Summary
Background
The centrosome is the major microtubule organizing center (MTOC) in dividing cells and in many post-mitotic, differentiated cells. In other cell types, however, MTOC function is reassigned from the centrosome to non-centrosomal sites. Here, we analyze how MTOC function is reassigned to the apical membrane of C. elegans intestinal cells.
Results
After the terminal intestinal cell division, the centrosomes and nuclei move near the future apical membranes, and the postmitotic centrosomes lose all, or nearly all, of their associated microtubules. We show that microtubule-nucleating proteins such as γ-tubulin and CeGrip-1 that are centrosome components in dividing cells become localized to the apical membrane, which becomes highly enriched in microtubules. Our results suggest that centrosomes are critical to specify the apical membrane as the new MTOC. First, γ-tubulin appears to redistribute directly from the migrating centrosome onto the lateral, then apical membrane. Second, γ-tubulin fails to accumulate apically in wild-type cells following laser ablation of the centrosome. We show that centrosomes localize apically by first moving toward lateral foci of the conserved polarity proteins PAR-3 and PAR-6, and then move together with these foci toward the future apical surface. Embryos lacking PAR-3 fail to localize their centrosomes apically, and have aberrant localization of γ-tubulin and CeGrip-1.
Conclusions
These data suggest that PAR proteins contribute to apical polarity in part by determining centrosome position and that the reassignment of MTOC function from centrosomes to the apical membrane is associated with a physical hand-off of nucleators of microtubule assembly.
Introduction
Microtubules are critical regulators of cell shape, polarity and transport and must be spatially organized to fulfill these distinct functions. In dividing animal cells, centrosomes serve as the major microtubule organizing center (MTOC), nucleating and coordinating microtubules into a radial array. The centrosome is a non-membrane bound organelle composed of two centrioles that are surrounded by a cloud of pericentriolar material (PCM). Microtubule minus ends are nucleated from PCM components including γ-tubulin and γ-tubulin ring complex proteins (γ-TuRCs) such as CeGrip-1/dgrip91/Spc98p [1].
The centrosome often remains the major MTOC in post-mitotic, differentiated cells that have simple, radial arrays of microtubules. However, many types of polarized cells, such as neurons [2,3], syncytial myotubes [4], and epithelia [5,6], have more complex arrangements of microtubules that appear to be organized by non-centrosomal MTOCs. In some cases, such as in Drosophila tracheal cells [7], C. elegans germ cells [8], and Xenopus epidermal cells [9], these non-centrosomal MTOCs contain the microtubule nucleator γ-tubulin and members of the γ-TuRC, and thus might nucleate microtubules like centrosomes in dividing cells. In contrast, non-centrosomal MTOCs might instead capture microtubules produced elsewhere [10,11]. For example, the non-centrosomal microtubules in neurons [12] and cochlear cells [13] are not associated with γ-tubulin and are thought to be released from the centrosome.
Numerous studies have focused on how centrosomes function as MTOCs in dividing cells, but comparatively little is known about the composition or specification of non-centrosomal MTOCs. Here, we use the C. elegans intestine as a model to study how MTOC function is reassigned from the centrosome to the apical surface of an epithelial cell. We show that centrosomes traffic to the apical surface along with the conserved polarity proteins PAR-3 and PAR-6. The microtubule nucleators γ-tubulin and CeGrip-1 appear to be handed-off from the centrosome as a new, non-centrosome based MTOC is established. Our mutant analysis and laser ablation studies show that both the centrosome and PAR-3 are critical for the transition in MTOC function to the apical membrane.
Results
Developing intestinal cells specify an apical MTOC
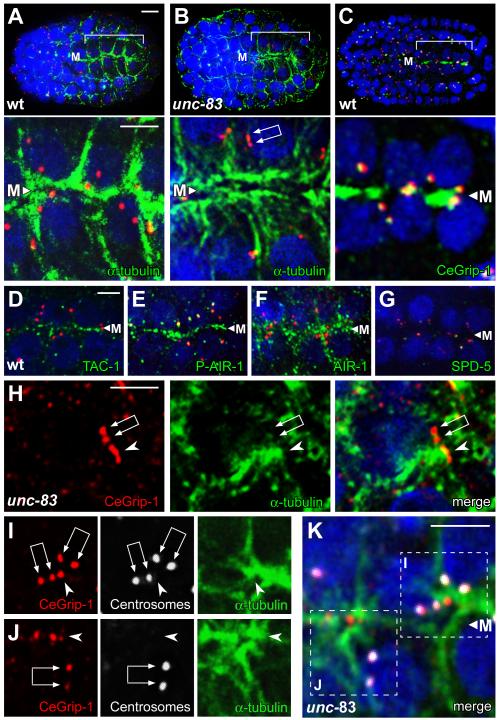
The C. elegans intestine arises clonally from an early embryonic cell called the E blastomere. The centrosome functions as the MTOC during the divisions of E and its descendants, and contains high levels of γ-tubulin and other PCM proteins such as CeGrip-1 [1], AIR-1/Aurora-A [14], ZYG-9/XMAP-215 [15], TAC-1/TACC3 [15], and SPD-5 [16] (Figure 1A, 1D, and data not shown). After four cell cycles, 12 of the 16 E descendants cease dividing, although the centrosome undergoes one additional duplication or splitting to form a centrosome pair in all the E16 cells (Figure 1B). These cells group together to form the E16 primordium, which resembles a cylinder elongated along a central anterior/posterior axis called the midline. During cell polarization, the nucleus and centrosome pair migrate from a position adjacent to the lateral membrane to the midline-facing surface, which differentiates as the apical membrane (Figure 1C; Figure S1A and S1B; [17]).
