Abstract
DNA methylation is a heritable epigenetic mark that controls gene expression, is responsive to environmental stresses, and, in plants, may also play a role in heterosis. To determine the degree to which DNA methylation is inherited in rice, and how it both influences and is affected by transcription, we performed genome-wide measurements of these patterns through an integrative analysis of bisulfite-sequencing, RNA-sequencing, and siRNA-sequencing data in two inbred parents of the Nipponbare (NPB) and indica (93–11) varieties of rice and their hybrid offspring. We show that SNPs occur at a rate of about 1/253 bp between the two parents and that these are faithfully transmitted into the hybrids. We use the presence of these SNPs to reconstruct the two chromosomes in the hybrids according to their parental origin. We found that, unlike genetic inheritance, epigenetic heritability is quite variable. Cytosines were found to be differentially methylated (epimutated) at a rate of 7.48% (1/15 cytosines) between the NPB and 93–11 parental strains. We also observed that 0.79% of cytosines were epimutated between the parent and corresponding hybrid chromosome. We found that these epimutations are often clustered on the chromosomes, with clusters representing 20% of all epimutations between parental ecotypes, and 2–5% in F1 plants. Epimutation clusters are also strongly associated with regions where the production of siRNA differs between parents. Finally, we identified genes with both allele-specific expression patterns that were strongly inherited as well as those differentially expressed between hybrids and the corresponding parental chromosome. We conclude that much of the misinheritance of expression levels is likely caused by epimutations and trans effects.
Keywords: bioinformatics, plant biology
DNA methylation is an epigenetic mark that can often lead to the repression of gene expression (1, 2). It is enriched in heterochromatin and, when present at regulatory sites, usually acts as a repressor of expression, most notably in transposons (3). However, it is also found over coding regions, where it likely does not directly affect transcription and is associated with moderately expressed genes (2, 4–6). In plants, DNA methylation occurs in three different contexts: CG, CHG, and CHH (where H is any nucleotide but G). In Arabidopsis, each context is maintained by different enzymes: MET1 for CG sites, CMT3 for CHG sites and DRM2 for CHH sites. CG and CHG sites are symmetric across the two DNA strands, which is thought to be important for the maintenance of methylation at these sites following DNA replication. In contrast, CHH sites are not symmetric, and their methylation is mediated by RNA-directed DNA methylation pathways (RdDM), which use siRNAs to initiate de novo methylation (3). Cellular methylation states tend to persist during cell division, and recent studies in Arabidopsis have also shown that DNA methylation is faithfully inherited across generations (7, 8). Nonetheless, we are only beginning to understand how different methylation patterns from inbred parents may “interact” during the generation of their hybrid progeny (9, 10).
In this study, we generated integrative maps of whole-genome cytosine methylation profiles [bisulfite sequencing (BS-seq)] and transcriptional profiles (RNA-seq), to characterize two rice subspecies, Oryza sativa spp japonica [Nipponbare (NPB)] and Oryza sativa spp indica (93–11) and their two reciprocal hybrid offspring. Using a combination of BS-seq, RNA-seq, and siRNA-seq, we were able to generate allele-specific patterns of methylation and transcription in the hybrids, and thus directly measure the degree to which these are altered between the corresponding parental and F1 chromosomes.
Results and Discussion
We sequenced bisulfite-treated DNA from two cultivated subspecies of Oryza sativa, NPB and 93–11, as well as the F1 hybrids resulting from reciprocal crosses. We achieved coverage levels of 17× for NPB, 10× for 93–11, and about 25× for each of the two crosses (Dataset S1).
SNPs are Much More Common Between Varieties, and Very Infrequent Across Generations.
Using the Michigan State University rice genome version 6.1 annotation (11) as a reference, we mapped the bisulfite-converted reads to the genome using BS Seeker (12). From these alignments we determined SNPs between the NPB and 93–11 parental plants. Although there is ambiguity because of cytosine conversion in bisulfite-treated DNA, we can distinguish conversions from mutations by analyzing the sequence of the complimentary base of each cytosine, and, as a result, were able to call all possible types of SNPs. For our SNP calls we required that we had at least three reads on each strand and over 90% agreement between them before making a call (Fig. 1A). Using these SNP calls we reconstructed two genomes, one for each parent. We then mapped F1 reads against these inferred genomes to identify the parental chromosome from which they were derived.
