Abstract
Neuregulin 1 (NRG1) and ErbB4, critical neurodevelopmental genes, are implicated in schizophrenia, but the mediating mechanisms are unknown. Here we identify a genetically regulated, pharmacologically targetable, risk pathway associated with schizophrenia and with ErbB4 genetic variation involving increased expression of a PI3K-linked ErbB4 receptor (CYT-1) and the phosphoinositide 3-kinase subunit, p110δ (PIK3CD). In human lymphoblasts, NRG1-mediated phosphatidyl-inositol,3,4,5 triphosphate [PI(3,4,5)P3] signaling is predicted by schizophrenia-associated ErbB4 genotype and PIK3CD levels and is impaired in patients with schizophrenia. In human brain, the same ErbB4 genotype again predicts increased PIK3CD expression. Pharmacological inhibition of p110δ using the small molecule inhibitor, IC87114, blocks the effects of amphetamine in a mouse pharmacological model of psychosis and reverses schizophrenia-related phenotypes in a rat neonatal ventral hippocampal lesion model. Consistent with these antipsychotic-like properties, IC87114 increases AKT phosphorylation in brains of treated mice, implicating a mechanism of action. Finally, in two family-based genetic studies, PIK3CD shows evidence of association with schizophrenia. Our data provide insight into a mechanism of ErbB4 association with schizophrenia; reveal a previously unidentified biological and disease link between NRG1-ErbB4, p110δ, and AKT; and suggest that p110δ is a previously undescribed therapeutic target for the treatment of psychiatric disorders.
Keywords: neuregulin 3, AKT, development, neuroleptic
Schizophrenia is a severe neuropsychiatric disorder with a complex genetic etiology (1). Polymorphisms in the secreted growth factor neuregulin 1 (NRG1) have been associated with risk for schizophrenia (2–4) and recently, common genetic variation and structural microdeletions in ErbB4, a receptor tyrosine kinase for NRG1, have also been associated with the disorder (5–9). NRG1-ErbB4 signaling plays a critical role in neural development and synaptic plasticity (10–12) and NRG1 and ErbB4 mutant mice exhibit behavioral alterations (2, 12–14) consistent with other murine models of schizophrenia (15). Schizophrenia-associated genetic variation in these genes is implicated in human brain structure and function (6, 16, 17), but the biological mechanisms accounting for these diverse effects and how they translate into illness are unclear.
Increased NRG1 and ErbB4 expression has been observed in schizophrenia postmortem brain tissue (5, 7, 18) and in neurons derived from induced pluripotent stem cells (IPSCs) of patients (19). Risk-associated polymorphisms in these genes affect expression of specific NRG1 and ErbB4 isoforms in human brain (7, 18, 20). In particular, risk polymorphisms in ErbB4 (rs7598440, rs839523, and rs707284) predict increased expression of the ErbB4 CYT-1 receptor (5, 7), one of two biologically occurring ErbB4 isoforms, the other being ErbB4 CYT-2 (21). Unlike ErbB4 CYT-2, ErbB4 CYT-1 includes a phosphoinositide 3-kinase (PI3K)-binding site and is capable of activating the PI3K pathway (21–24). Class IA PI3Ks are activated by receptor tyrosine kinases and exist as obligate heterodimers (25, 26) consisting of a p110 catalytic subunit (PIK3CA, PIK3CB, and PIK3CD encode the isoforms p110α, p110β, and p110δ, respectively) and a p85 regulatory subunit (PIK3R1, PIK3R2, and PIK3R3, which encode the p85α, p85β, and p55γ isoforms, respectively) (25). PI3K signaling in turn mediates NRG1-induced cell survival and neuronal and synaptic development (21–24). These findings implicate aberrant PI3K signaling as a potential pathogenic effect of schizophrenia-associated variation in ErbB4.
In this study, we have used a translational systems biology approach incorporating patient-derived lymphoblastoid B-cell lines (LCLs), human postmortem brain, and human molecular and clinical genetics combined with proof-of-concept pharmacological studies in rodents to test the hypothesis that abnormal PI3K signaling is related to schizophrenia-associated variation in ErbB4. Together our findings support this hypothesis, refine our knowledge of the signaling partners affected downstream of ErbB4, identify a previously unknown role of the lipid kinase, PIK3CD, in NRG1 signaling and schizophrenia, and implicate PIK3CD as a previously undescribed therapeutic target.
Results
Schizophrenia and ErbB4 Risk Genetic Variation Are Associated with ErbB4 CYT-1 Levels.
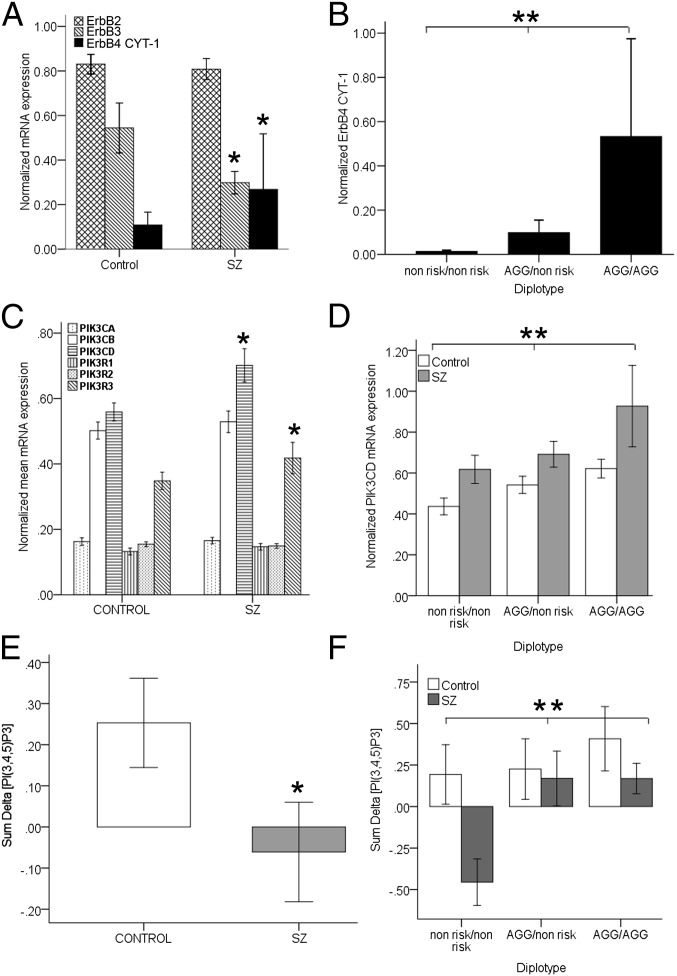
SNPs associated with expression of single genes are frequently associated with expression of other genes in the same biological pathway (27, 28). We tested whether ErbB4 SNPs have biological utility to dissect transcriptional networks downstream of their association with genetic risk for schizophrenia and expression of ErbB4. We established LCLs from schizophrenia patients and normal controls (n = 55) and using real-time quantitative RT-PCR (QPCR), determined expression traits for ErbB4 splice isoforms and downstream signaling partners. We found that human LCLs express JM-a/CYT-1 ErbB4 transcripts; thus the ErbB4 isoform aberrantly elevated in the brain in schizophrenia and regulated by a risk-associated haplotype in ErbB4 (5, 7) can be studied in this human peripheral cell model. We observed in schizophrenia that ErbB4 CYT-1 mRNA is increased [F(5, 50) = 3.81; P = 0.04, Fig. 1A] and that the ErbB4 risk haplotype (AGG; rs7598440, rs839523, rs707284) is associated with elevated ErbB4 CYT-1 transcription [F(5, 50) = 4.0; P = 0.01, Fig. 1B] in both controls and patients. These findings are consistent with and provide independent replication of analogous illness and genetic associations derived from human brain (5, 7). Interestingly, individuals homozygous for the nonrisk haplotype had undetectable levels of ErbB4 mRNA or protein (Fig. 1B). Human LCLs also expressed abundant ErbB2 and ErbB3, the two other receptors that mediate NRG1 signaling (Fig. 1A). ErbB3 levels were significantly reduced in patient LCLs [F(1, 55) = 3.80; P = 0.04, Fig. 1A], as also reported in postmortem human brain (29) (see below). ErbB3 levels, however, were not associated with the ErbB4 risk haplotype (P > 0.5) or correlated with ErbB4 levels (P > 0.9). Together, these findings illustrate the fidelity of the human LCL as a model for examination of the genetic regulation of NRG1-ErbB4 signaling and replicate, in an independent sample of living subjects, previous findings in postmortem brain in schizophrenia, including elevation of ErbB4 CYT-1 expression, the impact of ErbB4 risk polymorphisms on ErbB4 CYT-1 expression (7), and reduced ErbB3 expression (29).