Figure 1.
Centrosomes and γ-tubulin localization during MTOC reassignment
(A-C) Images from live recording of the E16 primordium, showing sister E16 cells from birth through polarization. Reporters show γ-tubulin (green, γ-tubulin:GFP), nuclei (red, histone:mCherry) and membranes (red, membrane:Cherry). Cells are oriented with respect to the midline (M) as diagrammed in the cartoons beneath each panel. Note the transition from round to columnar cell morphology associated with polarization. (D-F) Electron micrographs of centrosomes from stages corresponding to panels A, B, and C, respectively. Note the decrease in diameter of the PCM (indicated with purple arrows in panels D and E) as the nucleus (“nuc”) and centrosome localizes near the lateral membrane (L), and the apparent absence of PCM as they move near the midline in panel F. Green arrowheads indicate examples of microtubules contacting the PCM. Scale bar (A-C, 5μm, D-F, 200nm). See also Figure S1 and Movie S1.
Using electron microscopy, we found that centrosomes in the E16 primordium lose most of their PCM and associated microtubules as they reposition from lateral positions toward the future apical surface (Figures 1D-1F). In contrast, numerous microtubules appear near, or on, the apical plasma membrane during centrosomal repositioning (Figure 2A; Figure S2A and S2B). Immunostaining and live imaging experiments showed that many PCM proteins become highly enriched at the apical membrane of E16 cells, including γ-tubulin [18], CeGrip-1, TAC-1/TACC3, AIR-1/Aurora-A, and ZYG-9/XMAP-215 (Figure 1C and Figure 2C-2F; data not shown). The PCM protein SPD-5, which is required for centrosome maturation and localization of PCM proteins to the centrosome [16], appears to remain solely at the centrosome, albeit at a severely reduced level (Figure 2G). Thus, MTOC function appears to be reassigned from the centrosome to the apical surface sometime during centrosome repositioning, and the apical MTOC is associated with many, but not all, PCM components.
Figure 2.
Generation of the midline/apical MTOC
(A-K) All panels are optical sections through the midline (M) of an E16 primordium, immunostained as indicated; nuclei are stained with DAPI (blue). Centrosome pairs appear as two closely paired dots with variable spacing (double arrows in panels B, H, I, and J), or are unresolved as a larger dot. The upper panels in A-C show low magnification views of the entire embryo with the primordium bracketed, and the lower panels show high magnification views of same E16 cells; centrosomes are stained with IFA1 (red). In the unc-83 mutant (B), note the microtubules extending between the apical membrane and the nucleus. (C-F) The PCM proteins CeGrip-1, TAC-1, and AIR-1 are enriched at the midline/future apical surface. Although the kinase AIR-1 primarily co-localizes with microtubules during this stage, the phosphorylated, active form of AIR-1 is restricted to the apical surface and centrosomes (compare E and F; [30]). Panels H-J show examples of plumes (arrowheads) of CeGrip-1 near the centrosome pair; panels I and J are color separated images of the merged, boxed regions in K. Note the concentration of α-tubulin by the plume of CeGrip-1, and the relative absence of α-tubulin by the centrosome pair. Scale bar = 5μm (A-C), 2.5μm (D-K and insets in A-C). See also Figure S2, Figure S3, and Movie S2.
The apparent inactivation of the MTOC function of the centrosome, and the formation of a new apical membrane MTOC, could be independent events. However, the observation that PCM proteins are lost from centrosomes at about the time many of the same proteins appear at the apical membrane suggests centrosomes might have a role in MTOC reassignment. The apical position of the centrosome is itself not essential for intestinal cell function, as centrosomes do not remain apical in later stages of larval development. Moreover, mutants defective in the nuclear envelope protein UNC-83 are viable, but do not show the normal, close association of the centrosome with the apical membrane during embryogenesis [19]. Indeed, we found that the intestinal apical membrane in unc-83 mutants appears to have wild-type levels of γ-tubulin and CeGrip-1 (see below and Figure S3C, n>100), and is associated with large numbers of microtubules (Figure 2B; Figure S3A). Similarly, we observed defects in centrosome positioning, but an apparently normal apical MTOC, in mutants lacking expression of the dynein subunit DYRB-1/roadblock [20] (Figure S3B, S3D, S3F, γ-tubulin, n=125/125, CeGrip-1, n=85/86).
Plumes of microtubule nucleators originate near centrosomes during MTOC reassignment
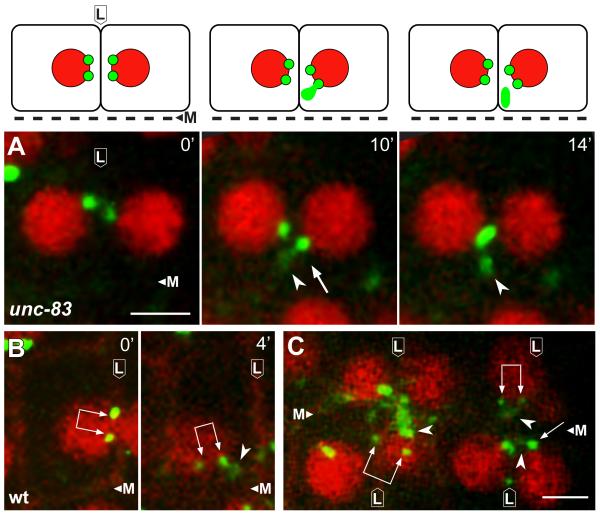
In examining the E16 primordium in fixed, immunostained unc-83 mutants, we noticed that some cells that had not yet accumulated apical proteins contained a prominent “plume” of CeGrip-1 (arrowhead in Figure 2H) that was near, but not coincident with, CeGrip-1 at the centrosome pair (double arrow in Figure 2H). Moreover, the plume of non-centrosomal CeGrip-1 appeared to be associated with higher levels of α-tubulin than did the nearby centrosomes (Figure 2H-2K). To analyze the behavior of other PCM proteins during the transition, we imaged γ-tubulin:GFP in live wild-type embryos and in unc-83 mutants (Figure 1A-1C; Movies S1-S3). These recordings showed that the level of γ-tubulin:GFP at the centrosome varies markedly in the dividing intestinal precursors, rising to peak intensity during mitosis (Figure 1A) when electron microscopy shows a large accumulation of PCM (Figure 1D). The centrosome and nucleus move toward the lateral membrane after mitosis (lateral migration; Figure 1B), and remain tightly apposed to this membrane for several minutes before continuing along the membrane toward the future apical membrane. The initially bright γ-tubulin:GFP signal at the centrosome diminishes after lateral migration (Figure 1B), as does the size of the PCM visible by electron microscopy (Figure 1E), and the GFP signal can often be resolved as two separate foci; these foci correspond to the centrosome pair.