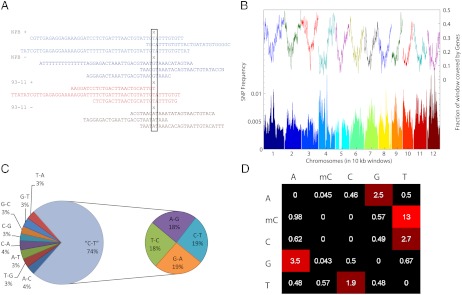
Fig. 1.
SNPs between NPB and 93–11. (A) Identification of SNPs required a minimum of three reads per strand in each parental strain. Although there is ambiguity because of cytosine conversion in bisulfite-treated DNA, we can resolve this by considering the sequence of the reverse strand, and, as a result, are able to call all possible types of SNPs. In the example, X indicates the location of the SNP and reads represent, in order, NPB forward strand, NPB reverse strand, 93–11 forward strand, and 93–11 reverse strand. (B) SNP distribution over the genome is plotted on the bottom in windows of 10,000 bases. Each color change represents a new chromosome. The top line represents the density of genes. (C) The fraction of the various types of SNPs found in NPB versus 93–11 is shown with the CT and GA SNPs grouped together, and they represent nearly 74% of all SNPs. (D) The relative frequency of each SNP between NPB and 93–11 is shown per 1,000 bases. Methyl-Cs convert to Ts at a rate that is nearly five times that of unmethylated Cs.
We found one SNP every 253 base pairs between the two parental strains (Fig. 1B), representing a level of divergence that is similar to previous japonica–indica comparisons (13). Not surprisingly, regions enriched with genes had lower densities of SNPs. Of these SNPs, the most frequent were C to T changes, or their complements corresponding to G to A mutations on the opposite strand. Combined, these types of mutations accounted for 73% of the SNPs that were found between varieties (Fig. 1C and Dataset S2), which represents a threefold enrichment compared to the expected value if all types of mutations were equally likely. We also observe that methylated cytosines mutate more than three times more frequently than nonmethylated cytosines (Fig. 1D), and they most often mutate to thymines. This is consistent with previous observations that the deamination of methylcytosine, which results in thymine, occurs at higher rates than other spontaneous mutations (14–16).
Next, we compared mutation rates between the parental strains and the hybrids. Not surprisingly, we found that SNPs were nearly absent across generations. If, as is likely, the few SNPs we call are false positives, we estimate the false discovery rate of our SNP calls to be less than 0.0003%.
We also found a small number of SNPs between the reference Michigan State University O. sativa japonica genome and our assembled version (1/90651 bp; 2,487 total) (Dataset S2), and considerably more SNPs by comparing our assembled version of the 93–11 genome with the reference Beijing Genome Institute’s O. sativa ssp. indica genome (1/51 bp), consistent with the fact that the 93–11 assembly is not as complete as that of the NPB cultivar (17). The fact that we only align reads that map to a unique position in the genome, have at most three mismatches with the reference genome, do not permit insertions or deletions, and require a minimum of three reads on both strands, allowed us to only call bases over about half of the genome in both NPB and 93–11, and thus we are reporting only a subset of all true SNPs.
Epimutations Occur Frequently and in Clusters Between NPB and 93–11.
Using the BS-seq reads, we were able to identify quantitatively the methylation status of cytosines in the parental genomes. By partitioning the F1 reads between their NPB or 93–11 chromosomes, we were also able to identify the allele-specific methylation status of cytosines in F1 plants. However, because the partitioning of reads between NPB and 93–11 parents can only occur at polymorphic sites, we were only able to do this for 6.2% of all cytosines with sufficient coverage.
When comparing sites where each library had similar levels of coverage, we found that NPB had a higher level of methylation than 93–11 at CG and CHG sites, but that 93–11 had more methylation at CHH sites (Fig. 2A). As one might expect, on a global level, the hybrids had methylation levels that were halfway in between the parental methylation levels. However, the methylation of individual chromosomes in the hybrids closely matched that of the corresponding parent.
