP hydrolysis profiles of MCM2–7 also do not conform to a simple Michaelis–Menten model, indicating that a combination of independent high- and low-affinity active sites exists within the enzyme (12, 19). Neither the structural consequences of nucleotide binding to the MCM2–7 ring nor a physical explanation for the asymmetric contributions of its constituent subunits has been delineated.
To investigate these issues, we used negative-stain electron microscopy (EM) and small-angle X-ray scattering (SAXS) to assess the nucleotide-dependent conformational dynamics of a naturally minimized MCM2–7 complex derived from Encephalitozoon cuniculi (Ecu). We find that, like its homologs, EcuMCM2–7 is an active ATPase that displays a multistage response to nucleotide concentration (12, 19). We also show that EcuMCM2–7 naturally adopts a left-handed, open-ring configuration through a systematic rotational offset between subunit pairs. Nucleotide binding results in ring constriction but not ring closure, even at high concentrations; these structural changes correlate with key transition points in ATPase activity, indicating that ring remodeling is coupled to ATP binding and turnover. Our data support the concept that MCM2–7 complexes naturally favor an open-ring state and that the nonequivalent biochemical properties of different MCM subunits may be linked to a predisposed structural relationship between neighboring protomers.
Results
Isolation and Characterization of the EcuMCM2–7 Complex.
E. cuniculi is a protozoan parasite with one of the smallest known eukaryotic genomes (2.9 MB), which nevertheless encodes nearly all known replicative factors, including all six core MCM subunits (20) (Fig. S1). However, because of its streamlined nature, many E. cuniculi proteins have been extensively minimized (21). For example, EcuMCM2–7 lacks many of the unstructured terminal and internal regions and thus, many of the kinase target sites found or predicted in other eukaryotic MCMs (Fig. 1A and Fig. S2 and Table S1). Correspondingly, genes for key regulatory factors such as Dbf4-dependent Cdc7 kinase do not seem to exist in E. cuniculi. Although E. cuniculi GINS homologs do exist, the organism does not seem to possess Cdc45, possibly using either Cdc45 from the host or an accessory factor that is too diverged in sequence to permit recognition. Nevertheless, all other features of eukaryotic MCM proteins, including the ATPase active site elements and Mcm6 winged helix domain, are conserved in EcuMCM2–7 (Fig. S1).
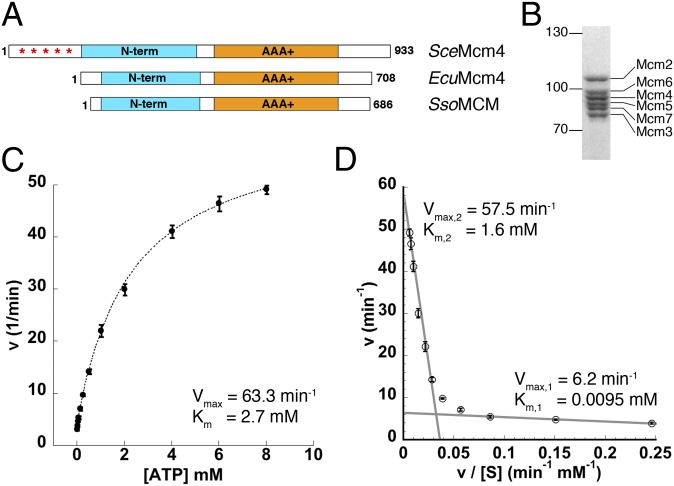
Fig. 1.
Purified EcuMCM2–7 is an active ATPase. (A) Schematic of the primary structure of (Middle) EcuMcm4 compared with (Top) SceMcm4 and (Bottom) Sulfolobus solfataricus MCM (SsoMCM). Asterisks denote the hyperphosphorylated N-terminal tail of SceMcm4. (B) Coomassie blue-stained SDS/PAGE of purified EcuMCM2–7. (C) Initial ATP hydrolysis kinetics for EcuMCM2–7 plotted as a function of ATP concentration. (D) Eadie–Hofstee plot of EcuMCM2–7 ATPase activity. Linear regions of low- and high-velocity turnover are indicated by gray lines.
Therefore, we reasoned that EcuMCM2–7 might serve as a simplified model system that recapitulates the basal functional properties of eukaryotic MCMs without the complicating effects of posttranslational modifications. We coexpressed and purified all six EcuMCM2–7 subunits from baculovirus-infected insect cells (Materials and Methods, SI Materials and Methods, and Fig. S3A). For affinity chromatography, all six subunits were produced as fusions to the maltose binding protein (MBP), which was subsequently removed during the final stages of purification. The resultant complex eluted as a single peak from a size-exclusion column at a volume consistent with an intact heterohexamer (Fig. S3B). The final product was >95% pure by Coomassie-stained SDS/PAGE (Fig. 1B) and capable of binding short ssDNA sequences in the presence of ATP, with an apparent KD comparable with the KD reported for other eukaryotic and archaeal MCMs (∼200 nM) (Fig. S3C) (12, 22–25).