The formation and lateral migration of the centrosome pair appears to occur normally in unc-83 mutants but apical migration is incomplete (Movie S2); centrosomes and nuclei initially approach the intersection of the lateral and apical surfaces, but fail to continue to the apical membrane. A plume of non-centrosomal γ-tubulin:GFP appears near the centrosome pair as it migrates from a lateral to apical position (Figure 3A; Movie S3), similar to the immunostained images of CeGrip1 in fixed embryos (Figure 2H-2K). Remarkably, live imaging showed that the plume of γ-tubulin:GFP appeared to originate directly from the paired centrosomes. We observed a similar plume of γ-tubulin:GFP near the centrosome pair in wild-type embryos and dyrb-1/roadblock mutants (Figure 3B and 3C; Movie S1 and data not shown). The plume was visible in approximately 83% of wild-type cells (n=66), however, the proximity of the centrosome pair to the membrane often made the origin of the plume difficult to resolve. We conclude that the PCM proteins CeGrip-1 and γ-tubulin localize to a transitional body, the plume, before accumulating at the future apical surface.
Figure 3.
A plume of γ-tubulin appears to emanate from the centrosome during MTOC reassignment
Panels are from live recordings of the E16 primordium in wild-type or unc-83 mutant embryos; lateral membranes (L) and midline (M) are indicated. Reporters show γ-tubulin (green, γ-tubulin:GFP), nuclei (red, histone:mCherry), and membranes (B and C, red, membrane:Cherry). Centrosome pairs are indicated by single or double arrows, depending on whether one or both centrosomes is visible in the frame. Note plumes of γ-tubulin (arrowheads) associated with centrosomes. Images in A are diagrammed above each panel. Scale bar (2.5 μm). See also Movie S3.
PAR-3 is required for proper centrosome positioning and apical γ-tubulin accumulation
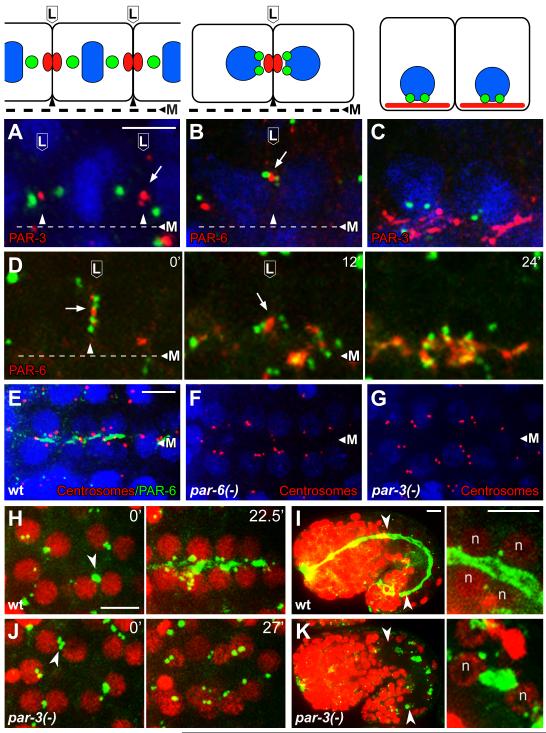
The migration of the centrosome pair to the lateral membrane following the E8 to E16 division is reminiscent of centrosome behavior in a subset of ascidian cells that divide asymmetrically during development. The ascidian centrosomes migrate toward a membrane-associated Centrosome Attracting Body that has been shown to contain members of the conserved PAR polarity complex, PAR-3, PAR-6, and aPKC [21]. In C. elegans, foci of PAR-3 and PAR-6 have been reported on or near the lateral membranes of E16 intestinal cells before these proteins localize apically (Figure 4A-4C, [22,23]). We found that the lateral migration of the centrosomes is oriented toward the PAR foci (Figure 4A, 4B, and 4D).
Figure 4.
