Abstract
The maize R2R3-MYB regulator C1 cooperates with the basic helix–loop–helix (bHLH) factor R to activate the expression of anthocyanin biosynthetic genes coordinately. As is the case for other bHLH factors, R harbors several protein–protein interaction domains. Here we show that not the classical but rather a briefly extended R bHLH region forms homodimers that bind canonical G-box DNA motifs. This bHLH DNA-binding activity is abolished if the C-terminal ACT (aspartokinase, chorismate, and TyrA) domain is licensed to homodimerize. Then the bHLH remains in the monomeric form, allowing it to interact with R-interacting factor 1 (RIF1). In this configuration, the R–RIF1 complex is recruited to the promoters of a subset of anthocyanin biosynthetic genes, such as A1, through the interaction with its MYB partner C1. If, however, the ACT domain remains monomeric, the bHLH region dimerizes and binds to G-boxes present in several anthocyanin genes, such as Bz1. Our results provide a mechanism by which a dimerization domain in a bHLH factor behaves as a switch that permits distinct configurations of a regulatory complex to be tethered to different promoters. Such a combinatorial gene regulatory framework provides one mechanism by which genes lacking obviously conserved cis-regulatory elements are regulated coordinately.
Keywords: gene regulation, promoter switch
The evolution of multicellular organisms was accompanied by an increase in the complexity of gene-regulatory mechanisms, reflected in the expansion of transcription factor families and in the intricacy of the interactions between regulatory proteins and cis-regulatory elements in what is known today as “combinatorial transcriptional control.” A premise of combinatorial control is that different arrangements of a discrete number of regulatory proteins can be used to regulate a much larger number of genes. Therefore, understanding how interactions between different regulatory proteins impact their ability to deploy the expression of specific gene sets is of fundamental biological importance.
The basic helix–loop–helix (bHLH) family of transcription factors is among the largest in multicellular organisms (1). The hallmark of the family is a bHLH domain, which consists of two functionally distinct regions. Generally, the basic region of the bHLH domain directly contacts DNA harboring an E-box sequence (CANNTG), and the HLH region provides the potential for homo- and heterodimerization. In addition to the HLH motif, bHLH factors often contain additional protein–protein interaction domains. For example, members of the MYC family of mammalian cell proliferation regulators, such as MAD or MNT, contain a leucine-zipper (LZ) region that contributes to the selective interaction with MAX, another bHLH-LZ protein (2). MAX can form homo- or heterodimers with several related proteins, including MAD (3) and MNT (4). Providing a textbook example of combinatorial transcriptional control, MYC–MAX and MAX–MAX complexes bind E-boxes, but only the MYC–MAX heterodimer activates cell-proliferation genes. In contrast, the formation of MAX–MAD and MAX–MNT heterodimers results in transcriptional repression through the recruitment of histone deacetylase complexes (5).
In plants, bHLH factors constitute one of the largest families of regulatory proteins (1). The Arabidopsis genome encodes for ∼162 bHLH proteins, about 10% of all of the known transcription factors in this plant (6–8). Maize R was the first plant bHLH transcription factor described (9). R belongs to a small gene family, which includes B, and R/B specify anthocyanin pigmentation in different plant tissues (10). They participate in the transcriptional regulation of anthocyanin pathway genes through the cooperation with the R2R3–MYB transcription factor C1 or its paralog PL1 (11). C1 and R/B interact physically through the MYB domain of C1 and the N-terminal region of R (12, 13), and C1 is responsible for tethering R through high-affinity P1-binding sites (haPBS) and low-affinity PBS (laPBS) cis-regulatory elements to flavonoid biosynthetic gene promoters, such as A1 (14, 15). R/B belong to the bHLH group IIIf (7), a subfamily that is shared with similar anthocyanin regulators in various plants as well as with the Arabidopsis GL3/EGL3 regulators of epidermal cell patterning (16). All these factors function by interacting with R2R3–MYB proteins, recognizing particular signature motifs in the corresponding MYB DNA-binding domains (13, 17). In addition, members of this group of bHLH proteins contain a conserved aspartokinase, chorismate mutase, TyrA (ACT)-like domain at the C termini, which participates in homodimer formation (18). The importance of the bHLH motif in the regulation of anthocyanin accumulation was revealed recently by the identification of R-interacting factor1 (RIF1), a maize nuclear protein with homology to breast cancer gene 2 (BRCA2)-interacting EMSY factor. RIF1 specifically interacts with the bHLH motif of R and links R function with promoter-specific histone functions (19). Surprisingly, however, thus far the canonical function of the bHLH region of R as a DNA-binding homo- or heterodimerization domain has not been demonstrated.
Here, we show that, for the bHLH region of R to dimerize, it must be extended at least eight residues to include a couple of evenly spaced leucine residues in what appears to be a short LZ motif. When this briefly extended bHLH dimerizes, it binds DNA, and we show by Systemic Evolution of Ligands by Exponential Enrichment (SELEX) that the R DNA-binding preference is to the G-box (CACGTG), a special type of E-box. However, if the bHLH region also includes the ACT domain, the R DNA-binding activity is lost, and the monomeric bHLH domain interacts with RIF1. We show that this configuration of the C1 R–RIF1 complex underlies the control of the expression of the A1 gene, in which recruitment to the A1 promoter is mediated primarily by the C1 DNA-binding activity. Mutations that abolish ACT-mediated dimerization restore the ability of R to dimerize through the bHLH and to recognize G-boxes. We further show that the topology of the complex responsible for the regulation of another anthocyanin biosynthetic gene, Bz1, primarily involves interactions of R with DNA, and in this case C1-mediated DNA interactions play a secondary role. In this configuration, RIF1 is not part of the regulatory complex, as shown by ChIP experiments. Our results provide a mechanism by which a dimerization domain in a bHLH factor behaves as a switch that permits different configurations of regulatory complexes to be tethered to different promoters, helping explain the elusive problem of how genes lacking conserved cis-regulatory elements can be coordinately regulated.
Results and Discussion
Briefly Extended bHLH Domain Mediates R Homodimerization and DNA Binding.