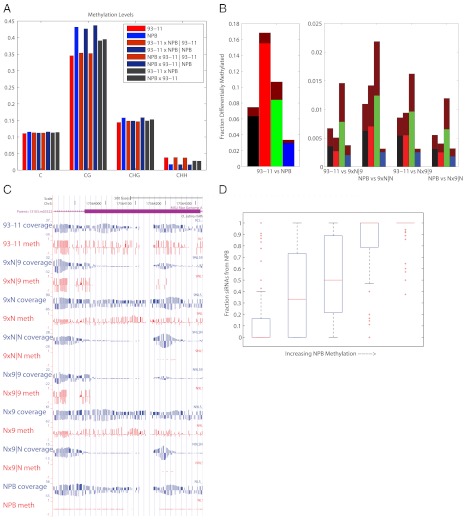
Fig. 2.
Epimutations. (A) NPB has slightly higher methylation levels at CG and CHG sites, whereas 93–11 has higher methylation levels at CHH sites. The methylation level of individual hybrid chromosomes is relatively unchanged compared to their parents. (B) The percentage of differentially methylated sites in each comparison in C (black), CG (red), CHG (green), and CHH (blue) contexts. The dark-red fraction of each bar indicates the percentage of epimutations that were also divergent between the parents. For the 93–11 versus NPB comparison, the red portion indicates the percentage that was overlapping in any of the transgenerational comparisons. (C) A typical epimutation cluster showing increased methylation in 93–11 and nearly absent methylation in NPB. There is also a region of differential methylation in the 93–11 chromosome of a hybrid and the 93–11 parent that cannot be attributed to a lack of coverage. 9 × N and N × 9 represent the reciprocal crosses between NPB and 93–11, and |N and |9 represent the NPB and 93–11 chromosomes within the hybrids. (D) Among epimutation clusters, as the fraction of methylation sites that are greater in NPB increases, so does the fraction of siRNAs that are found in NPB versus 93–11.
We used a binomial-based approach to determine whether sites were differentially methylated between samples, and refer to these events as “epimutations.” As discussed in detail in Materials and Methods, this was accomplished by requiring that the methylation level in both samples be outside of the 95% confidence interval of methylation of each other (Fig. S1). Further, we required that the methylation difference was greater than a threshold, defined as the average methylation level for each particular sequence context. In order to prevent coverage biases, we also restricted our analysis only to those sites that had comparable coverage in both samples.
Overall, we found that the epimutation rate was 7% (1/15 cytosines) between the NPB and 93–11 parental strains. Because cytosines represent about 20% of all bases, this leads to an overall epimutation rate of one in every 75 bases, which is considerably higher than the SNP rate of one in every 253 bases. The epimutation rates were higher at CG (17%) and CHG (11%) sites compared to CHH (3%) sites.
The epimutations between the parental strains are likely driven by both the cis genetic differences between the two cultivars as well as stochastic effects. To determine the rates of epimutation within the same local genetic background, we compared the methylation of the parental strains to the corresponding chromosomes in the hybrids. Here, we found that only about 0.8% (1/126) of cytosines have altered methylation (Fig. 2B and Dataset S3). Among these sites we found that CHG epimutations were the most common. Other studies have found CHG sites are associated with silent chromatin and H3K9 dimethylation, indicating that the repression of these sites may be variable between the parent and F1 plants (18).
We next asked whether epimutation events are isolated and independent, or whether they tend to occur in clusters. We searched for 100 bp–sized windows in which epimutations were enriched. Enriched windows were defined as those having at least five, and a minimum of 20%, of all cytosines that are differentially methylated between two samples. Using this approach we identified 16,865 epimutation clusters between NPB and 93–11. An example of such a cluster is shown in Fig. 2C. Of these clusters, 5,152 had epimutations with significantly greater methylation in 93–11 and 8,584 with greater methylation in NPB.
In contrast, there were fewer clusters of differential methylation between parents and F1 plants. We identified an average of 340 clusters meeting our criteria between NPB and the NPB chromosomes in the two F1 hybrids, and 75 between 93–11 and the 93–11 chromosomes in the two F1 hybrids. A reduced occurrence of clustered epimutations is expected because there are far fewer epimutations between parents and F1s than between the two parents. However, we found that as a fraction of the total, transgenerational epimutations were less clustered than those between the parents, with clusters representing 20% of all epimutations between parents, but only 5% in NPB F1 plants and 2% in 93–11 F1 plants.