Also similar to other MCMs (12, 19, 22, 26), the ATPase activity of EcuMCM2–7 was only minimally stimulated by DNA (Fig. S3D). Plotting initial hydrolysis rates vs. ATP concentration yielded an overall Vmax = 63.3 ± 2.9 min−1 and Km = 2.7 ± 0.3 mM, with no evidence of cooperativity (Fig. 1C). Eadie–Hofstee analysis of these data (v vs. v/[S]) revealed a strong deviation from linearity (Fig. 1D), subdividing the overall kinetic profile into low- and high-velocity modes akin to the multimodal kinetics reported for yeast and fly MCMs (12, 19). Overall, the ATPase activity of EcuMCM2–7 is similar to SceMCM2–7 (19) and much greater than the activity of DmeMCM2–7 (12). Unfortunately, efforts to measure helicase activity for EcuMCM2–7 resulted in only a modest amount of displaced product visible on radiolabeled gels, suggesting that, as with SceMCM2–7 (14), special buffer conditions may be required to observe unwinding.
EcuMCM2–7 Adopts a Left-Handed, Open-Ring Conformation.
Next, we examined the structure of EcuMCM2–7 in the presence and absence of the ATP analog ATPγS using negative-stain EM. Micrographs of apo EcuMCM2–7 exhibited evidence of hexameric particles (Fig. S4 A and B), but the high conformational heterogeneity in the sample precluded attempts at 3D reconstruction. By contrast, samples imaged with 10 mM ATPγS yielded more uniform particles (Fig. S4C), with 2D class averages showing a nearly exclusive abundance of either skewed, two-tiered species or open rings composed of six discrete lobes (Fig. 2A and Fig. S4D). 3D reconstructions (at 24 Å resolution) showed that these images correspond to side and top/bottom views, respectively, of a lock washer-shaped hexameric particle (Fig. 2B, SI Materials and Methods, and Fig. S4 D–F). In contrast to the mixture of fully open and notched-ring forms reported for DmeMCM2–7 (16), EcuMCM2–7 seems to adopt a single nonplanar, open-ring state (Fig. 2B). Although subunit mapping was not performed for EcuMCM2–7, findings for both yeast and fly MCM2–7 (14–16, 18) suggest that the opening in the E. cuniculi complex likely corresponds to the Mcm2/5 DNA gate.
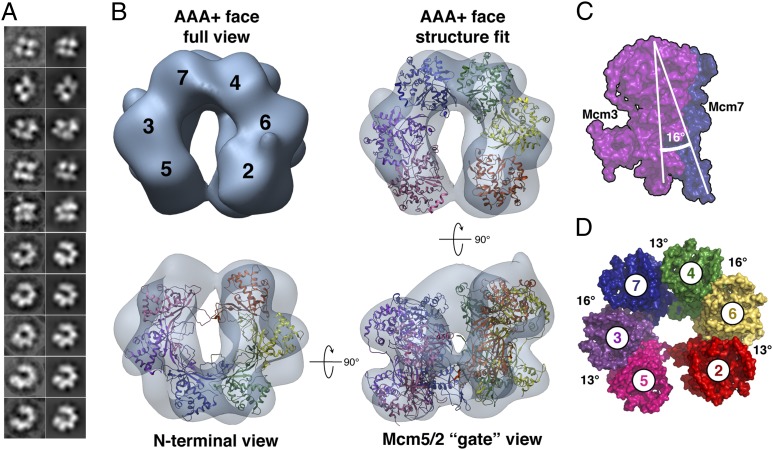
Fig. 2.
EcuMCM2–7 is a left-handed open ring. (A) Reference-free class averages (Left) and corresponding forward projections (Right) from ATPγS-bound EcuMCM2–7 3D reconstructions. (B) Final 3D reconstruction of ATPγS-bound EcuMCM2–7. Monomer crystal structures of SsoMCM (Protein Data Bank ID code 3F9V) (28) are fitted into the reconstruction. (C and D) Intersubunit angles in EcuMCM2–7. Atomic models of SsoMCM monomers, docked and colored as in B, are shown in surface representation.