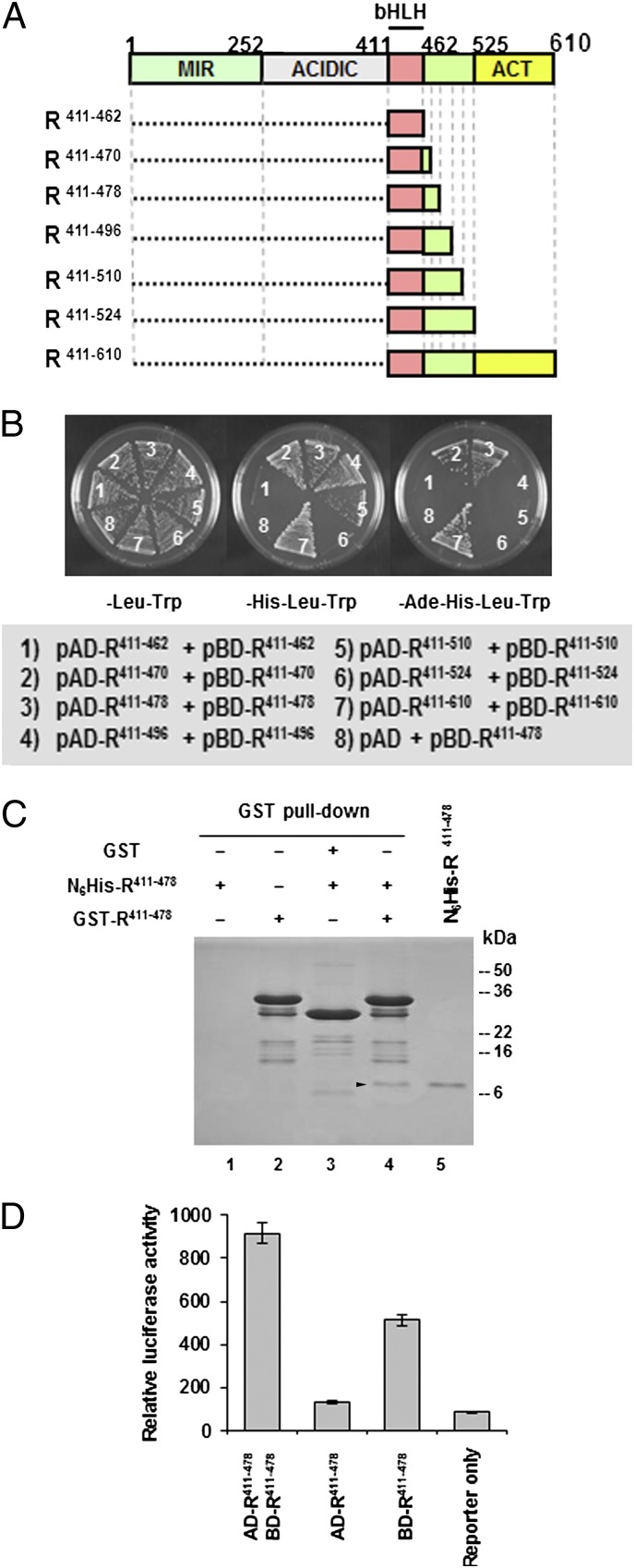
R can form homodimers through the C-terminal ACT domain (18), but the possibility that the bHLH also may mediate homodimer formation has not been formally investigated previously. To determine whether the bHLH region of R mediates homodimerization, we carried out yeast two-hybrid analyses using the corresponding region of R (residues 411–462) (Fig. 1A) fused to the GAL4 DNA-binding (pBD-R411–462) and GAL4 activation (pAD-R411–462) domains in the AH109 yeast strain. As indicated by the absence of growth in medium lacking adenine, histidine, leucine, and tryptophan (−Ade−His−Leu−Trp) (Fig. 1B), this region of R was not sufficient for homodimer formation. Interestingly, however, when similar experiments were conducted with a briefly extended bHLH region (e.g., extended toward the C terminus by eight or sixteen amino acids: bHLH411–470 and bHLH411–478) (Fig. 1A), robust growth in −Ade−His−Leu−Trp medium was observed (Fig. 1B, compare 1 with 2 and 3). If, however, the bHLH region was extended even further (bHLH411–496, bHLH411–510 and bHLH411–524) (Fig. 1A), the homodimerization ability decreased (Fig. 1B, compare 2 and 3 with 4–6). Remarkably, constructs containing the ACT domain provided a much stronger interaction than those that contained only the extended bHLH region (bHLH411–470 and bHLH411–478) (Fig. 1B, compare 2 and 3 with 7), highlighting the important role of this domain in R dimerization. To obtain a quantitative estimate of the interaction strength, β-gal activities of all the yeast strains carrying the various constructs were assayed. In agreement with the selection data, only the combinations that supported growth on −His−Leu-Trp or −Ade−His−Leu−Trp media showed β-gal activities above background (Table S1).
Fig. 1.
Dimerization mediated by the bHLH and ACT domains of R. (A) Schematic representation of the R functional domains. ACT, ACT-like domain; ACIDIC, acidic domain; bHLH, basic helix–loop–helix domain; MIR, MYB-interacting region. Boxes represent different R fragments used in yeast two-hybrid assay and bacterial expression. (B) Yeast two-hybrid assays for homodimerization of the bHLH domain with various C-terminal extensions. A briefly extended bHLH domain of R (R411–478) homodimerizes in yeast. R411–610 was used as a positive control in this assay, and the empty pAD-GAL4-2.1 vector was used as a negative control. The pAD-GAL4-2.1 and the pBD-GAL4 Cam plasmids contain the GAL4 activation and DNA-binding domains, respectively. All bHLH fragments excluding the ACT domain (R411–462, R411–470, R411–478, R411–496, R411–510, and R411–524) or including the ACT domain (R411–610) were fused to both pAD-GAL4-2.1 and pBD-GAL4 Cam plasmids and cotransformed into yeast strain AH109 containing the HIS3 and ADE2 reporter genes. The protein–protein interactions were detected by yeast growth in −His−Leu−Trp and −Ade−His−Leu−Trp selection media. (C) Stained SDS/PAGE gel of GST pull down using bacterially expressed GST-R411–478 and N6His-R411–478. In lane 4 the arrowhead indicates the pull-down product N6His-R411–478, which has the same molecular mass as the purified N6His-R411–478 (lane 5). (D) R411–478 homodimerization in tobacco protoplasts. R411–478 fused to the GAL4 activation domain (AD-R411–478) and to the DNA-binding domain (BD-R411–478) was coelectroporated into tobacco protoplasts with a reporter plasmid containing a firefly luciferase gene under the control of a minimal CaMV35S promoter with five tandem repeats of GAL-responsive elements and the rbcS terminator. The reporter plasmid coelectroporated only with AD-R411–478 or BD-R411–478 served as negative control. A plasmid containing a β-glucuronidase (GUS) gene under the control of CaMV35S promoter was used as a control. The luciferase activity was normalized against GUS activity.
To validate the observed interactions further, the various bHLH fragments (Fig. 1A) were expressed in Escherichia coli as N-terminal GST and histidine-tagged (N6His-) fusions and were purified by glutathione- or Ni-affinity chromatography, respectively. GST-R411–478 and N6His-R411–478 were used in GST pull-down experiments (SI Materials and Methods), taking advantage of their size difference (∼35 kDa and 9 kDa, respectively). After incubation of N6His-R411–478 with GST-R411–478-coated beads, followed by washes and elution, a protein band corresponding to N6His-R411–478 was recovered on an SDS/PAGE gel (Fig. 1C, arrowhead in lane 4). The GST-coated control beads did not pull down N6His-R411–478 (Fig. 1C, lane 3) indicating that the interaction was mediated by R411–478. Finally, the ability of R411–478 to dimerize in plant cells was confirmed by the activation of a luciferase reporter construct harboring GAL4-binding sites by coexpression of the pAD-R411–478 and pBD-R411–478 constructs in tobacco protoplasts but not by pAD-R411–478 (Fig. 1D). pBD-R411–478 alone also resulted in the activation of the reporter, suggesting that pBD-R411–478 can interact with an endogenous transactivator. Taken together, these results provide conclusive evidence that the briefly extended bHLH from R (residues 411–478p Fig. 1A) can mediate robust homodimer formation.