Fig. 1.
(A) Increased expression of ErbB4 CYT-1 mRNA [*F(5, 50) = 3.81; P = 0.04] and decreased ErbB3 in patients with schizophrenia (n = 23) compared with normal control subjects (n = 32) [*F(1, 55) = 3.80; P = 0.045]. (B) Association between ErbB4 risk haplotype (AGG; rs7598440; rs839523; rs707284) and ErbB4 CYT-1 levels in the whole cohort [F(5, 50) = 4.00; P = 0.012] (AGG/AGG, n = 13; AGG/nonrisk, n = 28; nonrisk/nonrisk, n = 14). (C) Increased expression of PIK3CD (**β = 0.41; t = 3.20, P ≤ 0.002) and PIK3R3 (*β = 0.26; t = 1.97, P ≤ 0.025) in patients with schizophrenia (n = 23) compared with normal control subjects (n = 32). (D) PIK3CD expression in normal and patient-derived LCLs is associated with an ErbB4 risk haplotype (**β = −0.32; t = −2.57, P ≤ 0.01). (E and F) NRG1-stimulated intracellular [PI(3,4,5)P3] production is (E) reduced in schizophrenia (*β = −0.27; t = −1.89, P ≤ 0.032; n = 29 vs.19) and (F) associated with ErbB4 risk haplotype [*β = −0.41; t = −3.01, P ≤ 0.004; whole sample (AGG/AGG, n = 11; AGG/nonrisk, n = 23; nonrisk/nonrisk, n = 13)]. Values are mean ± SEM.
PIK3CD Levels Are Associated with Schizophrenia and ErbB4 Genetic Variation.
The evidence that ErbB4 CYT-1 is associated with risk genetic variation in ErbB4 and schizophrenia in both brain and LCLs suggests that the PI3K signaling pathway, to which the receptor is coupled, is where the downstream effects of NRG1-ErbB4 alterations are mediated. In LCLs, we examined quantitative expression of class IA PI3K gene transcripts and found two, the catalytic subunit, PIK3CD, and the regulatory subunit, PIK3R3 that were increased in schizophrenia and associated with ErbB4 risk genotype. In a multiple-regression analysis, 23% of the variance in PIK3CD expression was explained by two factors [full model F(3, 52) = 5.0, P = 0.004], schizophrenia and ErbB4 haplotype, with a 40% increase in schizophrenia (P = 0.002, Fig. 1C) and greater expression in all subjects with the ErbB4 AGG risk haplotype (P = 0.01, Fig. 1D). The increased expression of PIK3CD in patients was seen in each genotype group, indicating an effect of illness that is independent of genotype (Fig. 1D). A similar compound effect on PIK3R3 mRNA expression was also seen, with 17% of the variance [full model F(3, 52) = 3.4, P = 0.025] explained by schizophrenia (P = 0.05, Fig. 1C) and by the ErbB4 haplotype (β = −0.26; t = −2.0; P = 0.04; mean ± SD; AGG/AGG, 0.41 ± 0.28; AGG/nonrisk, 0.36 ± 0.16; nonrisk/nonrisk, 0.31 ± 0.1).
PIK3CD is the predominant p110 catalytic subunit expressed in B lymphocytes (30) and high levels of expression are also reported in the embryonic mouse nervous system (31). Interestingly, PIK3CD associates in vitro, nonselectively with p85 regulatory subunits (25) and targeted deletion of the PIK3CD gene in mice produces compensatory changes in the p85 and p55 regulatory genes (32). Thus, we hypothesized that the change in PIK3R3 was a secondary effect, i.e., that expression of PIK3CD would predict expression of PIK3R3 and account for its relative increase. In human LCLs, we found positive correlations between PIK3R3 and PIK3CD transcripts (Spearman’s rho = 0.41, P = 0.002) independent of ErbB4 genetic variation, and the effect of ErbB4 genetic variation and schizophrenia on PIK3R3 expression was explained by PIK3CD alone [model F(4, 51) = 3.9, P = 0.007; β = 0.43; t = 3.1; P = 0.003], suggesting that altered PIK3R3 expression is secondary to changes in PIK3CD. These correlational data combined with evidence implicating PIK3CD but not PIK3R3 genetically in schizophrenia (see below) suggest that the primary PI3K abnormality in the pathway concerns PIK3CD. Our results indicate an influence of ErbB4 risk-associated genetic variation and disease on quantitative expression of PIK3CD in the same directionality as ErbB4 CYT-1. To test whether these effects are specific to the PI3K pathway, we also examined expression of genes in the ERK-MAPK pathway (GRB2, SOS1, and MAPK1) to which ErbB4 CYT-1 also couples (22). No diagnostic or genotype effects were observed (Fig. S1).
NRG1-Mediated Intracellular Phosphatidylinositol,3,4,5 Triphosphate ([PI(3,4,5)P3]) Signaling Is Decreased in Schizophrenia and Associated with ErbB4 Risk Polymorphisms.