Conversion of 2D EM projections into 3D structures by single-particle reconstitution results in ambiguous chirality (27). Docking studies using the crystal structure of an archaeal MCM monomer (28) initially showed that six subunits could be reasonably fitted into both left- and right-handed EcuMCM2–7 reconstructions. However, cross-correlation values somewhat disfavored the right-handed conformation (Fig. S5A) and fitting into the right-handed open ring resulted in clashes between protomers as well as disruption of AAA+ interfaces (Fig. S5 B–D). By contrast, the left-handed model (i) exhibited virtually no clashes, (ii) yielded the expected alignment of the five composite ATPase sites present in an open ring (Fig. S5 B–D), and (iii) revealed an additional feature in the EM density consistent with the C-terminal winged helix domain of Mcm6 (Fig. S5 E–G). Taken together, these results suggest that ATPγS-bound EcuMCM2–7 adopts a left-handed, open-ring state, and they support our assumption that the ring is breached at the Mcm2/5 interface. Absolute hand determination by tilt-pair analysis (SI Materials and Methods and Fig. S6A) showed that DmeMCM2–7 also adopts a left-handed, open-ring conformation vs. the right-handed structure that was reported previously (16); inspection of 3D reconstructions of MBP-modified DmeMCM2–7 confirmed this reassignment (SI Materials and Methods and Fig. S6 B–D and S7).
Computational monomer fitting reveals that the lock washer shape of EcuMCM2–7 arises from a 13–16° twist of each subunit with respect to its partner (Fig. 2 C and D). At first glance, this arrangement is reminiscent of the left-handed lock washer adopted by the Rho transcription termination factor, a homohexameric RecA-family ssRNA translocase. In Rho, however, ring opening is accomplished by a uniform ∼15° rotational offset (29). By contrast, rotations between adjoining subunits in EcuMCM2–7 alternate, with a greater angular rise occurring at the interfaces ascribed to Mcm6/4 and Mcm7/3. These staggered differences in orientation subdivide the hexamer into what seems to be a trimer of heterodimers (Fig. 2D and Movie S1).
Nucleotide Compacts but Does Not Close the EcuMCM2–7 Ring in Solution.
Although EM established that EcuMCM2–7 forms open rings in the presence of nucleotide, we were concerned that the observed predominance of this state might be influenced by surface adsorption effects. We, therefore, used SAXS (30) to determine the size and shape of the particle in solution. EcuMCM2–7 samples showed no signs of aggregation, which was evidenced by inspection of scattering at low angles (Fig. S8A). Scattering curves obtained under apo conditions and in the presence of 10 mM ATPγS were roughly similar but differed noticeably in the low and mid q range (Fig. 3A). We also noted that ATPγS led to decrease in the apparent radius of gyration (Rg; from 61.8 to 59.1 Å) (Fig. 3A, Inset). Several tests verified that the changes in scattering were not caused by complex dissociation or subunit unfolding, and thus, they could be interpreted as structural effects on the EcuMCM2–7 ring because of nucleotide binding (SI Materials and Methods). Even at 10 mM ATPγS, EcuMCM2–7 retains some conformational heterogeneity, which could be caused by unstructured terminal regions or flexing of the open ring in solution. This finding is evidenced by both the relatively uniform shape of the scattering curve (vs. more featured theoretical curves generated from atomic models) (Fig. 3B) and the inflated molecular weight estimated from the particle volume (SI Materials and Methods and Fig. S8C). Although these properties precluded an ab initio 3D reconstruction by SAXS, the data permitted comparison with our EM studies.
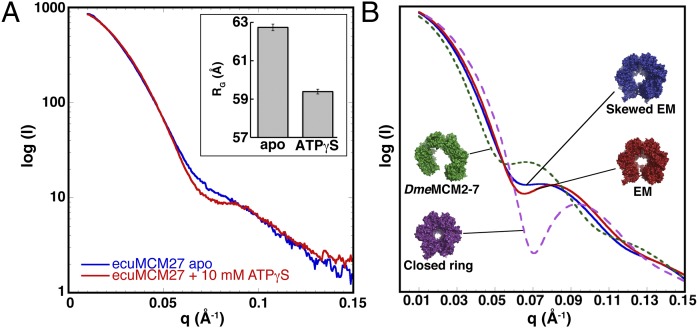
Fig. 3.
Nucleotide binding does not close the EcuMCM2–7 ring. (A) SAXS profiles for EcuMCM2–7 in the absence (blue) and presence (10 mM; red) of ATPγS. (Inset) Rg values calculated from each experiment. (B) Surface representation and theoretical scattering curve calculated from EcuMCM2–7 homology models docked into the EM reconstruction (red). Manually adjusted models and curves also are shown for a planar open ring based on DmeMCM2–7 (green dashed line), a more spiral, skewed EM-based state (blue line), and a fully closed hexamer (magenta dashed line).