PAR proteins in centrosome positioning and γ-tubulin localization
(A-C) Intestinal primordial cells in immunostained embryos at (A) metaphase of the E8 to E16 division, (B) shortly after division when paired centrosomes migrate to lateral membranes, and (C) during apical polarization; cartoons of the cells are shown in the upper panels. The images show centrosomes (green; SPD-5 in panel A and IFA1 in, B, C), and PAR-3 or PAR-6 as indicated; nuclei are stained with DAPI (blue). Note that the paired centrosomes in panel B have moved toward the lateral focus of PAR-6. (D) Image sequence from live E16 cells beginning at a stage similar to panel B and showing centrosomes (green, γ-tubulin:GFP) and PAR-6 (red, PAR-6:Cherry); time in minutes at upper right. Note that the centrosome pairs in both cells move apically with the focus of PAR-6, and that PAR-6 spreads after reaching the apical surface. (E-G) Images of the E16 primordium in immunostained wild-type embryos and embryos depleted of both maternal and zygotic PAR-6 or PAR-3 (n=25 and 27 embryos, respectively). Note that most of the paired centrosomes (red, IFA) are apical after depletion of PAR-6 but not after depletion of PAR-3. (H-K) γ-tubulin (green, γ-tubulin:GFP) and nuclei (red, histone:Cherry, ‘n’) in wild type embryos (H,I) and embryos depleted of PAR-3 (J,K). Note the failure of centrosomes and γ-tubulin to localize apically in the PAR-3-depleted embryo. The left panels in I and K show low magnifications of entire embryos during later morphogenesis, with the intestines indicated by arrowheads; the right panels show high magnification views of some of the intestinal cells. γ-tubulin is apical in the wild-type embryo, but ectopic in the PAR-3-depleted embryo. Scale bar (A-C, 2.5μm, E-K, 5μm). See also Figure S4, Movie S4, and Movie S5.
The plume of γ-tubulin:GFP initially appears to bridge the centrosome pair with the PAR foci on the lateral membrane, then separates from the centrosomes and co-localizes with the PAR foci (Movie S4). Centrosomes, the plume of γ-tubulin:GFP, and the PAR foci move together toward the midline (Figure 4D; Movie S4). At the midline, γ-tubulin:GFP and the PAR proteins both spread across the apical surface and increase in intensity (Figure 1C, Figure 4C and 4D). These results suggest that the apical reassignment of MTOC function involves an intermediate stage where PCM proteins such as γ-tubulin and CeGrip-1 are handed off to a membrane-associated complex that includes PAR proteins.
To test possible roles for the PAR proteins in centrosome repositioning and MTOC reassignment, we used a stage-specific targeted degradation strategy to deplete PAR proteins after completion of their essential roles in early embryogenesis, as previously described [24]. Briefly, a homozygous par mutant is rescued transiently by the corresponding PAR protein, which is fused to a peptide tag that mediates stage-specific degradation (see Experimental Procedures). We found that PAR-6 appears dispensable for centrosome positioning and apical localization of CeGrip-1, however, the CeGrip-1 appears more diffusely apical in par-6 mutants than in wild-type embryos (Figure 4E, 4F; Figure S4A-4B). In contrast, PAR-3 is essential for centrosome positioning and apical γ-tubulin localization: Wild-type embryos show apically positioned centrosomes and apical γ-tubulin within 30 minutes after the E8 to E16 division (Figure 4H). In par-3(-) embryos, centrosomes migrate to the lateral membranes, as in wild-type embryos (arrowhead, Figure 4J, t=0′). However, the centrosomes then appear to distribute randomly, and after 30 minutes the embryos show no apical γ-tubulin at 30 minutes (Figure 4G, Figure 4J, t=27′; Movie S5). Later embryos develop large patches of γ-tubulin and CeGrip-1 at aberrant locations in intestinal cells (Figure 4K, n=40; Figure S4D); in the few cases where the genesis of a γ-tubulin patch could be determined, it originated at or near the centrosome (data not shown). Thus, centrosome mispositioning in the absence of PAR-3 is associated with improper localization of γ-tubulin and CeGrip-1. These results support earlier findings that PAR-3, but not PAR-6 is required for some aspects of epithelial cell polarization [22,23].
Microtubules are required for centrosome positioning and to promote efficient apical differentiation
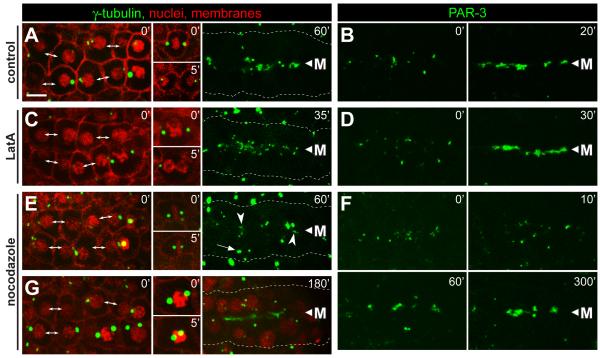
We used the microtubule inhibitor nocodazole and the microfilament inhibitor latrunculin A (LatA) to test whether the cytoskeleton was required for apical localization of γ-tubulin and PAR-3. Embryos were selected for analysis in which the majority of the E8 cells had just completed the division to E16. In control, mock treated embryos, γ-tubulin and PAR-3 localized to the midline/apical surface within 20 to 60 minutes (Figure 5A and 5B, n=3). LatA treated embryos showed a similar localization of both γ-tubulin (Figure 5C, n=3) and PAR-3 (Figure 5D, n=4) in cells that had completed the E8 to E16 division. In contrast to the LatA results, E16 cells treated with nocodazole showed a strong delay in the apical localization of both γ-tubulin (Figure 5E, 5G, n=9; Movie S6) and PAR-3 (Figure 5F, n=7). In addition, nocodazole inhibited the migration of centrosomes and nuclei to the apical surface (Figure 5G, n=9; Movie S6).
Figure 5.
Inhibition of microfilaments and microtubules during epithelial polarization.