PI3K catalyses formation of the second messenger, [PI(3,4,5)P3] (26). To directly assess the biochemical consequences of differential PIK3CD levels in schizophrenia and of genetic variation in ErbB4, we used a flow cytometric immunoassay to measure NRG1-induced intracellular [PI(3,4,5)P3] levels in human LCLs. We observed that overall 29% of the variance in NRG1-induced [PI(3,4,5)P3] production was explained by four factors (diagnosis, ErbB4 risk haplotype, PIK3CD mRNA, and PIK3CD protein levels), each of which contributed at least in part independently to variation in NRG1-induced [PI(3,4,5)P3] levels [full model F(4, 44) = 5.07, P = 0.005]. In schizophrenia, NRG1 induction of [PI(3,4,5)P3] levels was significantly reduced (Fig. 1E). Furthermore, the ErbB4 risk haplotype differentially predicted NRG1-induced [PI(3,4,5)P3] levels (Fig. 1F), as did PIK3CD mRNA (P = 0.009; β = −0.37; t = −2.7) and protein expression (P = 0.05; β = −0.20; t = −1.49). No differences were observed in “baseline” [PI(3,4,5)P3] between the groups (P > 0.5). In the patients alone, a dramatic 76% of the variance in NRG1-induced [PI(3,4,5)P3] production was explained by three factors, ErbB4 genotype, PIK3CD mRNA, and PIK3CD protein levels [full model F(3, 17) = 15.43, P < 0.00001].
The multiple-regression model identified complex and competing relationships of these factors. Genotype independently predicted a relatively enhanced response to NRG1 stimulation in LCLs from patients homozygous or heterozygous for the ErbB4 risk-associated haplotype (P < 0.0001; β = −0.90; t = −6.2; Fig. 1F), consistent with the effect of this haplotype on ErbB4 CYT1 and PIK3CD expression and its effects also in normal subjects (Fig. 1 A, B, and F). However, in addition, inverse correlations were observed in schizophrenia between both PIK3CD mRNA and protein levels and [PI(3,4,5)P3] production, independent of genotype (Fig. S2). These findings, although unexpected, are consistent with published reports in cancer, in which pathologically elevated levels of PIK3CD result in dampened PI3K signaling (33, 34). In schizophrenia, elevated PIK3CD expression and blunted NRG1-induced [PI(3,4,5)P3] production are consistent with this biological effect. These biochemical data suggest that the genetic association with elevated ErbB4 CYT1 and PIK3CD expression reflects a complex dysregulation of this pathway in schizophrenia and that pathological levels of PIK3CD in patients negatively impacts PI3K signaling.
PIK3CD also mediates B-cell antigen receptor (BCR)-mediated [PI(3,4,5)P3] production (35). If the molecular effect of the ErbB4 schizophrenia-associated haplotype is specifically related to NRG1/ErbB4-stimulated PIK3CD activation, then there should be no association with CD19/BCR-induced activation of PIK3CD. We observed no association [full model F(2, 25) = 2.0, P > 0.15; diagnosis, P > 0.13; haplotype, P > 0.31]. Hence, schizophrenia and the ErbB4 risk haplotype appear related specifically to an NRG1-ErbB4-CYT-1-PIK3CD pathway, rather than there being a more generalized abnormality of PI3K signaling.
PIK3CD Expression in Human Brain Is Associated with ErbB4 Risk Polymorphisms and Down-Regulated by Antipsychotic Medication in the Rat.
Given the cellular and context specificity of molecular networks and the potential concern with LCLs that gene expression patterns can be related to clonal or transformed cells (36), we assessed expression phenotypes related to the ErbB4-PI3K pathway in postmortem human brain. Consistent with the LCL data (Fig. 1), the ErbB4 risk haplotype predicted increased PIK3CD mRNA expression in hippocampus and in dorsolateral prefrontal cortical gray matter (DLPFC) of normal individuals, two brain regions prominently implicated in the pathophysiology of schizophrenia (Fig. S3A), suggesting that genetic regulation of levels of ErbB4 and PIK3CD are normally tightly regulated in brain and the periphery. We also replicated in brains of separate individuals the finding in LCLs (Fig. 1) of increased PIK3R3 expression in schizophrenia (Fig. S3B) and of a decrease in ErbB3 mRNA in DLPFC [F (1, 105) = 5.35; P = 0.023, mean ± SD; controls, 2.95 ± 1.3, n = 73; schizophrenia, 2.34 ± 0.9, n = 32].
In contrast, patients with schizophrenia showed no association between ErbB4 risk genotype and PIK3CD levels in either the hippocampus or the DLPFC (P > 0.5) and no increase in PIK3CD expression (Fig. S3C). We hypothesized that this may be due to effects of antipsychotic medications (this confounder likely does not apply to the LCLs in view of their transformation and multiple passages). Consistent with this possibility, PIK3CD mRNA levels tended to correlate negatively with lifetime neuroleptic exposure (r = −0.268; P = 0.07) in patients with schizophrenia. To further examine this experimentally, we administered haloperidol, a standard antipsychotic drug, to rats for 28 d and found that treatment led to specific reductions of PIK3CD gene expression in brain (Fig. S3D). It is particularly noteworthy that haloperidol treatment did not affect PIK3R3, ErbB4, or ErbB3 mRNA expression (Fig. S4). Unlike PIK3CD, these transcripts all show consistent alterations in the brain and LCLs of patients with schizophrenia. These data, as well as providing a possible explanation for the normal expression of PIK3CD mRNA in brain of subjects with schizophrenia, suggest that reduction of p110δ activity may be relevant for the actions of antipsychotic drugs.
These results in human and rat brain, together with the evidence that schizophrenia and the ErbB4 risk haplotype are both associated with increased PIK3CD expression in patient-derived LCLs, suggest that rationally designed drugs specifically inhibiting the PIK3CD protein (p110δ) may represent a targeted therapeutic approach for the treatment of schizophrenia. We next explored this possibility.
Pharmacological Inhibition of p110δ (PIK3CD) Blocks the Disruptive Effects of Amphetamine in a Mouse Model of Psychosis and Increases AKT Thr308 Phosphorylation in Mouse Brain.