We first created a theoretical scattering curve from the MCM homology models docked into our ATPγS-bound EM volume (Fig. 3B and SI Materials and Methods). The resultant curve compared reasonably well with the experimental scattering data obtained in the presence of ATPγS and exhibited features such as a dip at mid q values. By contrast, neither the curve calculated from a closed-ring hexamer model nor the curve derived from the open-ring lockwasher state seen for DmeMCM2–7 resemble the observed scattering (Fig. 3B). Interestingly, the apo EcuMCM2–7 scattering could only be approximated by taking the ATPγS-bound EM-based homology model and imposing larger offsets (+5°) between adjoining subunits to increase the pitch of the cracked ring (Fig. 3B). Together, these findings suggest that the open-ring state observed for ATPγS-bound EcuMCM2–7 by EM is the dominant form of the complex in solution and that, in absence of ATP, EcuMCM2–7 may adopt a more extended spiral conformation.
EcuMCM2–7 Structural Transitions Correlate with Distinct Phases of ATPase Activity.
Given a flexible, multistate system, Rg can be used to quantitatively report on how the equilibrium between these states changes over a range of conditions (31). To understand in more detail how the EcuMCM2–7 structure responds to nucleotide, we collected SAXS measurements at several ATPγS concentrations (Fig. 4A). The resulting data showed a smoothly varying change in the mid q region of the curve that filled in the more discrete transition seen between 0 and 10 mM ATPγS (Fig. S8B). Rg values calculated from these curves also changed, reaching a minimum at ∼59.5 Å (±0.5) as ATPγS levels approached and exceeded 1 mM (Fig. 4A, Inset). Thus, nucleotide binding compacts the EcuMCM2–7 ring in a concentration-dependent manner.
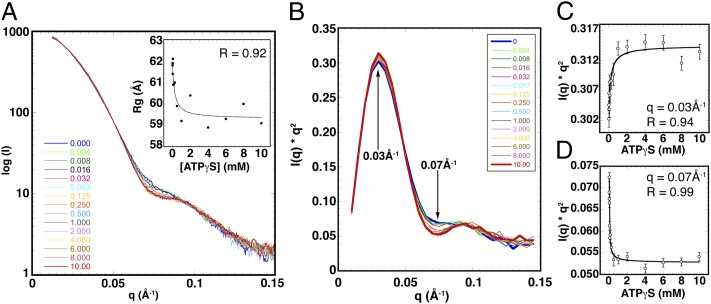
Fig. 4.
The EcuMCM2–7 ring contracts in a two-step process. (A) SAXS profiles for ATPγS titrated (0–10 mM) against EcuMCM2–7. (Inset) Rg values plotted against ATPγS concentration and fitted to a binding hyperbola (dotted line). (B) Smoothed I × q2 vs. q (Kratky) plot of scattering curves shown in A. (C and D) Scattering intensity values at low (C) and medium (D) q values plotted against ATPγS concentration and fitted to the binding hyperbola (solid line).
Plotting Rg against ATPγS concentration (Fig. 4A, Inset and SI Materials and Methods) and fitting the ATPγS-dependent Rg measurements to the Michaelis–Menten hyperbolic equation (SI Materials and Methods) yield an apparent equilibrium constant for ring compaction (KRq, app) of 0.23 ± 0.14 mM ATPγS. This value is nearly identical to the concentration of nucleotide required to switch the ATPase activity of EcuMCM2–7 from its low- to high-velocity mode (Fig. S8D), suggesting that nucleotide-induced structural and kinetic transitions in EcuMCM2–7 are tightly coupled. Given that the shift to a high-velocity ATPase rate in DmeMCM2–7 occurs at a similar concentration of substrate (12), this linkage may be broadly preserved.
Nucleotide Affects EcuMCM2–7 Structure in a Multistep Manner.
Although the change in Rg seen for EcuMCM2–7 was consistent with our structural and kinetic data, the magnitude of the effect on particle diameter was modest and exhibited a degree of variation beyond 1.0 mM ATPγS. Like Rg, scattering curves arise from the average ensemble of all particles in solution. Hence, scattering from a monodisperse sample can be used to directly assess the equilibrium between structural states. Replotting our SAXS data as I × q2 vs. q (also known as a Kratky plot) (30) confirms that the EcuMCM2–7 complex is remodeled by nucleotide (Fig. 4B). The Kratky peaks sharpen at high ATPγS concentrations, indicating that nucleotide binding constrains the conformational heterogeneity of EcuMCM2–7.
Two regions of the Kratky curve change as nucleotide is added (Fig. 4B): a maximum at q = 0.03Å−1 that reflects relatively large-scale domain movements and a minimum at q = 0.07Å−1 that corresponds to smaller-scale structural adjustments. When Kratky plot values at these two points are plotted against ATPγS concentration, both sets of data can be fit to the same hyperbolic function, which was used for assessing the changes in Rg (Fig. 4 C and D and SI Materials and Methods). From this finding, we can again determine pseudoequilibrium constants from the inflection points of the binding curves, now corresponding to large- and small-scale structural transitions (Fig. 4 C and D and SI Materials and Methods). Notably, the apparent equilibrium constant for the low q scattering change is virtually identical to the constant obtained for Rg (Kk1, app = 0.24 ± 0.11 mM). The congruence between these two constants and the ATP concentration required to shift EcuMCM2–7 into a high ATPase phase indicates that it is the occupancy of the particle’s lower-affinity ATPase sites that triggers the global structural alteration.