Each column shows examples of live, E16 primordia from wild-type embryos expressing the reporters indicated at top; the reporters used are as in Figure 1. Embryos were either mock treated (control), or exposed to latrunculin (LatA, 10μM) or nocodazole (10μg/mL) as indicated. The left panels for each column show the primordium immediately after exposure to the inhibitor. At the stage selected for γ-tubulin analysis, the primordium includes newly separated sister cells (double-headed arrows), as well as some cells that have not finished the E8 to E16 division; these latter cells arrest division (A,E,G) or fail cytokinesis (C) after treatment with inhibitor (inset panels). In the stage selected for PAR-3 analysis, PAR-3 foci were visible on the lateral membranes of E16 cells (left panels, t=0). (A-D) In control embryos and LatA-treated embryos, centrosomes, γ-tubulin and PAR-3 show robust apical localization before 60 minutes (see also timed sequence in Figure 4H). In LatA-treated cells that failed cytokinesis, γ-tubulin did not localize to the apical surface within the same time period (data not shown). (E-G) Embryos treated with nocodazole show only irregular apical localization by 60 minutes, but much better localization of γ-tubulin and PAR-3 by 180 minutes and 200 minutes, respectively. Centrosomes (E, 60′ arrow, G) and nuclei (G) fail to move apically following nocodazole treatment. Note the spreading of PAR-3 foci away from the midline 10 minutes after nocodazole treatment, suggesting that these foci normally traffic toward the midline on microtubules. Scale bar = 5μm. See also Movie S6.
The centrosome is required for apical accumulation of γ-tubulin
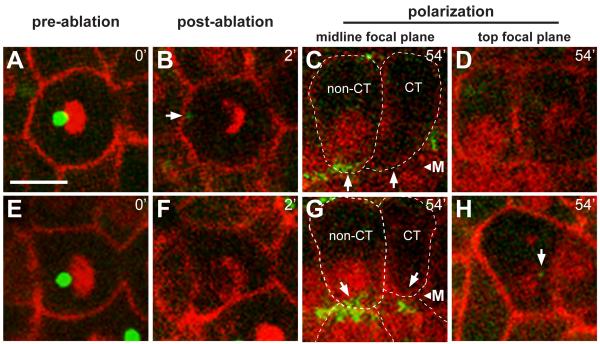
To test whether the centrosome is required in apical differentiation, we used a laser microbeam to ablate centrosomes in individual intestinal cells (Figure 6A and 6E). γ-tubulin:GFP was used to identify the centrosome, and to monitor subsequent apical differentiation; at the time of ablation only about 3.7% of the total cytoplasmic γ-tubulin:GFP is concentrated at the centrosome (see Experimental Procedures). The laser appeared to destroy centrosomes effectively: (1) The ablated centrosomes showed a disintegration of PCM, as observed in other systems [25], with little (4/19 experiments) or no (15/19 experiments) recovery of γ-tubulin (Figure 6B), (2) centrosome ablation at metaphase caused an apparent cell cycle arrest with a monopolar spindle (Movie S7), and (3) cells neighboring the targeted cell continued to divide and/or differentiate normally. We ablated centrosomes at telophase of the E8 to E16 division, and imaged cells until neighboring control cells showed apical γ-tubulin accumulation. The level of γ-tubulin was then measured in both types of cells. In all experiments (n=19), apical γ-tubulin was not detectable in cells following centrosome ablation (Figure 6C and 6G). Moreover, nuclear migration was defective, suggesting that centrosomes are essential for the normal, apical localization of intestinal nuclei (Figure 6C, 6D, 6G, and 6H).
Figure 6.
Centrosomes are required for accumulation of γ-tubulin at the apical surface
Live images of two wild type embryos (A-D and E-H) before (t=0) and after (t=2 minutes) laser ablation of the centrosome, and again after 54 minutes; reporters as in Figure 1. γ-tubulin normally accumulates at the midline before 1 hour, as is visible in the non-centrosome ablated (non-CT) cells, but has not accumulated in the cells with the ablated centrosomes (CT). Note that the nuclei in CT cells have not localized to the midline, but are instead at different focal planes (top focal plane, D and H). The arrow in panel H indicates a non-apical, small cytoplasmic focus of γ-tubulin that likely corresponds to a fragment of the ablated centrosome. CT cells have levels of cytoplasmic γ-tubulin that are comparable to non-CT cells, but lack apical accumulation; the apical surface of CT cells showed a statistically insignificant enrichment of γ-tubulin (−.0018±.012, n=19) compared to non-CT cells (.2081±.082, n=38, two tailed t-test; p ≤ 1.83×10-17). Scale bar = 5μm. See also Movie S7.
Discussion
Following the E8 to E16 division, C. elegans intestinal cells transition from a centrosomal MTOC with a radial array of microtubules to an apical membrane MTOC with a linear array. The transition occurs rapidly, with apical enrichment of microtubules and the loss of centrosomal microtubules evident within 30 minutes. The speed and reproducible timing of these events makes the intestine an attractive model to study the MTOC transition. Using electron microscopy and immunostaining for specific PCM components, we showed that most of the PCM is lost from the centrosome during this transition. At about the same time, some PCM proteins, including the microtubule nucleators γ-tubulin and CeGrip-1, appear near membranes. Remarkably, these nucleators appear to be stripped from the centrosome in the form of a cytoplasmic plume, likely causing or contributing to the loss of MTOC function at the centrosome. In contrast, the cytoplasmic plume is associated with microtubules, suggesting that it either retains MTOC function, or maintains the linkage with existing microtubules.