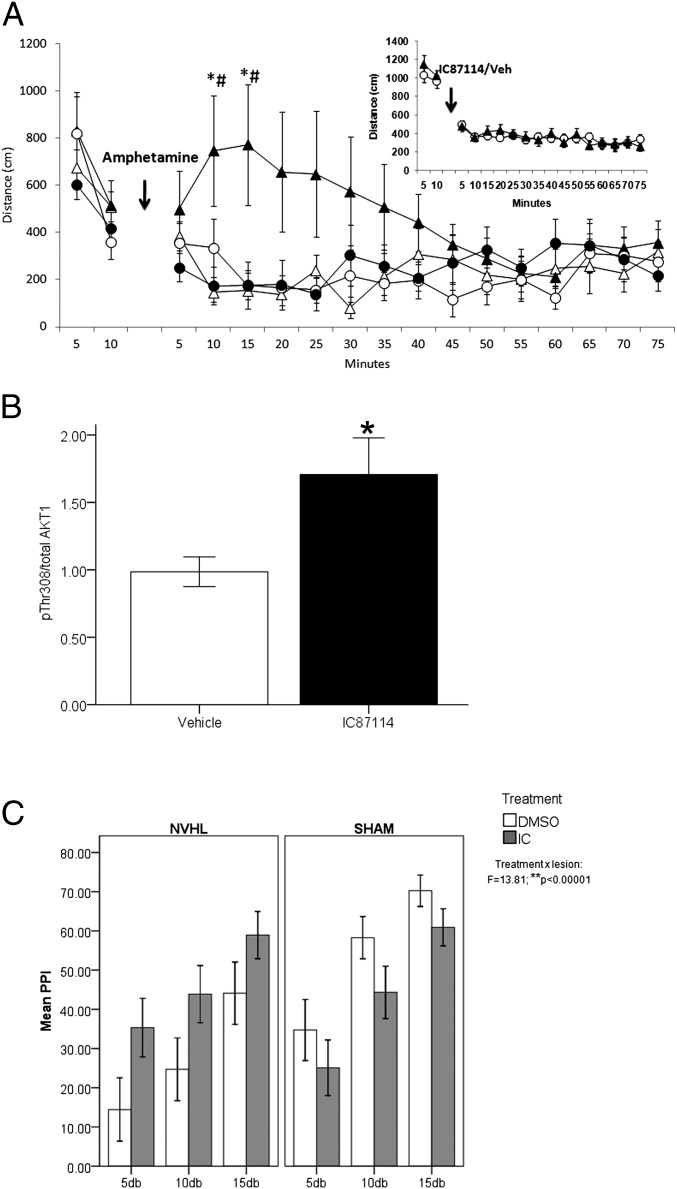
Amphetamine-induced hyperlocomotion in rodents is a traditional preclinical model for screening antipsychotic drug potential and has been widely used as a rodent model of psychosis-like behavior, related to a hyperdopaminergic state (15, 37). The rodent amphetamine model of psychosis has considerable face validity and all current antipsychotic drugs demonstrate efficacy in this model (37). To test the therapeutic potential of specific inhibition of the p110δ catalytic subunit, we treated animals with a selective small-molecule inhibitor of p110δ (IC87114) or vehicle before administration of amphetamine (0.75 mg/kg and 1.5 mg/kg). IC87114 specifically antagonizes p110δ over a concentration range of 0.1–10 μM and the IC50 of the compound is 100 nM (38, 39). IC87114 treatment dramatically blocked amphetamine-induced hyperlocomotion, suggestive of antipsychotic potential (Fig. 2A). Moreover, this result was at a dose that had no effect on spontaneous locomotor activity in the absence of amphetamine (Fig. 2A and Inset in Fig. 2A), which is in contrast to that seen with antipsychotic drugs including haloperidol and clozapine that cause marked hypokinesia (15, 40). Blood–brain barrier permeability analysis using lipophilicity and relative hydrophobicity measures revealed that IC87114 readily permeates the blood–brain barrier (Cbrain/Cblood > 2.0).
Fig. 2.
(A) The effect of i.p. injection of the p110δ inhibitor [IC87114] (0.1 mg/kg) 30 min before amphetamine injection (0.75 mg/kg or 1.5 mg/kg) on locomotion in the mouse showed a significant interaction with amphetamine dose and time [treatment × dose × time (F(14, 308) = 3.04; P = 0.0002): solid circle, IC87114 pretreatment, 1.5 mg/kg amphetamine, n = 7; solid triangle, vehicle pretreatment, 1.5 mg/kg amphetamine, n = 7; open triangle, vehicle pretreatment, 0.75 mg/kg amphetamine, n = 7; open circle, IC87114 pretreatment, 0.75 mg/kg amphetamine, n = 7]. Posthoc tests revealed that pretreatment with IC87114 abolished the hyperlocomotor effects of 1.5 mg/kg amphetamine (*P < 0.05 vs. IC87114 pretreatment; #P < 0.05, 1.5 mg/kg amphetamine vs. 0.75 mg/kg amphetamine). No effects of IC87114 were observed on normal baseline locomotor activity (Inset: solid triangle, IC87114, n = 19; open circle, vehicle, n = 18). (B) Acute IC87114 treatment in mouse (0.1 mg/kg) increases phosphorylation of AKT at Thr308 (*P ≤ 0.05). Striatum, n = 4 per group. (C) Effect of pretreatment with IC87114 (0.1 mg/kg) 30 min before apomorphine injection on PPI in the NVHL rat model showed a highly significant interaction with lesion status [Treatment × lesion: F(1, 112) = 13.81, P < 0.00001]. Posthoc tests revealed significant improvement of PPI in NVHL rats treated with IC87114 compared with vehicle [F(1, 56) = 7.3, P < 0.009] and no significant effect of IC87114 on sham-operated rats [F(1, 55) = 7.3, P = 0.085]. Main effects of lesion status (F = 9.33; P = 0.003) and prepulse intensity (F = 20.78; P < 0.00001) were also observed on PPI, whereby PPI progressively increased with higher prepulse intensities and NVHL rats had reduced PPI compared with controls. n = 9–11 rats per group. Mean ± SEM.
Dopamine D2 receptors, the main target of clinically effective antipsychotic drugs (41), modulate activity of the protein kinase AKT (42, 43), which has recently been proposed as a potential mechanism of their therapeutic effects. Antipsychotic drugs antagonize a negative regulator of AKT activity, the D2/β-arrestin 2 complex (44), and result in increased phosphorylation of AKT at the Thr308 residue (45), a site necessary and sufficient to activate AKT. Furthermore, impaired AKT signaling has been reported in the brains and LCLs of patients with schizophrenia (45, 46). Thus, the ability of antipsychotics to increase AKT signaling may be relevant to a molecular mechanism of therapeutic action. The effect of p110δ modulation on AKT activity in vivo in brain has not been previously addressed. We found that acute treatment with IC87114 significantly increased Thr308 phosphorylation in the mouse brain (Fig. 2B). These data suggest that targeted inhibition of p110δ may compensate for the effects of hyperdopaminergia by increasing active levels of AKT, supporting a potential role for p110δ-AKT signaling in the treatment of psychosis, without the untoward side effects of D2 blockade.
Pharmacological Inhibition of p110δ Reverses Prepulse Inhibition (PPI) Deficits in a Rat Neurodevelopmental Model of Schizophrenia.