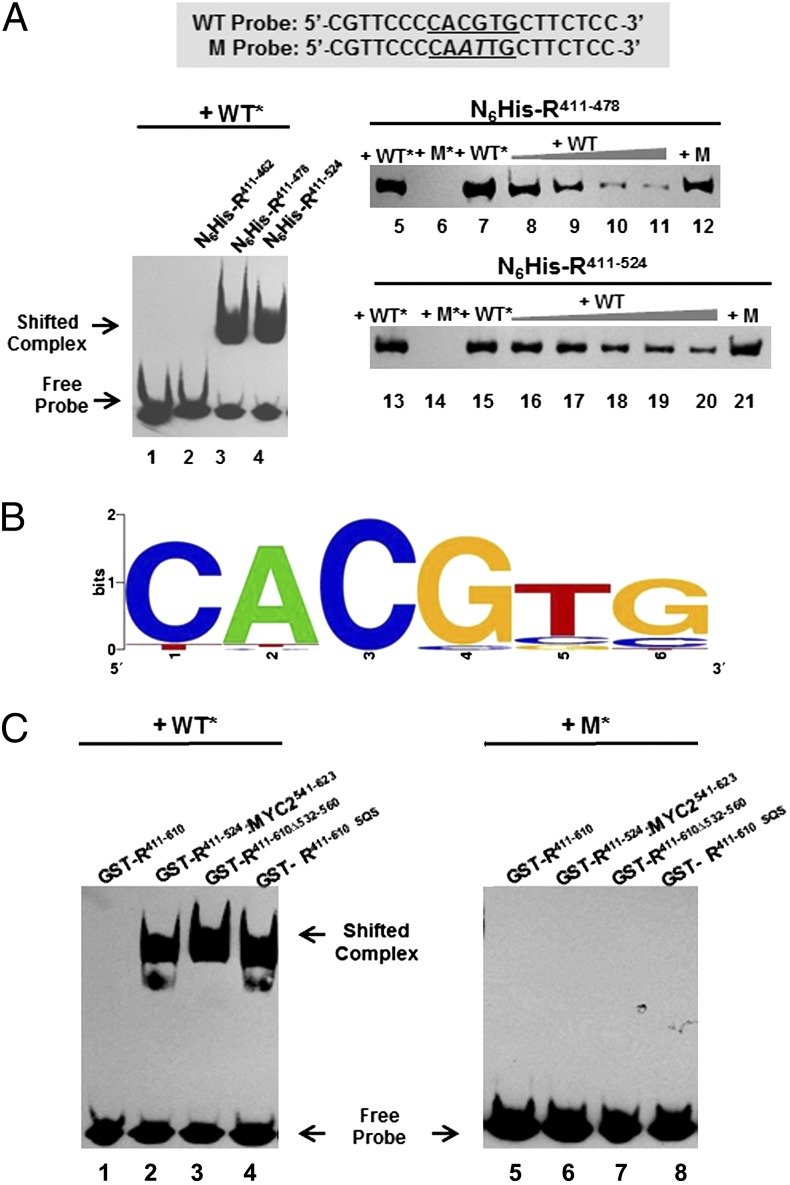
We next investigated whether the ability of R bHLH to form homodimers conferred proteins carrying this domain the capacity to bind DNA. Binding of purified recombinant N6His-R411–478 was tested by EMSA on a biotin-labeled, double-stranded 20-bp oligonucleotide (Fig. 2A, WT probe) harboring an E-box with the core motif CACGTG (a G-box). As a control, EMSA also was performed on a mutant probe (Fig. 2A, M probe) lacking a functional G-box. Binding of N6His-R411–478 to the WT probe, but not to the M probe, was observed (Fig. 2A, N6His-R411–478, compare lanes 5 and 6). Increasing concentrations of the unlabeled WT probe, but not of the M probe, competed with the binding of N6His-R411–478 to the WT probe (Fig. 2A, lanes 7–11). Interestingly, and suggesting that the interaction with DNA further stabilizes homodimer formation, N6His-R411–524 also showed robust and specific DNA-binding activity similar to that of N6His-R411–478 (Fig. 2A, N6His-R411–524, lanes 13–21), despite the lack of dimerization in yeast two-hybrid assays (Fig. 1B). N6His-R411–462, which lacks the extended bHLH region and showed no dimerization in yeast two-hybrid assay (Fig. 1B), was unable to bind the G-box (Fig. 2A; compare lane 2 with lanes 3 and 4).
Fig. 2.
The DNA-binding specificities of R bHLH-domain fragments with or without the ACT domain as determined by EMSA and SELEX. (A) Binding of various lengths of R bHLH-domain proteins to the G-box or mutated G-box probes in EMSA. The sequences of the WT and mutant (M) probes are shown. The G-box sequence is underlined. The mutation in the G-box sequence of the M probe is indicated by an italicized letter. In lanes 1–4, the biotin-labeled WT G-box probe (WT*) was incubated either alone or with N6His-R411–462, N6His-R411–478, or N6His-R411–524. In lanes 5 and 6, N6His-R411–478 was incubated with WT* probe and biotin-labeled mutant G-box probe (M*) in which the G-box is destroyed by a CG–AT substitution. In lanes 7–11, dissociation of N6His-R411–478 and the WT* probe is shown in the presence of 0–400× of unlabeled WT probe. No dissociation of N6His-R411–478 and the WT* probe is detected in the presence of additional 300× of unlabeled M probe (lane 12). In lanes 13–21, N6His-R411–524 was subjected to essentially the same experiment described for N6His-R411–478 in lanes 5–12. (B) The consensus DNA-binding sequence of N6His-R411–478 as determined by SELEX. By analyzing 57 unique sequences derived from SELEX, the core-binding motif for N6His-R411–478 was established and displayed using WebLogo. The size of the characters represents the frequency of occurrence. (C) Suppression of the G-box–binding activity of the R bHLH domain by the ACT domain. Autoradiographs of EMSA show the interactions of WT* probe (Left) and M* probe (Right) with purified GST-tagged R411–610 (lanes 1 and 5), R411–524:MYC2541-623 (lanes 2 and 6), R411–610Δ532–560 (lanes 3 and 7), and R411–610SQS (lanes 4 and 8).
To determine which E-box is recognized preferentially by the extended bHLH region of R, we conducted SELEX experiments on a double-stranded oligonucleotide population containing 26 bp of random sequence. The consensus obtained from the analysis of 57 selected sequences shows that R preferentially recognizes G-boxes (Fig. 2B). Taken together, these results demonstrate that the briefly extended bHLH region of R mediates homodimer formation, with a marked preference for the G-box DNA motif.
ACT Domain Suppresses G-Box–Mediated DNA-Binding Activity of R.