Microtubule nucleators in the plume do not transfer directly to the apical membrane, and instead first localize by the lateral membrane near foci of PAR-3 and PAR-6; the plume appears to connect the centrosome pair with these foci, and all move in concert toward the midline or future apical membrane. The nucleators do not localize apically in par-3(-) mutants, suggesting that the foci might be critical for movement. The asymmetric movement of PAR-3 is a hallmark of polarity in 1-cell C. elegans embryos, and driven by a PAR-3-modulated contraction of the actomyosin cytoskeleton [26]. In contrast, E16 intestinal cells treated with the microfilament inhibitor LatA show apparently normal apical localization of PAR-3 and γ-tubulin. Treatment with microtubule inhibitors greatly delayed the apical localization, suggesting that microtubules in the plume might contribute to movement. We do not know whether the slow apical localization observed in these cells occurs through a few, inhibitor-resistant microtubules, or through a microtubule-independent process.
After arriving at the future apical surface, γ-tubulin and other former PCM components appear to spread across the apical membrane; spreading occurs with a marked increase in the level of these proteins at the apical membrane, and with a concomitant increase in apical microtubules. Because the microtubule inhibitor delayed spreading, as well as the initial apical localization, of γ-tubulin, we speculate that spreading might involve a bootstrap sequence wherein microtubule motor proteins transport additional nucleators apically. Consistent with a recruitment mechanism, we showed that par-3(-) embryos develop large, non-apical foci of γ-tubulin on some lateral membranes. Because lateral membranes are specialized for adhesion, it is possible that this restricts the spread of the γ-tubulin foci compared to the contact-free apical surface.
Our results show that centrosomes play a critical role in apical differentiation that is separate from their well-studied role in cell division, because ablation of post-mitotic centrosomes prevents the localization of a microtubule nucleator, γ-tubulin, to the apical surface. These results raise the question of why an MTOC does not form de novo at the apical surface, and instead appears to require a centrosomal origin. In dividing cells, centrosomes nucleate large, radial arrays of microtubules during a comparatively brief period of the cell cycle, yet centrioles at the core of the centrosome are not disassembled and re-formed [27]. Instead, centrioles appear to provide a critical platform that regulates subsequent centriole, and thus centrosome, assembly, and limits the number of centrosomes within a cell. By analogy, the plume might contain a pre-formed platform for microtubule nucleation, and be necessary if there is an immediate requirement for microtubules to transfer PAR foci apically.
Apically-positioned centrosomes have long been noted in animal epithelia, where they often nucleate the assembly of cilia [28,29]. However, the intestinal epithelia cells in C. elegans show a transient, apical localization of centrosomes, and these cells lack cilia. Instead, our results suggest that apical localization is associated with a transfer of MTOC function from the post-mitotic centrosome to the future apical membrane. Future studies should reveal whether centrosomes play a related role in the organization of apical MTOCs in other systems, independent of their role in cilia assembly.
Experimental Procedures
Worm Strains
Nematodes were cultured and manipulated as previously described [31]. Unless otherwise indicated, experiments were performed using one-day old N2 adults. The following strains and alleles were used during the course of the above research: JJ2200 [γ-tubulin:GFP; histone:Cherry, derived from TH27 and RW10757 (provided by R.W.)], unc-83(ku18) [19], unc-83(e1408) [19], JJ2292 [unc-83(e1408); γ-tubulin:GFP; histone:Cherry, derived from JJ2200], JJ2330 [γ-tubulin:GFP; histone:Cherry; membrane:Cherry, derived from JJ2200 and OD70 (pie-1prom:mCherry:PH(PLC1delta1))], JJ2069 [γ-tubulin:GFP; PAR-6:Cherry, derived from TH27 and TH110 [[32], provided by A.H.], JJ1556 [PAR-3:GFP] [24], par-6(zu170); unc-101; zuIs43[pie-1prom:gfp:par-6:zf1] [22], provided by J.N., par-6(tm1425)/unc-101; him-8 [22], provided by J.N., JJ1743 [par-6(tm1425)/hIn1; him-8], par- 3(tm2716); unc-32; zuIs20 [par-3:zf1:gfp, unc-119] [23], provided by J.N., par- 3(tm2716)/hT2 [bli-4(e937) let-?(q782) qIs48]; him-8) [23], provided by J.N., JJ2190 [γ-tubulin:GFP; histone:Cherry; par- 3(tm2716)/hT2 [bli-4(e937) let-?(q782) qIs48]; him-8, derived from JJ2200], dyrb-1(tm2645) [20], provided by B.B., JJ2331 [γ-tubulin:GFP; histone:Cherry; dyrb-1(tm2645), derived from JJ2200]
PAR protein depletion
PAR proteins were depleted from embryos as previously described [22-24]. For PAR-6 depletion, par-6(zu170); unc-101; zuIs143[pie-1prom:gfp:par-6:zf1] hermaphrodites was mated with either par-6(tm1425)/unc-101; him-8, or par-6(tm1425)/hIn1; him-8 males. The non-Unc progeny were selected and allowed to self-fertilize. For PAR-3 depletion, par- 3(tm2716); unc-32; zuIs20 [par- 3:zf1:gfp, unc-119] hermaphrodites were mated with either par- 3(tm2716)/hT2 [bli-4(e937) let-?(q782) qIs48]; him-8) or JJ2190 [γ-tubulin:GFP; histone:Cherry; par- 3(tm2716)/hT2 [bli-4(e937) let-?(q782) qIs48]; him-8] males. Progeny lacking the balancer were allowed to self-fertilize. Embryos were incubated for 3-6 hours from fertilization and fixed and stained with antibodies against PAR-6 and GFP or PAR-3 and GFP, respectively, to identify the embryos that were depleted for PAR-6 or PAR-3. For live imaging of PAR-3 depleted animals, embryos were filmed until the mutant phenotype was observable in about ¼ of embryos. The wild-type embryos express a PAR-3 rescuing transgene marked with GFP, so both γ-tubulin and PAR-3 are visible in the green channel (Figure 4I).