By contrast, the apparent equilibrium constant for the scattering change at intermediate q is an order of magnitude lower (Kk2, app = 0.032 ± 0.010 mM). This difference implies that local conformational transitions occur at low concentrations of nucleotide, which in turn, mediate the high-affinity, low-velocity phase of ATPase activity reported by Eadie–Hofstee analysis (Fig. S8D). Although the exact nature of these local effects cannot be defined from SAXS data alone, this correspondence suggests that they reflect the association of nucleotide with tight-binding active sites. Because Rg remains virtually unchanged at these substrate concentrations (Fig. 4A, Inset), these early nucleotide binding events may reflect rearrangements in loops and secondary structure around the catalytic center rather than en bloc domain or subunit movements. The fit to the two-site binding model, although imprecise, was in agreement with this interpretation (SI Materials and Methods and Fig. S8 F–H). Overall, our results show that the effects of nucleotide binding on EcuMCM2–7 architecture are nonuniform and that they likely arise from the differential occupancy of at least two classes of active sites in the complex.
Discussion
To begin to define the nucleotide-dependent architectural dynamics of the eukaryotic MCM2–7 helicase, we undertook a structural study of a naturally minimized version of the complex from E. cuniculi. EcuMCM2–7 lacks the large, unstructured, and posttranslationally modified regions that help regulate loading and activation in other eukaryotic helicases (Fig. S1) (10, 32), and it could serve as a model system for probing basic structure/activity relationships of eukaryotic MCMs. EcuMCM2–7 was isolated as a stable assembly, with ATPase and DNA binding properties comparable with those properties of other MCM proteins (Fig. 1 and Fig. S3).
Our SAXS studies suggest that the complex ATPase behavior seen in archaeal and eukaryotic MCMs is linked to a set of ATP-dependent large- and small-scale structural changes, which occur at different nucleotide concentrations (Fig. 4). The likewise biphasic relationship between ATPase activity and ATP levels (Fig. 1D) reflects the contributions of both tight-binding, low-velocity sites and weakly binding, high-velocity sites. Remarkably, the nucleotide-dependent transition point for the global structural changes coincides with the entry of the EcuMCM2–7 into a fast ATP turnover regimen. This finding implies that, as ATP fills a set of secondary sites in the EcuMCM2–7 ring, the shift to a more compact ring state helps switch the enzyme into a high-velocity ATPase state (Fig. 5A).
Fig. 5.
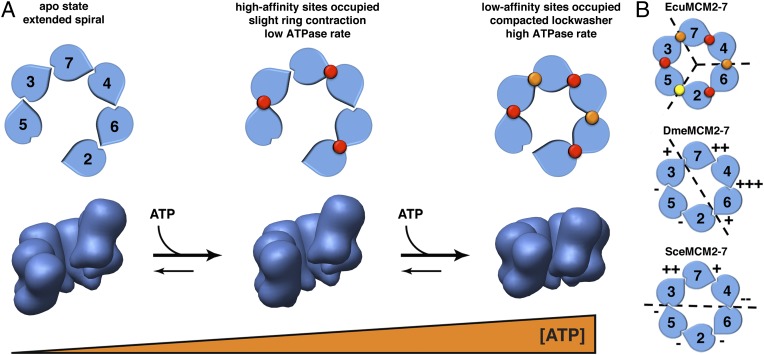
Model for ATP-mediated restructuring and activation of EcuMCM2–7. (A) Schematic for a two-step, nucleotide-dependent change in the ATPase activity and structural organization of EcuMCM2–7. Occupancy of high- and low-affinity ATPase sites is denoted by red and orange circles, respectively. (B) Apparent functional and/or structural subdivisions of eukaryotic MCM assemblies based on available data to date (dashed lines); + and − symbols denote the relative importance of select MCM active sites for ATPase activity (12, 15). High- and low-affinity sites in EcuMCM2–7 are indicated as in A, whereas the closed Mcm5/2 site is marked with a yellow circle. The EcuMCM2–7 data show that a structural asymmetry between different pairs of subunits may underlie the different classes of ATPase sites seen biochemically. Yeast and fly mutational data have uncovered differences in the relative contributions of individual subunits to ATPase activity that we suggest may be rooted in an asymmetric structural pattern around the MCM ring.