Consistent with our previous inability to detect DNA binding of R to a number of different E-box sequences, N6His-R and GST-R failed to bind the WT probe. However, the results presented here demonstrate that the extended bHLH region of R binds a canonical G-box (Fig. 2 A and B). To solve this apparent paradox, we investigated whether regions in R might interfere with the DNA-binding activity of the bHLH region. In contrast to N6His-R411–524, DNA-binding activity was not detected for GST-R411–610 (Fig. 2C, lane 1) despite its ability to form homodimers (Fig. 1B). However, if the dimerization function of the ACT domain is abolished by a deletion of residues 532–560 (18), DNA binding to the WT probe is restored (Fig. 2C, GST-R411–610Δ532–560, lane 3). A three-amino acid substitution (S560A, Q562A, S564A) in the ACT domain of R411–610 (GST-R411–610 SQS), which significantly reduces the ability of the ACT domain to dimerize (18), restored its DNA-binding activity (Fig. 2C, lane 4). Finally, we replaced the ACT domain of R with that of AtMYC2 (SI Materials and Methods), resulting in GST-R411–524/MYC2541–623. The ACT domain of AtMYC2 does not homodimerize, and, consistent with a specific role of the R ACT domain in suppressing bHLH-mediated DNA-binding activity, GST-R411–524/MYC2541–623 binds the WT probe (Fig. 2C, lane 2). GST-R411–524/MYC2541–623, GST-R411–610Δ532–560, and GST-R411–610 SQS do not bind to the M probe (Fig. 2C, lanes 6–8). These results indicate that R homodimer formation through the ACT domain prevents the bHLH dimeric arrangement that is essential for DNA-binding activity.
RIF1 Interacts with Monomeric R bHLH.
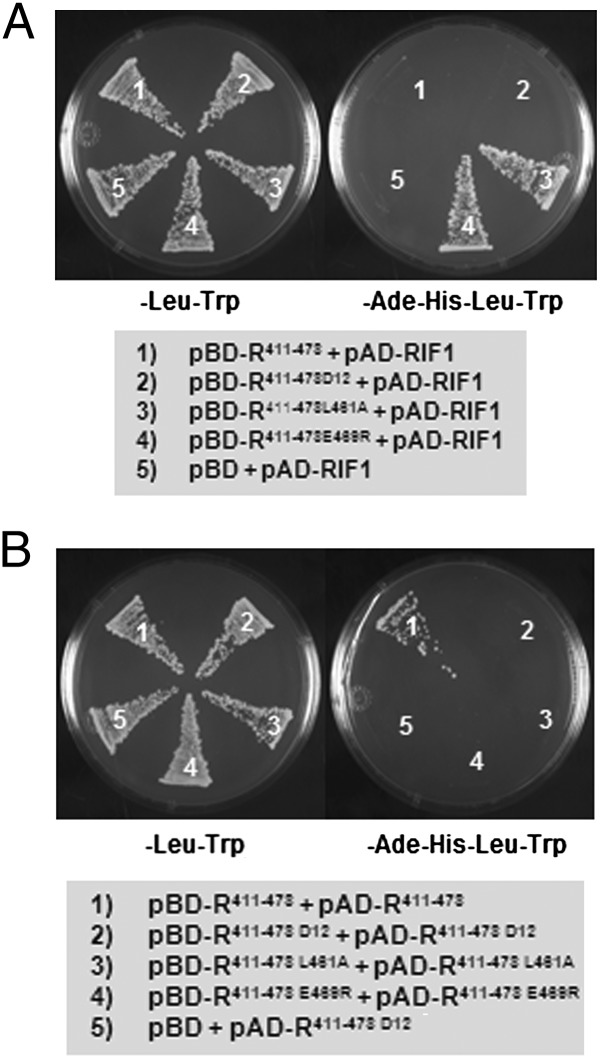
RIF1 was identified previously as an Emsy N-terminal–domain protein that specifically recognizes the bHLH motif of R and is essential for the regulatory activity of R on chromatin templates (19). RIF1 interacts with the 411–462 bHLH region (19). Because R411–462 does not form homodimers (Fig. 1B), these results indicate that RIF1 can interact with the monomeric form of the R bHLH. However, when the briefly extended bHLH (pBD-R411–478) was tested, no interaction with RIF1 (pAD-RIF1) was observed (Fig. 3 A, 1). These results suggest that the formation of a bHLH-mediated homodimer interferes with the binding of RIF1 with R. These findings should allow us to identify mutations that interfere with R bHLH dimerization but that fail to affect the R bHLH–RIF1 interaction. Computer-aided, structural homology modeling of the bHLH of R against the well-studied, classic MAX bHLH-LZ placed key residues in the extended bHLH (residues 411–478) in position for a leucine zipper-like (LZL) functionality, which perhaps contributes to stabilizing the dimerization of the bHLH (Fig. S1). Consistent with such a prediction, the mutations L461A and E469R that disrupt LZL structure (Fig. S1) abolished the dimerization of R411–478 (Fig. 3 B, 3 and 4). Interestingly, these mutations that interfere with bHLH homodimerization restored the interaction of pBD-R411–478 with pAD-RIF1 (Fig. 3 A, 3 and 4). RD12, an allele of R which harbors an insertion of three amino acids in the second helix of the bHLH domain as a consequence of the excision of a Ds transposon, also was tested for dimerization and interaction with RIF1. RD12 significantly reduced anthocyanin accumulation and flavonoid gene expression (19, 20). The three-amino acid insertion in the bHLH domain of RD12 abolished not only dimerization (Fig. 3 B, 2) but also its interaction with RIF1 (Fig. 3 A, 2). Taken together, these results demonstrate that RIF1 recognizes only a bHLH monomer and that bHLH dimerization blocks the R–RIF1 interaction.
Fig. 3.
The ability of RIF1 to interact with the dimeric R411–478 and its monomeric mutants. (A) Yeast two-hybrid assay for pAD-RIF1 with the dimer-forming R411–478 and its monomeric mutants (R411–478D12, R411–478L461A, and R411–478E469R) fused to pBD-GAL4-Cam plasmid. (B) The monomeric nature of the R411–478 mutants confirmed by yeast two-hybrid assay. The empty pBD-GAL4-Cam vector was used as a negative control. The yeast two-hybrid assay was done using the strain AH109 containing the HIS3 and ADE2 reporter genes. Growth in −Ade−His−Leu−Trp medium is indicative of interaction.
ACT Domain Plays a Switch Function in the Regulatory Specificity of R.
Our results so far provide evidence that the extended bHLH region of R can dimerize and bind G-box DNA motifs but can do so only when the ACT domain is unable to homodimerize. Indeed, the ACT domain is essential for the regulation of anthocyanin accumulation by R (18), and our results indicate that the interaction of R and RIF1 occurs only when the bHLH is in its monomeric form. Thus, the ACT domain appears to serve a regulatory switch function, and we hypothesize that it allows R to participate in at least two mechanisms in regulatory complexes: (i) It is tethered to some promoters through C1 and recruits RIF1, as we have shown for A1 (19) (ACT domain switch ON), or (ii) it binds directly to G-box–containing promoters and activates transcription (ACT domain switch OFF).