Given the limitations of pharmacological-based models of neuropsychiatric disorders (37), we further examined the therapeutic potential of IC87114, using a neurodevelopmental animal model of schizophrenia. The rat neonatal ventral hippocampal lesion (NVHL) model is a well-characterized animal model of schizophrenia that is not based on a dopaminergic construct (47, 48) and exhibits postadolescent onset of multiple behavioral and neurobiological phenotypes related to schizophrenia (48, 49). One of the adult-onset schizophrenia-like characteristics of NVHL rats is deficits in sensorimotor gating as evidenced by altered PPI of the acoustic startle response, a phenotype that is exaggerated by the dopamine agonist, apomorphine (49). Here, we confirm that adult NVHL rats have significantly reduced PPI compared with sham-operated controls [Fig. 2C; F(1, 112) = 9.33, P = 0.003]. The effect of i.p. administration of IC87114 (0.1 mg/kg) on PPI showed a highly significant interaction with lesion status [Fig. 2C; treatment * lesion, F(1, 112) = 13.81, P < 0.00001]. Posthoc tests on mean PPI across stimulus strength revealed a highly significant improvement of PPI in NVHL rats [F(1, 56) = 7.3, P < 0.009] and no significant effect on sham-operated rats [F(1, 55) = 7.3, P = 0.085]. These data demonstrate that p110δ inhibition has broad-spectrum antipsychotic potential.
PIK3CD Genetic Association with Schizophrenia.
The evidence that PIK3CD is involved in the biochemistry of NRG1/ERBB4 signaling related to SZ, in the current pharmacological treatment of the condition, and represents a potential treatment target raises the possibility that it may also play a primary etiopathogenic role. We therefore investigated PIK3CD for genetic association with schizophrenia. Association was examined in two independent family-based samples with an affected proband and one case–control dataset (Table S1). We genotyped 19 SNPs spanning a 92.72-kb region [chromosome (chr)1: 9,618,018–9,710,740] encompassing PIK3CD (77.17-kb gene; chr1, 9,634,390–9,711,563). The SNPs comprised 13 tag SNPs from HAPMAP (rel 22/PhaseII) and 6 SNPs selected in potentially functional domains including known promoters, 5′- and 3′-untranslated regions (UTRs), and conserved noncoding sequences. Single-marker analysis in the Clinical Brain Disorders Branch (CBDB) sibling study (CBDB SS) families revealed nominal evidence for association with schizophrenia to SNPs in the PIK3CD 5′-promoter region (rs6540991, P = 0.05; rs6660363, P = 0.04; and rs4601595, P = 0.05), the 3′-intronic region (rs9430220, P = 0.009), and the 3′-UTR (rs1141402, P = 0.021; rs1135427, P = 0.03; and rs12037599, P = 0.05; Table S1). Interestingly, the 5′ and 3′ markers showing association and replication (see below) are in weak linkage disequilibrium (LD) with each other (D′ range, 0.26–0.41; r2 range, 0.04–0.1), suggesting two independent signals within the gene. Next, we sought to replicate the association in two additional datasets. Association to the same alleles was confirmed for rs6540991 (P = 0.001) and rs9430220 (P = 0.05) in the National Institute of Mental Health Genetics Initiative (NIMH-GI) African-American family sample (Table S1). Furthermore, case–control analysis confirmed association of the above SNPs, rs6540991 (P = 0.02) and rs9430220 (P = 0.03) and to the same alleles, in a replication dataset comparing unrelated cases from the SS family data (plus 100 additional cases) to a set of independent unrelated controls (Table S1). Twenty-five SNPs in PIK3R3 were also examined for association (Table S2) and none was observed, supporting our hypothesis that alterations in PIK3R3 in schizophrenia are at least in part secondary or compensatory to a primary PIK3CD and ErbB4 involvement in the disorder that includes a role in its genetic risk architecture.
Discussion
Our data identify a genetically regulated signaling pathway associated with schizophrenia that involves NRG1-ErbB4 and the PI3K enzyme, p110δ and demonstrate that targeted inhibition of p110δ increases AKT Thr308 phosphorylation, blocks the behavioral effects of amphetamine in a mouse pharmacological model of psychosis, and reverses PPI deficits in a neurodevelopmental rat model of schizophrenia, implicating it as a previously undescribed therapeutic target. Our results extend previous observations suggesting that a mechanism behind the genetic association of ErbB4 with schizophrenia involves augmented expression of a PI3K-linked, ErbB4 CYT-1 isoform (5,7), and we show that this change impacts expression and function of a specific downstream PI3-kinase target, p110δ and the biochemical function of the NRG1-ErbB4-PI3K-AKT signaling pathway.
In schizophrenia, impaired NRG1-mediated PI3K signaling {i.e., [PI(3,4,5)P3] production} is observed in the context of elevated levels of ErbB4 CYT-1 and PIK3CD expression. This apparent paradoxical relationship is potentially consistent with data showing that augmented expression of ErbB4 suppresses PI3K-dependent tumor growth (50) and that elevated levels of PIK3CD have opposing effects on PI3K signaling compared with other PI3K isoforms (33, 34), consistent with the suggestion that PIK3CD may function as a tumor suppressor. In keeping with these observations, we show that targeted inhibition of p110δ in brain results in increased AKT regulatory phosphorylation. Furthermore, studies of the ErbB4 interactome suggest that ErbB4 activates fewer pathways than other epidermal growth factor (EGF) family receptors (51) and CYT-1 isoforms exhibit quantitative signaling differences compared with CYT-2, attributable to their high susceptibility to internalization, monoubiquitination, and degradation (52). In schizophrenia, we propose that the increased expression of ErbB4 CYT-1 receptors and the increased fraction of CYT-1 to CYT-2 in the brain, combined with increased PIK3CD, create a molecular shift toward an attenuated NRG1-ErbB4-PI3K signaling system. This proposal is consistent with evidence showing impaired NRG1-mediated phosphorylation of AKT1 in patient-derived cells included in this study (53) and AKT1 dysfunction in schizophrenia (45, 46). It is also noteworthy that reduced ErbB3 gene expression is observed in brain and LCLs of patients with schizophrenia (ref. 29 and this study). ErbB3 has six PI3K docking sites (54) and its reduction likely contributes to the attenuated PI3K signaling observed in the disease. The biological basis of ErbB3 reduction in schizophrenia is at present unclear. We found no evidence of genetic association between the ErbB4 risk haplotype and ErbB3 expression or correlations between expression of the genes, suggesting that changes in ErbB3 are likely secondary to other changes in the NRG1-ErbB4 pathway.
A finding of gain-of-function of NRG1-ErbB4 signaling in postmortem brain tissue of patients with schizophrenia (55) is less readily reconcilable with these data. However, the readout in this earlier study pertained to a different ErbB4 physiology (namely, interaction with PSD-95 and NMDA receptors), and moreover, no genetic analyses were conducted.