Intriguingly, we found that significant numbers (approximately 45%) of transgenerational epimutations (between parent and F1 plants) were also identified as epimutations between the two parental varieties (Fig. 2B). Conversely, about 15% of the epimutations that were found between parental cultivars were also found to be epimutated across generations. This suggests that regions of discordant methylation between the two inbred parents are often aberrantly inherited in the F1 plants, a result that is consistent with recent findings in Arabidopsis hybrids (10). We found similar results for epimutation clusters, because 20% of those found in NPB F1 plants and nearly 35% of the clusters found in 93–11 F1 plants had some overlap with NPB vs. 93–11 epimutation clusters.
We next asked whether epimutation clusters are correlated with siRNAs, and whether this could explain the overlap we observe between interparental and transgenerational epimutations. The RdDM pathway is associated with feedback loops between the production of siRNAs and the recruitment of DRM methyltransferases (3, 19). However, the activation of this feedback is likely dependent on the dosage of siRNAs. As a result, we expect that some of the epimutation clusters are associated with differential siRNA production between the parental cultivars. At these loci the hybrid plants would produce siRNAs at a level that is intermediate between the two parents. In some cases this level may be sufficient to methylate both chromosomes in the hybrid, leading to the hypermethylation of the locus with respect to the unmethylated parent. Conversely, in other cases, the siRNA dosage may be insufficient to methylate the locus in either chromosome, leading to the hypomethylation of the locus with respect to the hypermethylated parent.
To understand the relationship between epimutation clusters and siRNAs we sequenced small RNAs from the parental and F1 samples. From the sequenced reads we selected 24-nt small RNAs that mapped uniquely to the genome, because we expect these to be enriched for siRNAs instead of miRNAs (20, 21). We found that 29% of the epimutation clusters had siRNAs associated with them. We also observed that epimutation clusters that were hypermethylated in NPB versus 93–11 also had more siRNAs in NPB versus 93–11. That is, within a cluster there was a strong correlation (rho = 0.7) between the fraction of epimutations where methylation was increased in NPB and the fraction of siRNAs belonging to NPB (Fig. 2D). Because of the paucity of SNPs in 24-nucleotide siRNAs we were unable to assign a sufficient number of siRNAs uniquely to the parental chromosomes, and thus were unable to analyze parent-specific siRNA levels in the hybrids.
Genes Expressed in an Allele-Specific Manner are Enriched for Epimutations.
To study the inheritance of gene-expression levels in the hybrid strains we measured transcript levels using RNA-seq. We obtained RNA-seq data from the same samples that were used for methylation profiling. Reads were mapped using TopHat (22) and, in the case of F1 plants, we assigned them to individual chromosomes using the SNP data discussed above. Based on the counts of SNP-containing reads, we computed the allele-specific expression levels in the F1 plants. We also used these data to estimate expression levels in both parents.
We first compared transcription between the NPB and 93–11 varieties. We defined genes as differentially expressed if (i) the difference in their counts was at least 100 reads per kilobase per million reads (RPKM) and had a p-value below 10-5 (based on a Poisson model), and (ii) they had at least a twofold change. We found 1,525 transcripts that met these criteria (Fig. 3A and Dataset S4 and S5).