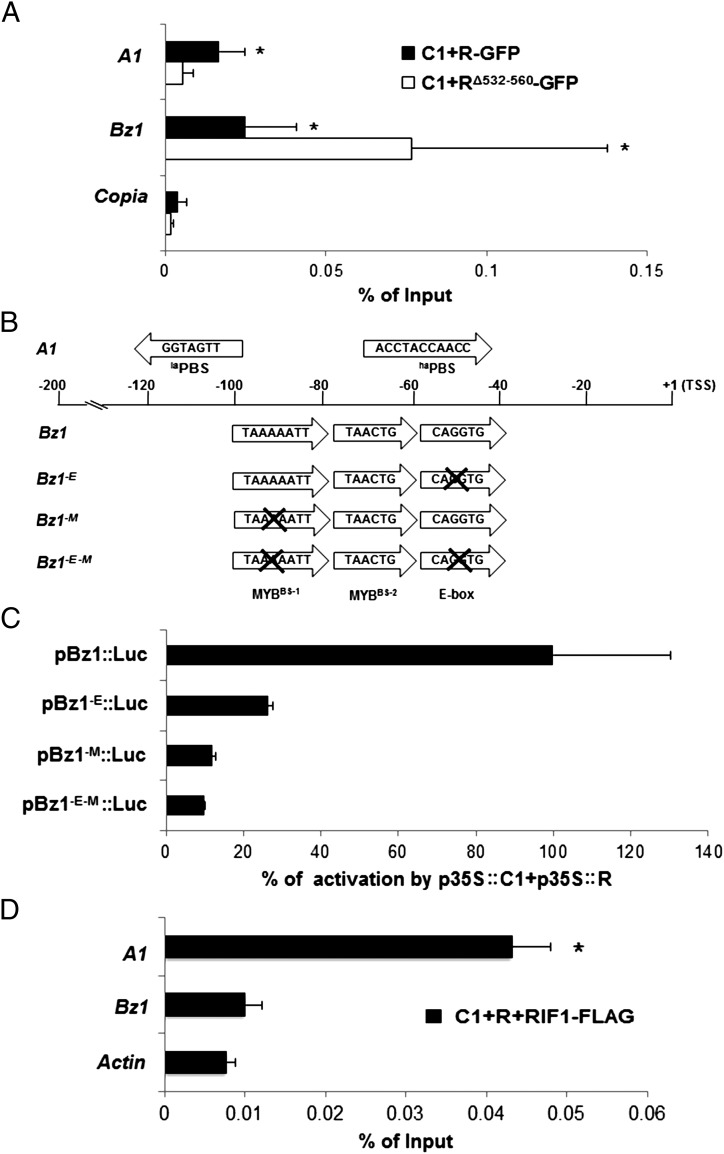
To test this dual R mechanism of action, we investigated how R controls the expression of Bz1. Bz1 encodes an anthocyanin biosynthesis enzyme that acts in the pathway several steps after A1 and which also is controlled by C1 and R. Previously Bz1 had been proposed to harbor canonical G-boxes in its promoter that were important for its expression (21), suggesting it as a good candidate for an ACT domain switch OFF target. We performed ChIP followed by quantitative PCR (qPCR) to show recruitment of the R–C1 or RΔ532–560–C1 complex to the A1 and Bz1 promoters. When R and C1, both driven by the 35S promoter, were cotransformed into maize protoplasts, a significant recruitment of the complex was observed on the A1 promoter, consistent with previous results (19). However, in the absence of the ACT domain (RΔ532–560), no recruitment of R to the A1 promoter was observed (A1 in Fig. 4A). When the presence of the Bz1 promoter was assessed in the same immunoprecipitated material, binding of the R–C1 complex was evident (Bz1 in Fig. 4A). However, in contrast to A1, the deletion of the ACT domain significantly enhanced binding to Bz1 (Fig. 4A, compare C1+RΔ532–560-GFP in A1 and Bz1).
Fig. 4.
ChIP and transient expression experiments on A1 and Bz1 promoter. (A) ChIP assays with anti-GFP antibody from chromatin obtained from maize protoplasts transformed with p35S:C1 (C1) and p35S:R-GFP (R-GFP) or p35S:RΔ532–560-GFP (RΔ532–560-GFP). The immunoprecipitates were analyzed by the presence of A1 or Bz1 promoter by qPCR. ChIP results were normalized to input DNA, and the percentage of input was calculated using the Copia and Actin genomic regions as control (only Copia is shown). Error bars indicate SEs of the three biological replicates. *P < 0.08. (B) Graphical representation of the key cis elements in the maize A1 and Bz1 promoters and the Bz1 promoter mutants. Sequence and arrangement of important MYBBS and E-box required for R-C1–mediated regulation present in both promoters are shown. The laPBS and haPBS in the A1 promoter are indicated. The positions of the cis elements relative to the TSS of each promoter are shown also. Arrows indicate the orientation of individual cis elements in each promoter. The E-box, MYBBS-1, and E-box/MYBBS-1 mutants of the Bz1 promoter are designated “Bz1-E,” “Bz1-M,” and “Bz1-E-M,” respectively. A cross-mark indicates mutation in the corresponding cis element. (C) Activation of Bz1 mutant promoters by C1 and R. Transient expression following bombardment of BMS maize cells with p35S::C1 + p35S::R, together with a Bz1 WT promoter/luciferase plasmid (pBz1::Luc), or a Bz1 promoter/luciferase plasmid containing a mutation in the E-box (pBz1-E::Luc), the MYBBS-1 (pBz1-M::Luc), or mutations in both the E-box and the MYBBS-1 (pBz1-E-M::Luc) (SI Materials and Methods). Each treatment was performed in triplicate. Error bars indicate the SD of the samples. (D) ChIP experiment showing differential binding of the C1 R–RIF1 complex to the A1 and Bz1 promoter. ChIP assays were carried out using anti-FLAG antibodies from chromatin obtained from maize protoplasts transformed with p35S::C1, p35S::R, and FLAG-tagged p35S::RIF1 (RIF1-FLAG). The immunoprecipitates were analyzed for the presence of A1 or Bz1 promoter by qPCR. ChIP results were normalized to input DNA, and percentage of input was calculated using the Actin genomic region as control. Error bars indicate the SEs of the three biological replicates. *P < 0.05.
The differential recruitment of the R–C1 or RΔ532–560–C1 complex to A1 and Bz1 promoters prompted us to investigate the identity and arrangement of cis-regulatory elements in these promoters that are required for R–C1–mediated regulation. In the A1 promoter, a region critical for R–C1–mediated regulation resides between −123 bp and −88 bp relative to the transcription start site (TSS) (22). In Bz1, the sequences between −76 and −45 bp and an AT-rich block between −88 bp and −80 bp relative to the TSS were shown to be important for C1–R–mediated regulation (21). In silico analysis and visual inspection of these critical regions of the A1 and Bz1 promoters revealed no significant sequence similarities (Fig. 4B) beyond those previously described and whose function remains unclear (23). Both promoters contain multiple putative MYB-binding sites (MYBBS) fitting the CT/AAC consensus. However, the sequences of the MYBBS in A1 and Bz1 are far from identical. In addition, the Bz1 promoter harbors an E-box (CAGGTG) close to the MYBBS, whereas E-box–like sequences are not obvious in A1. To evaluate the extent to which the two MYBBS and E-box motifs in Bz1 are important for C1–R–mediated activation, we performed combinatorial site-directed mutagenesis, and activities of the promoter mutants were assayed by transient expression experiments in Black Mexican Sweet (BMS) maize cells. As shown in Fig. 4C, activation of the Bz1 promoter dropped significantly when the MYBBS-1 (TAAAAATT; pBz1−M::Luc) or the E-box (pBz1−E::Luc) was mutated individually. The E-box/MYBBS-1 double-mutant (pBz1−E−M::Luc) further decreased Bz1 promoter activation. However, mutations of the MYBBS-2 (TAACTG) had no effect on Bz1 promoter activation (Fig. S2). Moreover, in EMSA, GST-R411–478 specifically recognized the E-box sequence in the Bz1 promoter (Fig. S3). These results indicate that the binding of R to the E-box plays a critical role in activation of the Bz1 promoter. Accordingly the bHLH region of R must be in a dimeric form, precluding the interaction with ACT. To test this hypothesis, we performed ChIP with FLAG-specific antibodies on a FLAG-tagged version of RIF1 transiently introduced into maize protoplasts together with C1 and R. Consistent with the model proposed, RIF1 is recruited to the A1 much more efficiently than to the Bz1 promoter (Fig. 4D).