We also found evidence of genetic association of PIK3CD with schizophrenia. Out of context, the clinical genetic findings are statistically weak and, although replicable, would not survive agnostic correction for all SNPs in the genome. Nevertheless, the consistent and convergent biological data in peripheral cells and in human brain provide strong prior probability for a role of PIK3CD in schizophrenia and the genetic data should be viewed in this context. In aggregate, the findings implicate NRG1-ErbB4-PI3K signaling as being involved in the predisposition to, and pathophysiology of, schizophrenia; as such, they strengthen the evidence for a role of NRG1 and ErbB4 in the disorder and extend this evidence to a specific subunit of PI3K as a key downstream effector.
Finally, as potential therapeutic targets, NRG1 and ErbB4 are challenging because they serve important and essential functions in normal cell physiology and their pharmacological modulation may have deleterious effects. A more favorable strategy may be to identify downstream targetable molecules with more optimum therapeutic potential. Although we cannot attest that elevated expression of PIK3CD causes the pattern of altered NRG1/ErbB4 signaling in schizophrenia, the proof-of-concept that elevated PIK3CD is a targetable pathogenic component is our findings that IC87114 ameliorates schizophrenia-related neurobehavioral phenotypes in two independent animal models. Although the exact mechanism of therapeutic action of IC87114 in these animal models is at present unclear, the evidence that a p110δ inhibitor increases AKT phosphorylation in brain and has broad antipsychotic potential, combined with independent evidence of PIK3CD association with schizophrenia, suggests that PIK3CD also is involved in schizophrenia (and its treatment) independent of its association with NRG1-ErbB4 signaling. Rationally designed drugs directly targeting p110δ may therefore promise greater efficacy and fewer side effects than current neuroleptics. In summary, our results provide a foundation for future investigations of p110δ inhibition for the treatment of neuropsychiatric disorders.
Materials and Methods
Human Study Populations.
Blood collection and lymphocyte transformation were approved by the NIMH institutional review board, and all donors gave written, informed consent. All subjects were drawn from individuals participating in the CDBD SS, an ongoing investigation of neurobiological abnormalities related to genetic risk for schizophrenia (D.R.W., principal investigator). Thirty-four normal controls (18 females and 16 males, age 32.92 y at the time of blood collection) and 25 individuals with schizophrenia (11 females and 14 males, age 37.6 y,Caucasian of self-reported European ancestry) were used. For information on clinical genetic populations, LCL transformation, culture, RNA extraction, and genotyping see SI Materials and Methods.
Human Brain Tissue Collection, Quality Control, RNA Extraction, and Reverse Transcription.
Postmortem brain tissue was collected at the Clinical Brain Disorders Branch, NIMH, with informed consent from the legal next of kin. Seventy-two normal controls [22 females and 50 males: 46 African-American, 21 American Caucasian, 4 Hispanic, and 1 Asian, mean age 41.5 ± (SD) 15.2 y, postmortem interval (PMI) 30.2 ± 14.1 h, pH 6.59 ± 0.32] and 31 patients with schizophrenia (13 females and 18 males: 18 African-Americans and 11 Caucasians, mean age 48.5 ± 17.7 y, PMI 35.1 ± 17.6 h, pH 6.49 ± 0.24) were available for this study (see ref. 7 for details of collection). See SI Materials and Methods for RNA extraction and reverse transcription.
Quantitative Real-Time RT-PCR.
Gene expression levels were measured by quantitative real-time RT-PCR, using an ABI Prism 7900 sequence detection system with 384-well format (Applied Biosystems) as described previously (7, 18). For detailed information on QPCR and assays see SI Materials and Methods.
Western Blot Analysis of PIK3CD in Human LCLs.
B lymphoblast protein samples were separated by standard gel electrophoresis and probed with the primary antibodies PI3Kinase p110δ (Abcam; ab32401) or SC-348 (Santa Cruz Biotechnology) for ErbB4 analysis. For detailed information see SI Materials and Methods.
Flow Cytometric Analysis of Intracellular [PI(3,4,5)P3] in Human LCLs.
Intracellular staining was used to determine relative [PI(3,4,5)P3] concentrations at the single-cell level, using the Cytofix/Cytoperm kit (BD Biosciences) and FACScan (BD Bioscience). Cells were stimulated with NRG1α (296-HR), a 65-aa residue recombinant protein from the EGF domain (177–241) (100 ng/mL; R&D Systems), or CD19/BCR. For detailed information see SI Materials and Methods.
NCGC00168114 (IC87114) Synthesis.
For detailed information of IC87114 chemical synthesis see SI Materials and Methods.
Rat Haloperidol Treatment Study.
All animal procedures were performed in accordance with the National Institutes of Health guidelines for use and care of laboratory animals. Male Sprague–Dawley rats (weight 250 g) were randomly assigned to drug treatment groups (eight per dose) and administered i.p. injections of haloperidol (0.08 or 0.6 mg/kg/d) or vehicle (0.02% lactic acid) once daily for 28 d. For further information see SI Materials and Methods.
IC87114 Pretreatment in a Mouse Amphetamine Model of Psychosis.
Eight-week-old C57BL/6J male mice were purchased (Jackson Laboratory) and maintained on a 12-h light/dark cycle, with free access to food and water. Testing was conducted at 2–3 mo, during the light phase of the circadian cycle. For examination of the effects of IC87114 pretreatment on baseline locomotor activity and amphetamine-induced hyperlocomotion, mice were tested in an experimental apparatus consisting of four Plexiglas Digiscan automated open fields. IC87114 was dissolved in 0.25% DMSO in physiological saline (vehicle) and injected i.p. (0.1 mg/kg). Vehicle-treated mice were injected with 0.25% DMSO in physiological saline. i.p. injection of d-amphetamine sulfate (0.75 mg/kg or 1.5 mg/kg; Sigma-Aldrich) was administered 30 min following IC87114 pretreatment. For details of testing see SI Materials and Methods.
IC87114 Pretreatment and PPI in the Rat NVHL Model of Schizophrenia.
Timed pregnant female Sprague–Dawley rats were obtained at gestation day 15 (Charles River), individually housed, and maintained in a standard 12-h light/dark schedule. Male pups at 7–8 d of age (P7–8) were subjected to bilateral NVHL or sham procedure (SHAM), as previously described (49). Animals were weaned and subjected to PPI testing at P57–60. Thirty minutes before testing, animals were given injections of either IC87114 (0.1 mg/kg) or vehicle (0.50% DMSO in saline). For detailed information see SI Materials and Methods.
IC87114 and AKT Signaling in Vivo.