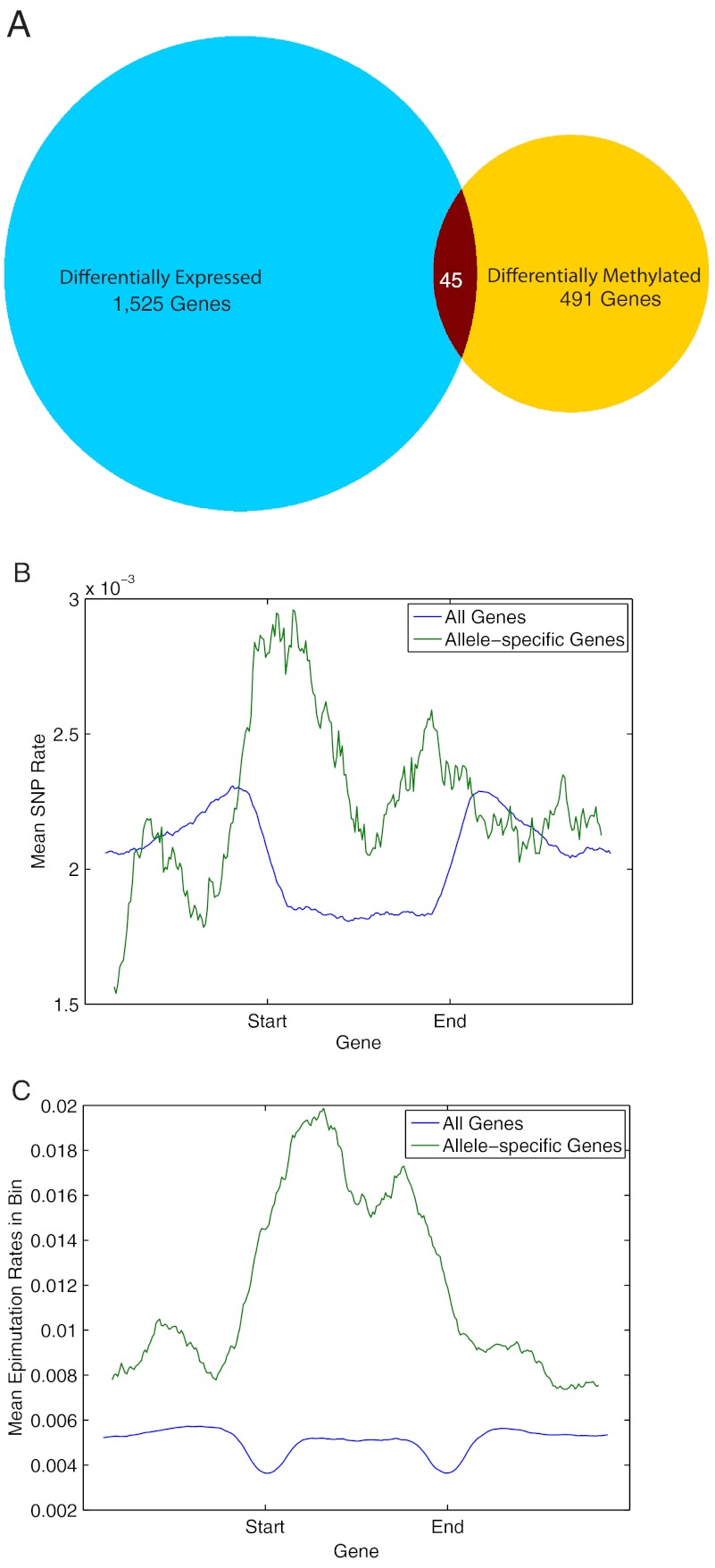
Fig. 3.
Transcript level changes. (A) Venn diagram showing the number of genes differentially methylated or differentially expressed between NPB and 93–11. (B) A metaplot of SNP density is displayed. Each gene was divided into 100 bins and the average SNP density is plotted in 100 bins upstream of the gene to 100 bins downstream of the gene. In allele-specific genes, SNP density increases over the genic region, whereas for all genes, it decreases. (C) Genes are similarly broken into bins as in B, and the epimutation density is plotted over all genes and allele-specific genes. As in the case of SNP density, the epimutation density increases over allele-specific genes compared to all genes.
To assess the association between methylation and transcription we measured both methylation and epimutations within transcribed loci and their immediate 1-kb upstream region. In order to find genes that were enriched for differential methylation, we used a binomial-based test to determine whether a significant number of sites were differentially methylated. We found that 491 out of 25,640 (Dataset S6 and S7) genes had significant enrichment for interparental epimutations (p-value < 0.01). Of these, 45 were also differentially expressed, indicating that there is a significant enrichment for differential methylation among the differentially expressed genes (p = 0.0024).
We next set out to determine how transcriptional differences between the parents were inherited in the hybrid plants, by analyzing RNA-seq reads corresponding to SNPs. We calculated F1 allele-specific expression estimates and used a t test to identify genes that were expressed in an allele-specific manner between the parents as well as between the corresponding chromosomes in the F1 hybrids. As in our previous comparison, we also required that the expression fold change between the two alleles be at least twofold and have a difference of at least 100 RPKM. We found 85 genes that fit these criteria (Dataset S8), and thus had significantly different expression levels between NPB and 93–11 chromosomes.