Conclusion
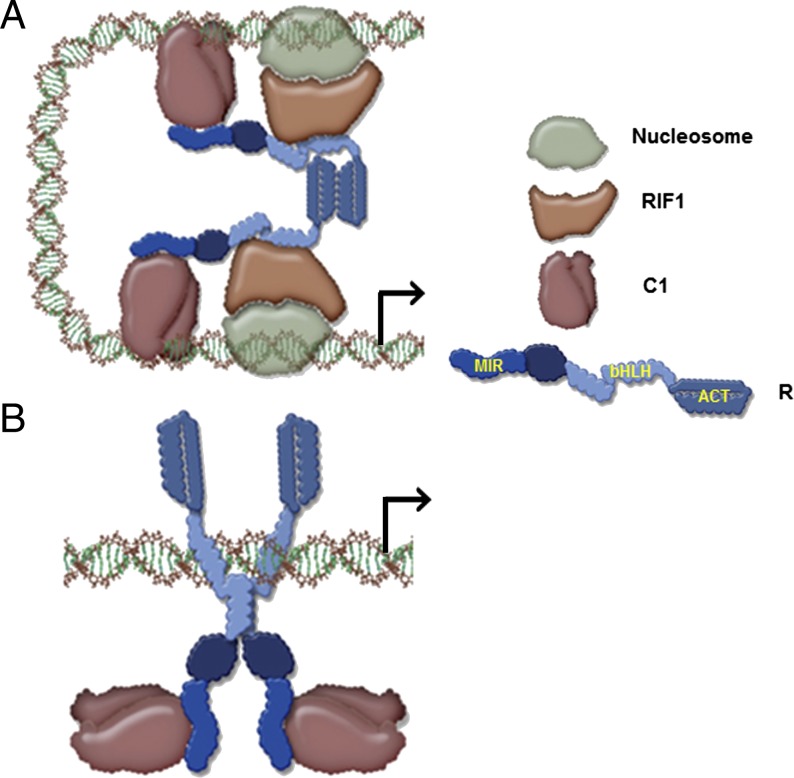
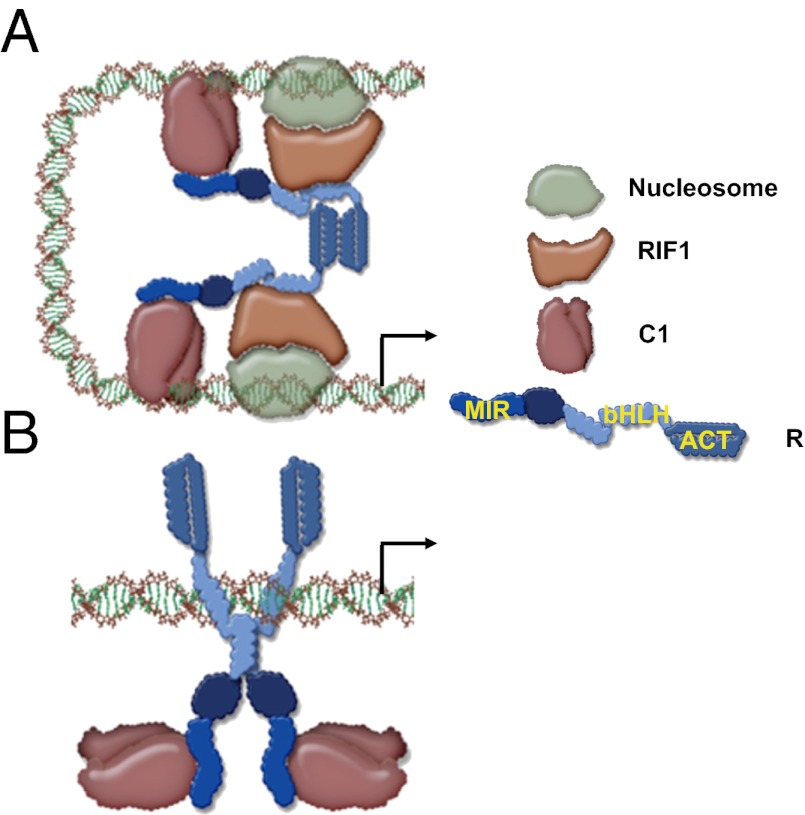
Our results provide a mechanism by which bHLH transcription factors participate in the coordinate regulation of multiple genes that lack obvious conservation of regulatory sequences. A1 provides one example of a promoter in which R is tethered to DNA through its interaction with the R2R3-MYB factor C1 (Fig. 5A). The activation of A1 in its normal chromatin setting requires the action of RIF1 (19), which interacts with the monomeric form of the bHLH of R. This R-bHLH monomeric conformation is preserved by the dimerization of the ACT domain. If, however, the dimerization of the ACT is disrupted, then the bHLH is licensed to dimerize and, as a dimer, recognizes E-box cis-regulatory elements, such as those present in the Bz1 promoter (Fig. 5B). Even though in this configuration the DNA-binding activity of R contributes to a significant DNA tethering of the R–C1 complex, C1 continues to be essential for Bz1 activation because it provides a powerful transcriptional activation domain (24). Because RIF1 is not recruited significantly to the R–C1 regulatory complex on Bz1, it is possible either that a factor other than RIF1 takes over the chromatin function in this complex or that chromatin does not control Bz1 expression as it does A1. Based on these models, whether the ACT is a monomer or a dimer is of fundamental importance to determine which DNA-binding domain will make DNA contact. We do not yet understand how dimerization of the ACT is controlled. However, its structural homology with small molecule-binding regions in enzymes that are allosterically regulated (18) suggests that a small molecule might be involved. From this perspective, it is interesting that Bz1 functions downstream of A1 and that in many other plants early and late flavonoid biosynthesis genes can be differentiated based on their regulation (25). Hence, it is possible that a flavonoid pathway intermediately downstream of A1 signals R through the ACT to change its conformation and activate late-pathway genes. Additional experiments will be required to test this hypothesis.
Fig. 5.
Proposed model for promoter switching by R. (A) ACT domain ON model shows C1 making the DNA contacts and recruiting R through its N-terminal MYB-interacting region (MIR). The ACT C-terminal region of R forms a dimer; therefore the bHLH motif remains as a monomer and can bind to RIF1, which recognizes chromatin components. This model explains the expression of A1. (B) ACT domain OFF model shows R binding to DNA by forming homodimers through the bHLH because the ACT is not homodimerizing. C1 continues to interact with the MIR region of R, providing transcriptional activatory function.
Materials and Methods
Additional details of materials and methods are given in SI Materials and Methods, Tables S1 and S2, and Figs. S1–S3.
Maize Protoplast Transformation and ChIP Experiments.