Cytoskeletal Inhibitor Experiments
Embryos were added to a coverslip and washed sequentially with the following: poly-lysine, .1% trypan blue, 2 washes of embryonic growth media (EGM, [33]), and 3 washes of EGM + inhibitor or .2% DMSO. In the third wash, the coverslip was inverted over a microscope slide with Teflon squares (Thermo/Fisher Scientific). The embryos were supported by 22.5 uM beads (Whitehouse Scientific). The eggshell and viteline membrane were permeabilized using a Micropoint tuneable dye laser (coumarin 441nm). Embryos were filmed for one hour and then at subsequent timepoints as indicated. Following filming, embryos were incubated overnight and checked the next day for the production of gut granules, indicating that the embryos were viable. Embryos that did not produce gut granules or whose cells stained with the vital dye trypan blue were not included in the dataset. In the γ-tubulin:GFP experiments, inhibitor penetration was assessed by determining whether dividing intestinal cells arrested (nocodazole) or failed cytokinesis (latrunculin A) immediately after eggshell permeabilization (see Figure 5, insets for examples). For PAR-3:GFP experiments, permeabilization was assayed by monitoring the uptake of trypan blue into intestinal granules.
Centrosome Ablation
For centrosome ablation, embryos expressing γ-tubulin:GFP; histone:Cherry, and membrane:Cherry were mounted on a 3% agarose pad and E8 stage embryos were identified and followed until they entered mitosis. Late telophase centrosomes were targeted with a Micropoint pulse nitrogen pumped tuneable dye laser (coumarin 441nm) with a power output of between 5.5-9.5%, a step size of 1, and between 3 and 5 rounds of laser pulses. Embryos were photographed immediately prior to ablation, just after ablation, and then for a number of time points after ablation until γ-tubulin:GFP had distributed to the apical surface of control cells. At the time of centrosome ablation, approximately
3.7% of the cytoplasmic γ-tubulin:GFP was concentrated at the centrosome. Further detailed Experimental Procedures are described in Supplemental Information.
Supplementary Material
Highlights.
-MTOC function transitions from the centrosome to the apical surface of C. elegans epithelia
-Apical localization of centrosomes and microtubule nucleators requires PAR-3
-Microtubule nucleators appear to emanate from the centrosome during MTOC transition
-The centrosome is required for the apical accumulation of microtubule nucleators
Acknowledgements
We thank Jeremy Nance, Anthony Hyman, Asako Sugimoto, Bruce Bowerman, Pierre Gonczy, Karen Oegema, and Robert Waterston for providing strains and antibodies. We also thank L. Holt, W. Marshall, U. Sheth, S. Dennis. J. Molk, and J. Rasmussen for critical review of the manuscript. Some of the nematode strains used in this work were provided by the Caenorhabditis Genetic Center, which is funded by the NIH National Center for Research Resources (NCRR). J.L.F and J.R.P. are supported by Howard Hughes Medical Institute and NIH RO1 GM09858. J.L.F. is supported by a Helen Hay Whitney Foundation Postdoctoral Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hannak E, et al. The kinetically dominant assembly pathway for centrosomal asters in Caenorhabditis elegans is gamma-tubulin dependent. J Cell Biol. 2002;157:591–602. doi: 10.1083/jcb.200202047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray D, Bunge MB. Serial analysis of microtubules in cultured rat sensory axons. J Neurocytol. 1981;10:589–605. doi: 10.1007/BF01262592. [DOI] [PubMed] [Google Scholar]
- 3.Baas PW, Deitch JS, Black MM, Banker GA. Polarity orientation of microtubules in hippocampal neurons: uniformity in the axon and nonuniformity in the dendrite. Proc Natl Acad Sci U S A. 1988;85:8335–9. doi: 10.1073/pnas.85.21.8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tassin AM, Maro B, Bornens M. Fate of microtubule-organizing centers during myogenesis in vitro. J Cell Biol. 1985;100:35–46. doi: 10.1083/jcb.100.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bré MH, Kreis TE, Karsenti E. Control of microtubule nucleation and stability in Madin-Darby canine kidney cells: the occurrence of noncentrosomal, stable detyrosinated microtubules. J Cell Biol. 1987;105:1283–96. doi: 10.1083/jcb.105.3.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bacallao R, Antony C, Dotti C, Karsenti E, Stelzer EH, Simons K. The subcellular organization of Madin-Darby canine kidney cells during the formation of a polarized epithelium. J Cell Biol. 1989;109:2817–32. doi: 10.1083/jcb.109.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brodu V, Baffet AD, Le Droguen PM, Casanova J, Guichet A. A developmentally regulated two-step process generates a noncentrosomal microtubule network in Drosophila tracheal cells. Dev Cell. 2010;18:790–801. doi: 10.1016/j.devcel.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 8.Zhou K, Rolls MM, Hall DH, Malone CJ, Hanna-Rose W. A ZYG-12-dynein interaction at the nuclear envelope defines cytoskeletal architecture in the C. elegans gonad. J Cell Biol. 2009;186:229–41. doi: 10.1083/jcb.200902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee C, Scherr HM, Wallingford JB. Shroom family proteins regulate gamma-tubulin distribution and microtubule architecture during epithelial cell shape change. Development. 2007;134:1431–41. doi: 10.1242/dev.02828. [DOI] [PubMed] [Google Scholar]