Allele-specific expression is potentially mediated by both genetic and epigenetic factors. We found that most (79%) genes had at least one SNP between NPB and 93–11, and 37% had more than five SNPs. In contrast, genes that were expressed in an allele-specific manner had a significantly higher SNP rate (Fig. 3B). We also found that of the 85 genes that were expressed in an allele-specific manner, a significant number, 15, were also differentially methylated (p = 10-26). A plot of epimutation densities over allele-specific genes also shows that the level of epimutations tends to rise significantly over the body of these genes (Fig. 3C). Thus, we observe that the genetic differences that underlie allele-specific genes also correlates with dramatic differences in their methylation state, and that the combination of these two effects is likely mediating the allele-specific expression differences we observed.
To disentangle genetic and epigenetic factors that mediate expression changes, we also looked for genes that were differentially expressed between the parents and their corresponding chromosomes in the hybrid. We found 1,619 genes that had transgenerational differential expression in at least one parent–F1 comparison using the same criteria described above. Of these, we found only a single case that was both differentially expressed and differentially methylated. Because most of these genes have similar methylation profiles and identical genetic backgrounds, their transcriptional changes may be mediated by trans effects in the hybrids. The fact that this group of genes is much larger than the allele-specific genes suggests that these trans effects dominate the genetically and epigenetically mediated allele-specific expression phenotypes.
Conclusion
Although it is thought that DNA methylation patterns are to a large degree heritable in plants, the frequency of epimutations and their underlying causes are still poorly understood. By sequencing both parental and hybrid strains we were able to measure the frequency of transgenerational epimutations and found that they occur in about one in every 125 cytosines across genetically identical chromosomes. This shows that DNA methylation is in fact quite variable across generations, and suggests that this variability across inbred strains may have important phenotypic consequences.
There are two primary mechanisms that could account for changes in DNA methylation across generations. The first are stochastic effects that may accrue as somatic cells in the parents lead to germ line cells and eventually somatic cells of their progeny. The second cause of epigenetic variability may be trans effects, as the heterozygous hybrids are genetically different from the inbred parents. Specifically, we suggest that these trans effects may be in part mediated by RdDM pathways that mediate the interaction between the production of siRNAs and DNA methylation. Differences in siRNA production between parents at specific loci would lead to the inheritance of average levels of siRNAs in the progeny, which could in turn lead to either the hyper- or hypomethylation of the two alleles, depending on whether the dosage is sufficient or insufficient to activate RdDM.
It is likely that both these mechanisms are at play in the F1 hybrids we have analyzed. The fact that epimutations are clustered between parents suggests that siRNAs are likely mediating these differences, and we find a strong association between siRNA dosage differences and epimutation levels in our data. However, because the fraction of transgenerational epimutation clusters is quite low, stochastic effects must also be playing a role in the transmission of DNA methylation patterns across generations.
To understand whether epimutations have observable molecular phenotypes, we also examined transcription using RNA sequencing. We found that genes that are expressed in a strongly allele-specific manner across samples are enriched both for genetic and epigenetic differences, suggesting that these two effects may be acting cooperatively to maintain these patterns. However, loci showing transgenerational gene-expression changes across identical chromosomes are not significantly enriched for epimutations, suggesting that trans effects may be mediating the bulk of these changes. However, a single epimutation within a key cis-regulatory site may be sufficient to disrupt the expression of a gene, and therefore it is not possible to rule out stochastic epimutations as a significant driver of transgenerational transcriptional variability.
The two varieties of rice that we studied demonstrate significant heterosis, a phenomenon whereby matings by more distant plants, intra- or interspecies, result in offspring with superior agronomic traits, such as tiller number and dry weight (9, 23–25). Previous studies of heterosis have revealed possible mechanisms to account for the increased vigor, but the mechanisms that underlie this complex phenotype are still poorly understood, and the role of DNA methylation in heterosis is unclear (26–29). Our results suggest that the majority of gene-expression changes in hybrids are not associated with cis-acting DNA methylation changes, and instead indicate that trans effects may mediate the majority of the transcriptional differences in hybrid offspring. However, it is possible that a subset of the gene-expression changes may also be caused by intergenerational epimutations in the hybrids.