Protoplasts from 10- to 12-d-old etiolated maize seedlings were prepared essentially as described previously (19). The fluorescence furnished by p35S::sGFP was used to calculate the transformation efficiency, which usually ranged from 30–50%. If efficiency was below 30%, two or three individual reactions were pooled together to yield one biological replicate. ChIP experiments were performed as described (19, 26) using ∼105 protoplasts per reaction. Immunoprecipitation was performed overnight at 4 °C with either 1 μL anti-GFP antibody (ab290; Abcam) or 1 μL anti-FLAG antibody (Sigma-Aldrich) with biological triplicates. Real-time PCR was performed to quantify the enrichment of DNA regions by using specific primer pairs (Table 2). Student’s t test was performed between A1 or Bz1 and copia+Actin, at P < 0.08 (Fig. 4A) and between A1 or Bz1 and Actin at P < 0.05 (Fig. 4D).
Transient Expression Assays in BMS Maize Cells.
Transient expression assays in BMS maize cells were performed as described (13, 14, 27). Luciferase assays were performed in a 96-well plate using the Dual Luciferase assay kit from Promega and a luminometer Centro LB960 (Berthold Technologies). A p35S::Renilla construct (28) was included in each bombardment as a normalization control. The fold activation was calculated as the ratio between each particular treatment and the treatment with the appropriate reporter construct without activator. The fold activation of the Bz1 WT promoter by C1 and R was set to 100%, and the activation of each mutant promoter is shown in relation to that of the WT Bz1.
Recombinant Protein Expression and Purification.
Different fragments of the bHLH domain of R were generated by PCR and cloned into either the pET41a-GST vector for the production of a GST fusion protein or into the pET41a-6xHis vector for the production of a six-histidine–tagged fusion protein. Both vectors were modified from plasmid pET41a(+) (Novagen) and were kind gifts from Y. I. Chi (University of Kentucky, Lexington, KY). The constructs were verified by DNA sequencing and transformed into E. coli BL21(DE3) cells. Cell cultures were induced by adding isopropyl-β-d-thio-galactoside to a final concentration of 1 mM for 3 h at 30 °C. The bacterial cells were lysed using CelLytic B (Sigma-Aldrich) according to the manufacturer’s instructions. The GST fusion proteins were bound to Glutathione Sepharose 4B columns (Amersham) and eluted using 50 mM Tris⋅HCl (pH 8.0) and 10 mM glutathione; N6His fusion proteins were bound to HIS-Select nickel affinity gel (Sigma-Aldrich) and eluted using 50 mM sodium phosphate (pH 8.0), 300 mM NaCl, and 250 mM imidazole.
Analysis of Protein–Protein Interaction.
For yeast two-hybrid experiments, different fragments of the bHLH domain of R, with or without the C-terminal extension (i.e., R411–462, R411–470, R411–478, R411–496, R411–510, R411–524, and R411–610) were PCR amplified and cloned in the plasmids pAD-GAL4-2.1 and pBD-GAL4 Cam (Stratagene). The plasmids containing the R bHLH fragments were cotransformed into yeast strain AH109 (Clontech). To determine whether the R bHLH mutants (R411–478 D12, R411–478L416A, and R411–478E469R) form homodimers, each mutant was cloned into both pAD-GAL4-2.1 and pBD-GAL4 Cam and cotransformed to AH109 for the yeast two-hybrid assay. To determine the interaction between the R bHLH mutants and full-length RIF1, the RIF1 was cloned into pAD-GAL4-2.1 to create pAD-RIF1. Each R bHLH mutant cloned in pBD-GAL4 Cam was cotransformed with pAD-RIF1 into yeast strain AH109. Transformants were selected on synthetic defined (SD) medium lacking leucine and tryptophan (−Leu−Trp). Colonies then were screened for growth on SD medium lacking tryptophan, leucine, and histidine (−His−Leu−Trp) or on SD medium lacking tryptophan, leucine, histidine, and adenine (−Ade−His−Leu−Trp). β-Gal reporter assay of the transformed yeast colonies was performed as described in the yeast protocol handbook (Clontech). To test protein–protein interaction in vitro, a GST pull-down assay was carried out as described previously (18), with the modifications described in SI Materials and Methods. To test protein–protein interaction in planta, protoplasts were isolated from tobacco-cell suspension cultures (Nicotiana tabacum L. cv. Xanthi b ‘Brad’), and electroporation with supercoiled plasmid DNA was performed as previously described (29). The reporter and effector plasmids used in the protoplast assay are described in SI Materials and Methods. Luciferase and β-glucuronidase (GUS) activities were measured 20–22 h after incubation at 28 °C. GUS activity was measured according to protocols described by Jefferson et al. (30) and was used for normalization of luciferase activity. All constructs were tested in at least three independent experiments.
EMSA.
Probes used for EMSA were either a WT G-box motif from the Arabidopsis DFR promoter (5′-CGTTCCCCACGTGCTTCTCC-3′) or a mutated G-box motif (M) in which the core recognition sequence, CACGTG, was replaced by CAATTG. Complementary oligonucleotides, biotin-labeled at the 5′-end of each strand, were annealed to produce double-stranded probes for EMSA. The DNA-binding reactions were carried out in 10 mM Tris⋅HCl (pH 7.5), 50 mM KCl, 1 mM DTT, 5% (vol/vol) glycerol, and 100 ng poly(deoxyinosinic-deoxycytidylic) acid (polydI:dC) at a final volume of 20 μL. Purified proteins were incubated with 0.25–0.5 nM DNA probe on ice for 30 min. The DNA–protein complexes were resolved on 6% (wt/vol) nondenaturing polyacrylamide gels and then were transferred to Hybond-N+ nylon membranes (Amersham). The band shifts were detected by a chemiluminescent nucleic acid detection module (Pierce) and autoradiography.
SELEX.
SELEX was conducted essentially as described (31). N6His-R411–478 was expressed in E. coli and was affinity purified using Ni-NTA beads under natural conditions. The DNA library for SELEX (32) harbors the sequences: 5′-ACTCGAGGAATTCGGTACCCCGGGT(N)26TGGATCCGGAGAGCTCCCAACGCGT-3′, where N indicates an equimolecular distribution of A, C, G, and T at a final volume of 20 μL. Around 50 ng of purified N6His-R411–478 protein was incubated with radioactively labeled DNA library (∼105cpm) in buffer (20 mM Hepes, 50 mM KCl, 1 mM DTT, 0.5 mM EDTA, 5% (vol/vol) glycerol, 0.5 μg/μL BSA, and 100 ng polydI:dC) for 30 min, and the N6His-R411–478–DNA complex was separated by PAGE (referred to as “EMSA”). The DNA purified from the complex appears as a shifted band in EMSA and is used for the next round of SELEX selection. After the last (sixth) round of selection, the DNA from the shifted protein–DNA complex was extracted, PCR amplified, cloned into a pCR 2.1-TOPO vector (Invitrogen), and then sequenced. The sequences extracted from sequencing were analyzed manually and by using on-line software Gibbs Motif Sampler (http://bayesweb.wadsworth.org/gibbs/gibbs.html) and are displayed with Weblogo (http://weblogo.berkeley.edu/logo.cgi).
Structural Homology Modeling of R bHLH Domain.