Considering that the EcuMCM2–7 complex appears to be subdivided into pairwise units (Fig. 2 C and D)—likely corresponding to Mcm2/6, Mcm4/7, and Mcm3/5—it is possible that the catalytic centers between MCM heterodimers create one class of ATP binding site, whereas the active sites formed within heterodimers form another class (Fig. 5A). Different MCM subunits are known to contribute unequally to ATPase and helicase activity (12, 15, 18), which may help MCMs tolerate several inactive subunits without significant loss of function (12, 15, 33). If this model were correct, EcuMCM2–7 would exhibit an alternating distribution of high- and low-affinity active sites akin to a trimer of dimers. Such a scheme would differ from the more asymmetric architectures implied by mutagenesis studies of yeast and fly MCMs (12, 15) (Fig. 5B), but it may align with proposed views of archaeal MCM function (33). These trends suggest that the observed subunit inequivalencies may arise from intrinsic structural differences that allow one class of active site (e.g., high-turnover centers) to contribute disproportionately to the overall rate of hydrolysis compared with the other. Higher-resolution studies of substrate-bound MCM particles will be needed to more fully probe these similarities and differences.
Using EM and SAXS, we found that EcuMCM2–7 spontaneously and predominantly forms a left-handed, lock washer-shaped structure (Figs. 2 and 3) and that this state is adopted by DmeMCM2–7 as well (Fig. S6 and S7). Although we did not map the subunits, work in other systems (14–16, 18), as well as analysis of the features in our EM map (Fig. S5 E–G), supports the assumption that the EcuMCM2–7 ring is breached between Mcm2 and Mcm5. The observation of open MCM2–7 rings in yeast, flies, and microsporidians suggests that this state is a dominant structural ground state for the system. Interestingly, the addition of nucleotide to EcuMCM2–7 caused a global contraction but not full closure of the helicase (Figs. 3 and 4). This finding is similar to the effect of nucleotide seen for DmeMCM2–7, but it contrasts with findings in budding yeast, where the addition of ATP was reported to close the Mcm2/5 gate (14, 15). It may be that MCMs of different organisms may have evolved varying responses to nucleotide binding, possibly because of the differential contributions of distinct subunits to ATPase activity across species (12, 15, 18).
The most parsimonious role for an open MCM ring is to facilitate the loading and/or recycling of the helicase onto or off of replication origins, which do not possess freely accessible DNA ends. Generally, hexameric helicases are thought to gain access to nucleic acid substrates either by direct assembling onto DNA or a ring-opening mechanism (1–5). Of the latter approach, two variations have been described. One approach is the use of a secondary factor that actively breaches the helicase; the bacterial DnaC/DnaI protein has been proposed to actively open DnaB family helicases to support loading (1, 4, 5). The other strategy is for the hexameric helicase itself to interconvert between open and closed states. The Rho-transcription termination factor is thought to act in such a manner, adopting an open-ring state in the absence of RNA, and it spontaneously transitions into an active, closed-ring form on engaging substrate (29, 34, 35). Combining our data with other observations in the field, we suggest that MCM2–7 comprises a third hybrid category: a naturally open hexamer, which relies on auxiliary proteins (e.g., GINS and Cdc45) to form a functional toroid on binding DNA. The propensity for MCM2–7 to form open rings may also be exploited by other factors to promote MCM2–7 dissociation and/or inactivation after replication is complete.
Why an open MCM2–7 ring should adopt a left-handed configuration is less clear. Interestingly, on binding ssRNA, the left-handed, open-ring state of Rho converts into a closed-ring structure that bears a right-handed spiral arrangement of internal pore loops (29, 35). The significance of this conversion is unknown, but it may introduce physical strain within the helicase ring that could be harnessed by cycles of ATP binding and hydrolysis; strain has been proposed to assist motor activity in the ring-shaped F0F1-ATPase (36). Additionally, formation of a left-handed conformation could serve as an intermediate state that promotes the melting of DNA on MCM2–7 activation (for instance, by pulling strands in a direction counter to the innate right-handed chirality of the duplex). Future studies will be needed to distinguish between these mechanisms and better define the order of ATP binding and action of individual subunits in molecular detail.
Materials and Methods
More extensive protocols can be found in SI Materials and Methods.
Expression and Purification of EcuMCM2–7.
All EcuMCM2–7 subunits were constructed as protease-cleavable 6xHis-MBP fusions, which allowed removal of both tags during purification. Recombinant bacmids of individual subunits were generated using the Bac-to-Bac protocol (Invitrogen). EcuMCM2–7 was expressed by coinfection of cultured insect cells (Sf9) and purified using a combination of affinity, ion exchange, and size-exclusion chromatography.
DNA Binding and ATPase Assays.
DNA binding was assayed by fluorescence anisotropy in the presence of 1 mM ATP, with EcuMCM2–7 concentration varied within the 0–5 μM range. Initial ATP turnover rates were determined using a pyruvate kinase/aldehyde dehydrogenase coupled assay (37), with EcuMCM2–7 concentration fixed at 1.0 μM and ATP concentration varied from 0 to 8 mM. All data were plotted and analyzed by nonlinear regression in Kaleidagraph.