In conclusion, we have mapped the degree to which DNA methylation varies between parental and hybrid strains. These observations reinforce the notion of epigenetic plasticity and suggest that it may play important roles in both environmental adaptation as well as hybrid vigor. In the future it will be important to elucidate the magnitude and molecular details of these mechanisms.
Materials and Methods
Plant Materials and Growth Conditions.
Oryza sativa ssp. japonica (NPB), O. sativa ssp. indica (93–11), and their reciprocal cross F1s (NPB × 93–11 and 93–11 × NPB) were used in this study. Rice seeds were surface sterilized with 40% (vol/vol) sodium hypochlorite solution and transferred on 1/2 Murashige and Skoog medium. After germination, rice seedlings were transplanted into soil and grown at 26 °C/20 °C under a 10-h light/14-h dark cycle in growth chamber. Fully expanded leaves from 6-wk-old plants were collected for library construction.
Library Construction.
BS-seq libraries were made from genomic DNA isolated from leaf tissues of NPB, 93–11, and their two reciprocal hybrid offspring, 93–11 × NPB and NPB × 93–11, by a previously published method using premethylated Illumina adapters (30).
RNA-seq libraries were made from total RNA isolated from leaf tissues using TRIzol reagent (Invitrogen). Total RNA (10 ug) for each sample was used for purification of the poly(A)-containing mRNA molecules, RNA amplification, and synthesis of double-stranded cDNAs that were ligated to adapters, thus preparing the libraries for sequencing on the Illumina GAII. The RNA-seq library preparation protocol was followed as described in Illumina’s standard protocol for RNA-seq libraries (Illumina). Libraries were sequenced on an Illumina GAII at the Delaware Biotechnology Institute. This generated 74,171,650 total reads from NPB; 71,535,918 total reads from Oryza sativa indica (93–11); 74,999,214 total reads from the reciprocal hybrid offspring 93–11 × NPB; and 78,592,119 total reads from the reciprocal hybrid offspring NPB × 93–11. The mRNA sequence data are available from National Center for Biotechnolgy Information’s (NCBI) Gene Expression Omnibus (GEO) and are accessible via GEO Series accession number GSE38480.
Small RNA libraries were constructed from total RNA isolated from leaf tissues using TRIzol reagent (Invitrogen). The sRNA library-preparation protocol is based on the sRNA-Sequencing Sample Preparation Guide (Illumina). The libraries were sequenced on an Illumina GAII at the Delaware Biotechnology Institute. The sRNA sequence data are available from NCBI’s GEO and are accessible via GEO Series accession number GSE38480.
Data Processing and Analysis.
Bisulfite-sequenced reads from all libraries were mapped to the Oryza sativa ssp. japonica (cv. Nipponbare) version 6.1 reference genome (11) using BS Seeker (12) allowing for up to three mismatches. Reads that mapped with equal efficiency to multiple sites were discarded. Coverage was about 20× for NPB and 12× for 93–11 and about 25× for the hybrids. Using these reads as a scaffold, custom NPB and 93–11 genomes specific to our samples were built. Genotypes were called only when coverage consisted of at least three reads per strand and over 90% of the reads were in agreement. These custom genomes were used to call SNPs.
DNA methylation was calculated as the percent of cytosines that failed to undergo bisulfite conversion. DNA methylation context was determined based on the sequence in NPB. For most analyses, the sites that were considered were limited to those that had comparable coverage levels in all libraries, which was defined to be at least five and not more than 25 reads.
Epimutations were determined by finding sites where the implied methylation level of each sample was outside of the 5% confidence intervals of the other sample based on a binomial distribution. The difference in implied methylation levels also needed to be greater than a threshold based on the mean context specific methylation (Fig. S1).
Transcripts enriched for differential methylation were identified by using a binomial test. The inputs for this test were the number of epimutations present, the number of cytosines in the gene with sufficient coverage in the respective libraries, and the overall epimutation rate for the genome. Transcripts with p-values below 0.01 were considered to be enriched for epimutations.