A structural model of the bHLH domain of Zea mays R (National Center for Biotechnology Information accession P13526) was generated by SWISS-MODEL 3.5 using the Alignment Interface (33). Alignments to the Homo sapiens MAX sequence (obtained from Protein Data Bank structures 1an2 and 1hlo) were made initially with Clustal × 1.83 (34). Alignments then were tailored in an educated manner to remove excessive spacing. Because MAX structures 1an2 and 1hlo covered different but overlapping regions of the bHLH domain, R was modeled to both. The resulting models were overlaid with the MAX structures and fused in DeepView 3.7 (SP5) (35).
Supplementary Material
Acknowledgments
We thank Dr. Y. I. Chi (University of Kentucky) and Dr. J.-C. Jang (Ohio State University) for the generous gift of plasmids. This research was supported by grants from the Kentucky Tobacco Research and Development Center, University of Kentucky (to L.Y.), by Grants DBI-0701405 and IOS-1125620 from the National Science Foundation, and by Agricultural and Food Research Initiative Competitive Grant 2010-65115-20408 from the US Department of Agriculture National Institute of Food and Agriculture (to E.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 11918 (volume 109, number 30).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205513109/-/DCSupplemental.
References
- 1.Feller A, Machemer K, Braun EL, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011;66:94–116. doi: 10.1111/j.1365-313X.2010.04459.x. [DOI] [PubMed] [Google Scholar]
- 2.Blackwood EM, Eisenman RN. Max: A helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 3.Ayer DE, Kretzner L, Eisenman RN. Mad: A heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- 4.Hurlin PJ, Quéva C, Eisenman RN. Mnt, a novel Max-interacting protein is coexpressed with Myc in proliferating cells and mediates repression at Myc binding sites. Genes Dev. 1997;11:44–58. doi: 10.1101/gad.11.1.44. [DOI] [PubMed] [Google Scholar]
- 5.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 6.Bailey PC, et al. Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell. 2003;15:2497–2502. doi: 10.1105/tpc.151140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heim MA, et al. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003;20:735–747. doi: 10.1093/molbev/msg088. [DOI] [PubMed] [Google Scholar]
- 8.Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ludwig SR, Habera LF, Dellaporta SL, Wessler SR. Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc Natl Acad Sci USA. 1989;86:7092–7096. doi: 10.1073/pnas.86.18.7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ludwig SR, Wessler SR. Maize R gene family: Tissue-specific helix-loop-helix proteins. Cell. 1990;62:849–851. doi: 10.1016/0092-8674(90)90259-h. [DOI] [PubMed] [Google Scholar]
- 11.Cone KC, Cocciolone SM, Burr FA, Burr B. Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant. Plant Cell. 1993;5:1795–1805. doi: 10.1105/tpc.5.12.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goff SA, Cone KC, Chandler VL. Functional analysis of the transcriptional activator encoded by the maize B gene: Evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev. 1992;6:864–875. doi: 10.1101/gad.6.5.864. [DOI] [PubMed] [Google Scholar]
- 13.Grotewold E, et al. Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc Natl Acad Sci USA. 2000;97:13579–13584. doi: 10.1073/pnas.250379897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grotewold E, Drummond BJ, Bowen B, Peterson T. The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell. 1994;76:543–553. doi: 10.1016/0092-8674(94)90117-1. [DOI] [PubMed] [Google Scholar]
- 15.Sainz MB, Grotewold E, Chandler VL. Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell. 1997;9:611–625. doi: 10.1105/tpc.9.4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernhardt C, Zhao M, Gonzalez A, Lloyd A, Schiefelbein J. The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development. 2005;132:291–298. doi: 10.1242/dev.01565. [DOI] [PubMed] [Google Scholar]
- 17.Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004;40:22–34. doi: 10.1111/j.1365-313X.2004.02183.x. [DOI] [PubMed] [Google Scholar]
- 18.Feller A, Hernandez JM, Grotewold E. An ACT-like domain participates in the dimerization of several plant basic-helix-loop-helix transcription factors. J Biol Chem. 2006;281:28964–28974. doi: 10.1074/jbc.M603262200. [DOI] [PubMed] [Google Scholar]
- 19.Hernandez JM, Feller A, Morohashi K, Frame K, Grotewold E. The basic helix loop helix domain of maize R links transcriptional regulation and histone modifications by recruitment of an EMSY-related factor. Proc Natl Acad Sci USA. 2007;104:17222–17227. doi: 10.1073/pnas.0705629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Wang L, Kermicle JL, Wessler SR. Molecular consequences of Ds insertion into and excision from the helix-loop-helix domain of the maize R gene. Genetics. 1998;150:1639–1648. doi: 10.1093/genetics/150.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roth BA, Goff SA, Klein TM, Fromm ME. C1- and R-dependent expression of the maize Bz1 gene requires sequences with homology to mammalian myb and myc binding sites. Plant Cell. 1991;3:317–325. doi: 10.1105/tpc.3.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tuerck JA, Fromm ME. Elements of the maize A1 promoter required for transactivation by the anthocyanin B/C1 or phlobaphene P regulatory genes. Plant Cell. 1994;6:1655–1663. doi: 10.1105/tpc.6.11.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lesnick ML, Chandler VL. Activation of the maize anthocyanin gene a2 is mediated by an element conserved in many anthocyanin promoters. Plant Physiol. 1998;117:437–445. doi: 10.1104/pp.117.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sainz MB, Goff SA, Chandler VL. Extensive mutagenesis of a transcriptional activation domain identifies single hydrophobic and acidic amino acids important for activation in vivo. Mol Cell Biol. 1997;17:115–122. doi: 10.1128/mcb.17.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mol J, Grotewold E, Koes R. How genes paint flowers and seeds. Trends Plant Sci. 1998;3:212–217. [Google Scholar]
- 26.Ferreyra ML, et al. Cloning and characterization of a UV-B-inducible maize flavonol synthase. Plant J. 2010;62:77–91. doi: 10.1111/j.1365-313X.2010.04133.x. [DOI] [PubMed] [Google Scholar]
- 27.Hernandez JM, et al. Different mechanisms participate in the R-dependent activity of the R2R3 MYB transcription factor C1. J Biol Chem. 2004;279:48205–48213. doi: 10.1074/jbc.M407845200. [DOI] [PubMed] [Google Scholar]
- 28.Elomaa P, et al. A bHLH transcription factor mediates organ, region and flower type specific signals on dihydroflavonol-4-reductase (dfr) gene expression in the inflorescence of Gerbera hybrida (Asteraceae) Plant J. 1998;16:93–99. doi: 10.1046/j.1365-313x.1998.00273.x. [DOI] [PubMed] [Google Scholar]
- 29.Pattanaik S, et al. Isolation and functional characterization of a floral tissue-specific R2R3 MYB regulator from tobacco. Planta. 2010;231:1061–1076. doi: 10.1007/s00425-010-1108-y. [DOI] [PubMed] [Google Scholar]
- 30.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chai C, Xie Z, Grotewold E. SELEX (Systematic Evolution of Ligands by EXponential Enrichment), as a powerful tool for deciphering the protein-DNA interaction space. Methods Mol Biol. 2011;754:249–258. doi: 10.1007/978-1-61779-154-3_14. [DOI] [PubMed] [Google Scholar]
- 32.Huang H, Mizukami Y, Hu Y, Ma H. Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res. 1993;21:4769–4776. doi: 10.1093/nar/21.20.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]