Eight-week-old C57BL/6J male mice were purchased (Jackson Laboratory) and maintained on a 12-h light/dark cycle, with free access to food and water. IC87114 (0.1 mg/kg) was dissolved in 0.25% DMSO in physiological saline and administered by i.p. injection. Vehicle-treated mice received an equal volume by i.p. injection of 0.25% DMSO solution in physiological saline. Mice were decapitated after 45–60 min and the brains removed immediately for dissection and protein extraction. Western blot analyses for total AKT1, pAKT-Thr308, and β-Actin were conducted. For detailed information see SI Materials and Methods.
Statistical Analyses.
Multiple linear regression and ANOVA (within SPSS version 15.0) were used for analysis of human biological and animal data. Clinical genetic association was conducted using unconditional logistic regression and the family-based association test (FBAT). For detailed information of statistical analyses see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Ron McKay for his helpful review of the manuscript, Dr. Christopher Austin for helpful discussion and guidance, and William Leister for analytical support. We are thankful for the tireless work of the clinical recruitment and evaluation teams of the Clinical Brain Disorders Branch, National Institute of Mental Health (NIMH), and to the patients who participated in and donated biological material for these studies. Research at the National Institutes of Health (NIH) Chemical Genomics Center is supported by the Molecular Libraries Initiative of the NIH Roadmap for Medical Research and the Intramural Research Program of the National Human Genome Research Institute, NIH. This work was also supported by a Medical Research Council (United Kingdom) Career Development Award held by A.J.L. at the University of Oxford and by funds from the Intramural Research Program of the NIMH, NIH to the Clinical Brain Disorders Branch.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 11902.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206118109/-/DCSupplemental.
References
- 1.Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: On the matter of their convergence. Mol Psychiatry. 2005;10:40–68, 5. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- 2.Stefansson H, et al. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison PJ, Law AJ. Neuregulin 1 and schizophrenia: Genetics, gene expression, and neurobiology. Biol Psychiatry. 2006;60:132–140. doi: 10.1016/j.biopsych.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Allen NC, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: The SzGene database. Nat Genet. 2008;40:827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- 5.Silberberg G, Darvasi A, Pinkas-Kramarski R, Navon R. The involvement of ErbB4 with schizophrenia: Association and expression studies. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:142–148. doi: 10.1002/ajmg.b.30275. [DOI] [PubMed] [Google Scholar]
- 6.Nicodemus KK, et al. Further evidence for association between ErbB4 and schizophrenia and influence on cognitive intermediate phenotypes in healthy controls. Mol Psychiatry. 2006;11:1062–1065. doi: 10.1038/sj.mp.4001878. [DOI] [PubMed] [Google Scholar]
- 7.Law AJ, Kleinman JE, Weinberger DR, Weickert CS. Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Hum Mol Genet. 2007;16:129–141. doi: 10.1093/hmg/ddl449. [DOI] [PubMed] [Google Scholar]
- 8.Walsh T, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 9.Shi J, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fazzari P, et al. Control of cortical GABA circuitry development by Nrg1 and ErbB4 signalling. Nature. 2010;464:1376–1380. doi: 10.1038/nature08928. [DOI] [PubMed] [Google Scholar]
- 11.Li B, Woo RS, Mei L, Malinow R. The neuregulin-1 receptor erbB4 controls glutamatergic synapse maturation and plasticity. Neuron. 2007;54:583–597. doi: 10.1016/j.neuron.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golding JP, Trainor P, Krumlauf R, Gassmann M. Defects in pathfinding by cranial neural crest cells in mice lacking the neuregulin receptor ErbB4. Nat Cell Biol. 2000;2:103–109. doi: 10.1038/35000058. [DOI] [PubMed] [Google Scholar]
- 13.Roy K, et al. Loss of erbB signaling in oligodendrocytes alters myelin and dopaminergic function, a potential mechanism for neuropsychiatric disorders. Proc Natl Acad Sci USA. 2007;104:8131–8136. doi: 10.1073/pnas.0702157104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen YJ, et al. Type III neuregulin-1 is required for normal sensorimotor gating, memory-related behaviors, and corticostriatal circuit components. J Neurosci. 2008;28:6872–6883. doi: 10.1523/JNEUROSCI.1815-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, Lipska BK, Weinberger DR. Genetic mouse models of schizophrenia: From hypothesis-based to susceptibility gene-based models. Biol Psychiatry. 2006;59:1180–1188. doi: 10.1016/j.biopsych.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 16.Hall J, et al. A neuregulin 1 variant associated with abnormal cortical function and psychotic symptoms. Nat Neurosci. 2006;9:1477–1478. doi: 10.1038/nn1795. [DOI] [PubMed] [Google Scholar]
- 17.Konrad A, et al. ErbB4 genotype predicts left frontotemporal structural connectivity in human brain. Neuropsychopharmacology. 2009;34:641–650. doi: 10.1038/npp.2008.112. [DOI] [PubMed] [Google Scholar]
- 18.Law AJ, et al. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5′ SNPs associated with the disease. Proc Natl Acad Sci USA. 2006;103:6747–6752. doi: 10.1073/pnas.0602002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brennand KJ, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan W, et al. Molecular cloning of a brain-specific, developmentally regulated neuregulin 1 (NRG1) isoform and identification of a functional promoter variant associated with schizophrenia. J Biol Chem. 2007;282:24343–24351. doi: 10.1074/jbc.M702953200. [DOI] [PubMed] [Google Scholar]
- 21.Elenius K, et al. A novel juxtamembrane domain isoform of HER4/ErbB4. Isoform-specific tissue distribution and differential processing in response to phorbol ester. J Biol Chem. 1997;272:26761–26768. doi: 10.1074/jbc.272.42.26761. [DOI] [PubMed] [Google Scholar]
- 22.Junttila TT, Sundvall M, Määttä JA, Elenius K. Erbb4 and its isoforms: Selective regulation of growth factor responses by naturally occurring receptor variants. Trends Cardiovasc Med. 2000;10:304–310. doi: 10.1016/s1050-1738(01)00065-2. [DOI] [PubMed] [Google Scholar]
- 23.Kainulainen V, et al. A natural ErbB4 isoform that does not activate phosphoinositide 3-kinase mediates proliferation but not survival or chemotaxis. J Biol Chem. 2000;275:8641–8649. doi: 10.1074/jbc.275.12.8641. [DOI] [PubMed] [Google Scholar]