RNA-seq reads were mapped using the default parameters of TopHat (22). Coverage was about 20× in NPB, 15× in 93–11, and about 12× in the two hybrids. Differential expression was determined using a Poisson test. Starting with the number of RPKM of one sample, we used the Poisson test to compute the probability of observing the RPKM values in the other sample and vice versa. If either yielded a p-value below 10-5 and there was at least a twofold change and an absolute difference of at least 100 RPKM, the gene was called differentially expressed.
siRNA reads were processed in order to trim the adapter sequence and then were mapped using Bowtie allowing for up to three mismatches and requiring matches to be unique (31). Matches were further size-selected so that only 24-nt reads were kept.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Jonathan Hetzel and William Wong for their assistance with sequencing. R.K.C. is supported by NIH Training Grant GM07104. S.F. is a Special Fellow of the Leukemia and Lymphoma Society. S.E.J. is an Investigator of the Howard Hughes Medical Institute. This project was supported by the US National Science Foundation Plant Genome Research Program No. 0701745.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE38480).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209297109/-/DCSupplemental.
References
- 1.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki MM, Bird A. DNA methylation landscapes: Provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 3.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 6.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmitz RJ, et al. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334:369–373. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker C, et al. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature. 2011;480:245–249. doi: 10.1038/nature10555. [DOI] [PubMed] [Google Scholar]
- 9.He G, et al. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell. 2010;22:17–33. doi: 10.1105/tpc.109.072041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greaves IK, et al. Trans chromosomal methylation in Arabidopsis hybrids. Proc Natl Acad Sci USA. 2012;109:3570–3575. doi: 10.1073/pnas.1201043109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouyang S, et al. The TIGR rice genome annotation resource: Improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen PY, Cokus SJ, Pellegrini M. BS seeker: Precise mapping for bisulfite sequencing. BMC Bioinformatics. 2010;11:203. doi: 10.1186/1471-2105-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nasu S, et al. Search for and analysis of single nucleotide polymorphisms (SNPs) in rice (Oryza sativa, Oryza rufipogon) and establishment of SNP markers. DNA Res. 2002;9:163–171. doi: 10.1093/dnares/9.5.163. [DOI] [PubMed] [Google Scholar]
- 14.Yebra MJ, Bhagwat AS. A cytosine methyltransferase converts 5-methylcytosine in DNA to thymine. Biochemistry. 1995;34:14752–14757. doi: 10.1021/bi00045a016. [DOI] [PubMed] [Google Scholar]
- 15.Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci USA. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: Implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 17.Yu J, et al. A draft sequence of the rice genome (Oryza sativa L. ssp.indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 18.Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS One. 2008;3:e3156. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wassenegger M. The role of the RNAi machinery in heterochromatin formation. Cell. 2005;122:13–16. doi: 10.1016/j.cell.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE. Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci USA. 2007;104:4536–4541. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasschau KD, et al. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Birchler JA, Auger DL, Riddle NC. In search of the molecular basis of heterosis. Plant Cell. 2003;15:2236–2239. doi: 10.1105/tpc.151030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birchler JA, Yao H, Chudalayandi S, Vaiman D, Veitia RA. Heterosis. Plant Cell. 2010;22:2105–2112. doi: 10.1105/tpc.110.076133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang HY, et al. A genome-wide transcription analysis reveals a close correlation of promoter INDEL polymorphism and heterotic gene expression in rice hybrids. Mol Plant. 2008;1:720–731. doi: 10.1093/mp/ssn022. [DOI] [PubMed] [Google Scholar]
- 26.Song GS, et al. Comparative transcriptional profiling and preliminary study on heterosis mechanism of super-hybrid rice. Mol Plant. 2010;3:1012–1025. doi: 10.1093/mp/ssq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hochholdinger F, Hoecker N. Towards the molecular basis of heterosis. Trends Plant Sci. 2007;12:427–432. doi: 10.1016/j.tplants.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 28.Ni Z, et al. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 2009;457:327–331. doi: 10.1038/nature07523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Springer NM, Stupar RM. Allelic variation and heterosis in maize: How do two halves make more than a whole? Genome Res. 2007;17:264–275. doi: 10.1101/gr.5347007. [DOI] [PubMed] [Google Scholar]
- 30.Feng S, Rubbi L, Jacobsen SE, Pellegrini M. Determining DNA methylation profiles using sequencing. Methods Mol Biol. 2011;733:223–238. doi: 10.1007/978-1-61779-089-8_16. [DOI] [PubMed] [Google Scholar]
- 31.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.