EM.
Dilute samples of EcuMCM2–7 (∼15 μg/mL), preincubated with 10 mM ATPγS (Sigma-Aldrich), were deposited onto holey carbon grids and stained by uranyl formate. Data were collected at magnification of 30,000×, resulting in a pixel size of 3.56 A. Data were processed using the APPION processing environment (38). The 2D analysis was carried out using IMAGIC (39) and XMIPP (40). 3D reconstruction was performed by iterative projection-matching approach using EMAN2 (41) and SPARX (42). UCSF Chimera (43) was used for atomic model docking and visualization of structures.
SAXS.
SAXS data were collected at beamline 12.3.1 at the Advanced Light Source of Lawrence Berkeley National Laboratories. EcuMCM2–7 concentration was fixed at 3.0 mg/mL, whereas ATPγS concentration was varied within the 0–10 mM range. Data were processed and analyzed using Primus (44). Theoretical models were generated using FoxS (45). Nonlinear regression analysis was carried out in Kaleidagraph.
Supplementary Material
Acknowledgments
We thank Gregory L. Hura and Susan Tsutakawa for assistance with SAXS analysis, Ivar Ilves and Richard U. Rymer for insights into MCM biochemistry and assays, Ernesto Arias Palomo for assistance with EM, and Eva Nogales for the access to her electron microscope. We also thank Michael Cianfrocco, John Rubinstein, and Giulia Zanetti for help with absolute hand determination and Raffaella Carzaniga and Lucy Collinson at the London Research Institute EM Unit for technical support. This work was supported by an American Cancer Society Postdoctoral Fellowship (A.Y.L.), a European Molecular Biology Organization Postdoctoral Fellowship (A.C.), the Miller Research Fellowship (F.B.), and National Institutes of Health Grants R37-30490 (to M.R.B.) and GM074747 (to J.M.B.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The single-particle 3D EM reconstruction reported in this paper has been deposited in the EMDataBank, www.emdatabank.org/ (accession no. EMD-5429).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209406109/-/DCSupplemental.
References
- 1.Davey MJ, O’Donnell M. Replicative helicase loaders: Ring breakers and ring makers. Curr Biol. 2003;13:R594–R596. doi: 10.1016/s0960-9822(03)00523-2. [DOI] [PubMed] [Google Scholar]
- 2.Kumar A, et al. Model for T-antigen-dependent melting of the simian virus 40 core origin based on studies of the interaction of the beta-hairpin with DNA. J Virol. 2007;81:4808–4818. doi: 10.1128/JVI.02451-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuck S, Stenlund A. Mechanistic analysis of local ori melting and helicase assembly by the papillomavirus E1 protein. Mol Cell. 2011;43:776–787. doi: 10.1016/j.molcel.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Funnell BE, Baker TA, Kornberg A. In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J Biol Chem. 1987;262:10327–10334. [PubMed] [Google Scholar]
- 5.Davey MJ, Fang L, McInerney P, Georgescu RE, O’Donnell M. The DnaC helicase loader is a dual ATP/ADP switch protein. EMBO J. 2002;21:3148–3159. doi: 10.1093/emboj/cdf308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 7.Koonin EV. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 1993;21:2541–2547. doi: 10.1093/nar/21.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evrin C, et al. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci USA. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Remus D, et al. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Randell JC, et al. Mec1 is one of multiple kinases that prime the Mcm2-7 helicase for phosphorylation by Cdc7. Mol Cell. 2010;40:353–363. doi: 10.1016/j.molcel.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heller RC, et al. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011;146:80–91. doi: 10.1016/j.cell.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bochman ML, Schwacha A. The Mcm2-7 complex has in vitro helicase activity. Mol Cell. 2008;31:287–293. doi: 10.1016/j.molcel.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 15.Bochman ML, Schwacha A. The Saccharomyces cerevisiae Mcm6/2 and Mcm5/3 ATPase active sites contribute to the function of the putative Mcm2-7 ‘gate.’. Nucleic Acids Res. 2010;38:6078–6088. doi: 10.1093/nar/gkq422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costa A, et al. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol. 2011;18:471–477. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyubimov AY, Strycharska M, Berger JM. The nuts and bolts of ring-translocase structure and mechanism. Curr Opin Struct Biol. 2011;21:240–248. doi: 10.1016/j.sbi.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davey MJ, Indiani C, O’Donnell M. Reconstitution of the Mcm2-7p heterohexamer, subunit arrangement, and ATP site architecture. J Biol Chem. 2003;278:4491–4499. doi: 10.1074/jbc.M210511200. [DOI] [PubMed] [Google Scholar]