- 24.Krivosheya D, et al. ErbB4-neuregulin signaling modulates synapse development and dendritic arborization through distinct mechanisms. J Biol Chem. 2008;283:32944–32956. doi: 10.1074/jbc.M800073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geering B, Cutillas PR, Nock G, Gharbi SI, Vanhaesebroeck B. Class IA phosphoinositide 3-kinases are obligate p85-p110 heterodimers. Proc Natl Acad Sci USA. 2007;104:7809–7814. doi: 10.1073/pnas.0700373104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 27.Wessel J, Zapala MA, Schork NJ. Accommodating pathway information in expression quantitative trait locus analysis. Genomics. 2007;90:132–142. doi: 10.1016/j.ygeno.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Torkamani A, Schork NJ. Pathway and network analysis with high-density allelic association data. Methods Mol Biol. 2009;563:289–301. doi: 10.1007/978-1-60761-175-2_16. [DOI] [PubMed] [Google Scholar]
- 29.Aston C, Jiang L, Sokolov BP. Microarray analysis of postmortem temporal cortex from patients with schizophrenia. J Neurosci Res. 2004;77:858–866. doi: 10.1002/jnr.20208. [DOI] [PubMed] [Google Scholar]
- 30.Vanhaesebroeck B, et al. P110delta, a novel phosphoinositide 3-kinase in leukocytes. Proc Natl Acad Sci USA. 1997;94:4330–4335. doi: 10.1073/pnas.94.9.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eickholt BJ, et al. Control of axonal growth and regeneration of sensory neurons by the p110delta PI 3-kinase. PLoS ONE. 2007;2:e869. doi: 10.1371/journal.pone.0000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okkenhaug K, Vanhaesebroeck B. New responsibilities for the PI3K regulatory subunit p85 alpha. Sci STKE. 2001;2001:pe1. doi: 10.1126/stke.2001.65.pe1. [DOI] [PubMed] [Google Scholar]
- 33.Fransson S, Martinsson T, Ejeskär K. Neuroblastoma tumors with favorable and unfavorable outcomes: Significant differences in mRNA expression of genes mapped at 1p36.2. Genes Chromosomes Cancer. 2007;46:45–52. doi: 10.1002/gcc.20387. [DOI] [PubMed] [Google Scholar]
- 34.Carén H, Fransson S, Ejeskär K, Kogner P, Martinsson T. Genetic and epigenetic changes in the common 1p36 deletion in neuroblastoma tumours. Br J Cancer. 2007;97:1416–1424. doi: 10.1038/sj.bjc.6604032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bilancio A, et al. Key role of the p110delta isoform of PI3K in B-cell antigen and IL-4 receptor signaling: Comparative analysis of genetic and pharmacologic interference with p110delta function in B cells. Blood. 2006;107:642–650. doi: 10.1182/blood-2005-07-3041. [DOI] [PubMed] [Google Scholar]
- 36.Ge B, et al. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat Genet. 2009;41:1216–1222. doi: 10.1038/ng.473. [DOI] [PubMed] [Google Scholar]
- 37.Nestler EJ, Hyman SE. Animal models of neuropsychiatric disorders. Nat Neurosci. 2010;13:1161–1169. doi: 10.1038/nn.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sadhu C, Dick K, Tino WT, Staunton DE. Selective role of PI3K delta in neutrophil inflammatory responses. Biochem Biophys Res Commun. 2003;308:764–769. doi: 10.1016/s0006-291x(03)01480-3. [DOI] [PubMed] [Google Scholar]
- 39.Knight ZA, et al. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogren SO, Goldstein M. Phencyclidine- and dizocilpine-induced hyperlocomotion are differentially mediated. Neuropsychopharmacology. 1994;11:167–177. doi: 10.1038/sj.npp.1380103. [DOI] [PubMed] [Google Scholar]
- 41.Beaulieu JM, Gainetdinov RR. The physiology, signaling, and pharmacology of dopamine receptors. Pharmacol Rev. 2011;63:182–217. doi: 10.1124/pr.110.002642. [DOI] [PubMed] [Google Scholar]
- 42.Beaulieu JM, et al. Regulation of Akt signaling by D2 and D3 dopamine receptors in vivo. J Neurosci. 2007;27:881–885. doi: 10.1523/JNEUROSCI.5074-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beaulieu JM, et al. An Akt/beta-arrestin 2/PP2A signaling complex mediates dopaminergic neurotransmission and behavior. Cell. 2005;122:261–273. doi: 10.1016/j.cell.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 44.Masri B, et al. Antagonism of dopamine D2 receptor/beta-arrestin 2 interaction is a common property of clinically effective antipsychotics. Proc Natl Acad Sci USA. 2008;105:13656–13661. doi: 10.1073/pnas.0803522105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emamian ES, Hall D, Birnbaum MJ, Karayiorgou M, Gogos JA. Convergent evidence for impaired AKT1-GSK3beta signaling in schizophrenia. Nat Genet. 2004;36:131–137. doi: 10.1038/ng1296. [DOI] [PubMed] [Google Scholar]
- 46.Tan HY, et al. Genetic variation in AKT1 is linked to dopamine-associated prefrontal cortical structure and function in humans. J Clin Invest. 2008;118:2200–2208. doi: 10.1172/JCI34725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lipska BK, Weinberger DR. To model a psychiatric disorder in animals: Schizophrenia as a reality test. Neuropsychopharmacology. 2000;23:223–239. doi: 10.1016/S0893-133X(00)00137-8. [DOI] [PubMed] [Google Scholar]
- 48.Lipska BK, Weinberger DR. Genetic variation in vulnerability to the behavioral effects of neonatal hippocampal damage in rats. Proc Natl Acad Sci USA. 1995;92:8906–8910. doi: 10.1073/pnas.92.19.8906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lipska BK, et al. Neonatal excitotoxic hippocampal damage in rats causes post-pubertal changes in prepulse inhibition of startle and its disruption by apomorphine. Psychopharmacology (Berl) 1995;122:35–43. doi: 10.1007/BF02246439. [DOI] [PubMed] [Google Scholar]
- 50.Tovey SM, Dunne B, Witton CJ, Cooke TG, Bartlett JM. HER4 in breast cancer: Comparison of antibodies against intra- and extra-cellular domains of HER4. Breast Cancer Res. 2006;8:R19. doi: 10.1186/bcr1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaushansky A, et al. System-wide investigation of ErbB4 reveals 19 sites of Tyr phosphorylation that are unusually selective in their recruitment properties. Chem Biol. 2008;15:808–817. doi: 10.1016/j.chembiol.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sundvall M, et al. Isoform-specific monoubiquitination, endocytosis, and degradation of alternatively spliced ErbB4 isoforms. Proc Natl Acad Sci USA. 2008;105:4162–4167. doi: 10.1073/pnas.0708333105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sei Y, et al. Neuregulin1-induced cell migration is impaired in schizophrenia: Association with neuregulin1 and catechol-o-methyltransferase gene polymorphisms. Mol Psychiatry. 2007;12:946–957. doi: 10.1038/sj.mp.4001994. [DOI] [PubMed] [Google Scholar]
- 54.Schulze WX, Deng L, Mann M. Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol Syst Biol. 2005;1:2005–.0008. doi: 10.1038/msb4100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hahn CG, et al. Altered neuregulin 1-erbB4 signaling contributes to NMDA receptor hypofunction in schizophrenia. Nat Med. 2006;12:824–828. doi: 10.1038/nm1418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.