- 10.Bartolini F, Gundersen GG. Generation of noncentrosomal microtubule arrays. J Cell Sci. 2006;119:4155–63. doi: 10.1242/jcs.03227. [DOI] [PubMed] [Google Scholar]
- 11.Mogensen MM. Microtubule release and capture in epithelial cells. Biol Cell. 1999;91:331–41. [PubMed] [Google Scholar]
- 12.Baas PW, Joshi HC. Gamma-tubulin distribution in the neuron: implications for the origins of neuritic microtubules. J Cell Biol. 1992;119:171–8. doi: 10.1083/jcb.119.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mogensen MM, Mackie JB, Doxsey SJ, Stearns T, Tucker JB. Centrosomal deployment of gamma-tubulin and pericentrin: evidence for a microtubule-nucleating domain and a minus-end docking domain in certain mouse epithelial cells. Cell Motil Cytoskeleton. 1997;36:276–90. doi: 10.1002/(SICI)1097-0169(1997)36:3<276::AID-CM8>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 14.Hannak E, Kirkham M, Hyman AA, Oegema K. Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans. J Cell Biol. 2001;155:1109–16. doi: 10.1083/jcb.200108051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bellanger JM, Gönczy P. TAC-1 and ZYG-9 form a complex that promotes microtubule assembly in C. elegans embryos. Curr Biol. 2003;13:1488–98. doi: 10.1016/s0960-9822(03)00582-7. [DOI] [PubMed] [Google Scholar]
- 16.Hamill DR, Severson AF, Carter JC, Bowerman B. Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev Cell. 2002;3:673–84. doi: 10.1016/s1534-5807(02)00327-1. [DOI] [PubMed] [Google Scholar]
- 17.Leung B, Hermann GJ, Priess JR. Organogenesis of the Caenorhabditis elegans intestine. Dev Biol. 1999;216:114–34. doi: 10.1006/dbio.1999.9471. [DOI] [PubMed] [Google Scholar]
- 18.Bobinnec Y, Fukuda M, Nishida E. Identification and characterization of Caenorhabditis elegans gamma-tubulin in dividing cells and differentiated tissues. J Cell Sci. 2000;113:3747–59. doi: 10.1242/jcs.113.21.3747. [DOI] [PubMed] [Google Scholar]
- 19.Starr DA, et al. unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development. 2001;128:5039–50. doi: 10.1242/dev.128.24.5039. [DOI] [PubMed] [Google Scholar]
- 20.O’Rourke SM, Dorfman MD, Carter JC, Bowerman B. Dynein modifiers in C. elegans: light chains suppress conditional heavy chain mutants. PLoS Genet. 2007;3:1339–1354. doi: 10.1371/journal.pgen.0030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patalano S, et al. The aPKC-PAR-6-PAR-3 cell polarity complex localizes to the centrosome attracting body, a macroscopic cortical structure responsible for asymmetric divisions in the early ascidian embryo. J Cell Sci. 2006;119:1592–603. doi: 10.1242/jcs.02873. [DOI] [PubMed] [Google Scholar]
- 22.Totong R, Achilleos A, Nance J. PAR-6 is required for junction formation but not apicobasal polarization in C. elegans embryonic epithelial cells. Development. 2007;134:1259–68. doi: 10.1242/dev.02833. [DOI] [PubMed] [Google Scholar]
- 23.Achilleos A, Wehman AM, Nance J. PAR-3 mediates the initial clustering and apical localization of junction and polarity proteins during C. elegans intestinal epithelial cell polarization. Development. 2010;137:1833–42. doi: 10.1242/dev.047647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nance J, Munro EM, Priess JR. C. elegans PAR-3 and PAR-6 are required for apicobasal asymmetries associated with cell adhesion and gastrulation. Development. 2003;130:5339–50. doi: 10.1242/dev.00735. [DOI] [PubMed] [Google Scholar]
- 25.Grill SW, Howard J, Schäffer E, Stelzer EH, Hyman AA. The distribution of active force generators controls mitotic spindle position. Science. 2003;301:518–21. doi: 10.1126/science.1086560. [DOI] [PubMed] [Google Scholar]
- 26.Munro E, Nance J, Priess JR. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev Cell. 2004;7:413–24. doi: 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 27.Kochanski RS, Borisy GG. Mode of centriole duplication and distribution. J Cell Biol. 1990;110:1599–605. doi: 10.1083/jcb.110.5.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeligs JD, Wollman SH. Mitosis in rat thyroid epithelial cells in vivo. II. Centrioles and pericentriolar material. J Ultrastruct Res. 1979;66:97–108. doi: 10.1016/s0022-5320(79)90127-8. [DOI] [PubMed] [Google Scholar]
- 29.Buendia B, Bré MH, Griffiths G, Karsenti E. Cytoskeletal control of centrioles movement during the establishment of polarity in Madin-Darby canine kidney cells. J Cell Biol. 1990;110:1123–35. doi: 10.1083/jcb.110.4.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toya M, Terasawa M, Nagata K, Iida Y, Sugimoto A. A kinase-independent role for Aurora A in the assembly of mitotic spindle microtubules in Caenorhabditis elegans embryos. Nat Cell Biol. 2011;13:708–14. doi: 10.1038/ncb2242. [DOI] [PubMed] [Google Scholar]
- 31.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schonegg S, Constantinescu AT, Hoege C, Hyman AA. The Rho GTPase-activating proteins RGA-3 and RGA-4 are required to set the initial size of PAR domains in Caenorhabditis elegans one-cell embryos. Proc Natl Acad Sci U S A. 2007;104:14976–14981. doi: 10.1073/pnas.0706941104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shelton CA, Bowerman B. Time-dependent responses to glp-1-mediated inductions in early C. elegans embryos. Development. 1996;122:2043–50. doi: 10.1242/dev.122.7.2043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.