- 19.Schwacha A, Bell SP. Interactions between two catalytically distinct MCM subgroups are essential for coordinated ATP hydrolysis and DNA replication. Mol Cell. 2001;8:1093–1104. doi: 10.1016/s1097-2765(01)00389-6. [DOI] [PubMed] [Google Scholar]
- 20.Biderre C, Pagès M, Méténier G, Canning EU, Vivarès CP. Evidence for the smallest nuclear genome (2.9 Mb) in the microsporidium Encephalitozoon cuniculi. Mol Biochem Parasitol. 1995;74:229–231. doi: 10.1016/0166-6851(95)02495-6. [DOI] [PubMed] [Google Scholar]
- 21.Katinka MD, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 22.Haugland GT, Rollor CR, Birkeland NK, Kelman Z. Biochemical characterization of the minichromosome maintenance protein from the archaeon Thermoplasma acidophilum. Extremophiles. 2009;13:81–88. doi: 10.1007/s00792-008-0198-y. [DOI] [PubMed] [Google Scholar]
- 23.McGeoch AT, Trakselis MA, Laskey RA, Bell SD. Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat Struct Mol Biol. 2005;12:756–762. doi: 10.1038/nsmb974. [DOI] [PubMed] [Google Scholar]
- 24.Pucci B, et al. Modular organization of the Sulfolobus solfataricus mini-chromosome maintenance protein. J Biol Chem. 2007;282:12574–12582. doi: 10.1074/jbc.M610953200. [DOI] [PubMed] [Google Scholar]
- 25.Bochman ML, Schwacha A. Differences in the single-stranded DNA binding activities of MCM2-7 and MCM467: MCM2 and MCM5 define a slow ATP-dependent step. J Biol Chem. 2007;282:33795–33804. doi: 10.1074/jbc.M703824200. [DOI] [PubMed] [Google Scholar]
- 26.Atanassova N, Grainge I. Biochemical characterization of the minichromosome maintenance (MCM) protein of the crenarchaeote Aeropyrum pernix and its interactions with the origin recognition complex (ORC) proteins. Biochemistry. 2008;47:13362–13370. doi: 10.1021/bi801479s. [DOI] [PubMed] [Google Scholar]
- 27.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 28.Brewster AS, et al. Crystal structure of a near-full-length archaeal MCM: Functional insights for an AAA+ hexameric helicase. Proc Natl Acad Sci USA. 2008;105:20191–20196. doi: 10.1073/pnas.0808037105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skordalakes E, Berger JM. Structure of the Rho transcription terminator: Mechanism of mRNA recognition and helicase loading. Cell. 2003;114:135–146. doi: 10.1016/s0092-8674(03)00512-9. [DOI] [PubMed] [Google Scholar]
- 30.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: Defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 31.Blobel J, Bernadó P, Svergun DI, Tauler R, Pons M. Low-resolution structures of transient protein-protein complexes using small-angle X-ray scattering. J Am Chem Soc. 2009;131:4378–4386. doi: 10.1021/ja808490b. [DOI] [PubMed] [Google Scholar]
- 32.Chuang LC, et al. Phosphorylation of Mcm2 by Cdc7 promotes pre-replication complex assembly during cell-cycle re-entry. Mol Cell. 2009;35:206–216. doi: 10.1016/j.molcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreau MJ, McGeoch AT, Lowe AR, Itzhaki LS, Bell SD. ATPase site architecture and helicase mechanism of an archaeal MCM. Mol Cell. 2007;28:304–314. doi: 10.1016/j.molcel.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 34.Skordalakes E, Berger JM. Structural insights into RNA-dependent ring closure and ATPase activation by the Rho termination factor. Cell. 2006;127:553–564. doi: 10.1016/j.cell.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 35.Thomsen ND, Berger JM. Running in reverse: The structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–534. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cherepanov DA, Mulkidjanian AY, Junge W. Transient accumulation of elastic energy in proton translocating ATP synthase. FEBS Lett. 1999;449:1–6. doi: 10.1016/s0014-5793(99)00386-5. [DOI] [PubMed] [Google Scholar]
- 37.Lindsley JE. Use of a real-time, coupled assay to measure the ATPase activity of DNA topoisomerase II. Methods Mol Biol. 2001;95:57–64. doi: 10.1385/1-59259-057-8:57. [DOI] [PubMed] [Google Scholar]
- 38.Lander GC, et al. Appion: An integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 40.Scheres SH, Núñez-Ramírez R, Sorzano CO, Carazo JM, Marabini R. Image processing for electron microscopy single-particle analysis using XMIPP. Nat Protoc. 2008;3:977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 42.Hohn M, et al. SPARX, a new environment for Cryo-EM image processing. J Struct Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 43.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 44.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 45.Schneidman-Duhovny D, Hammel M, Sali A. FoXS: A web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010;38:W540–W544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.