Abstract
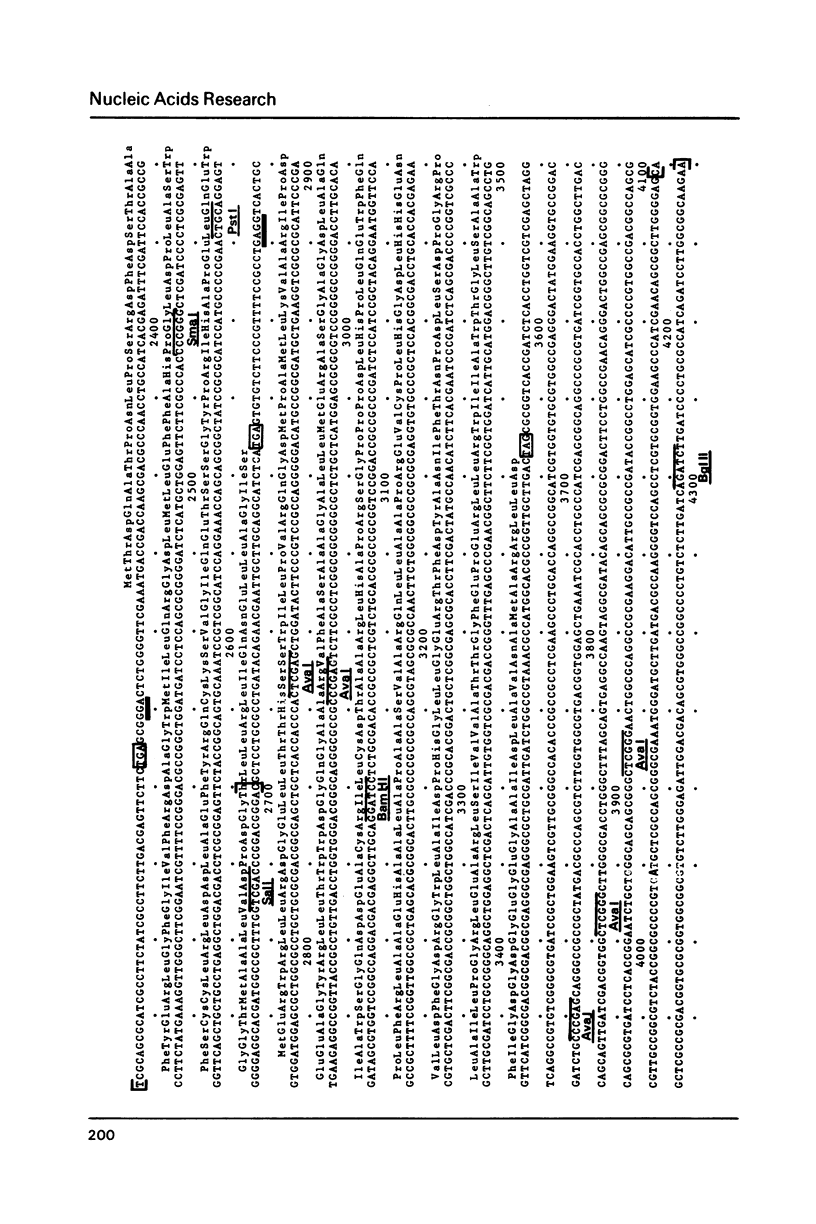
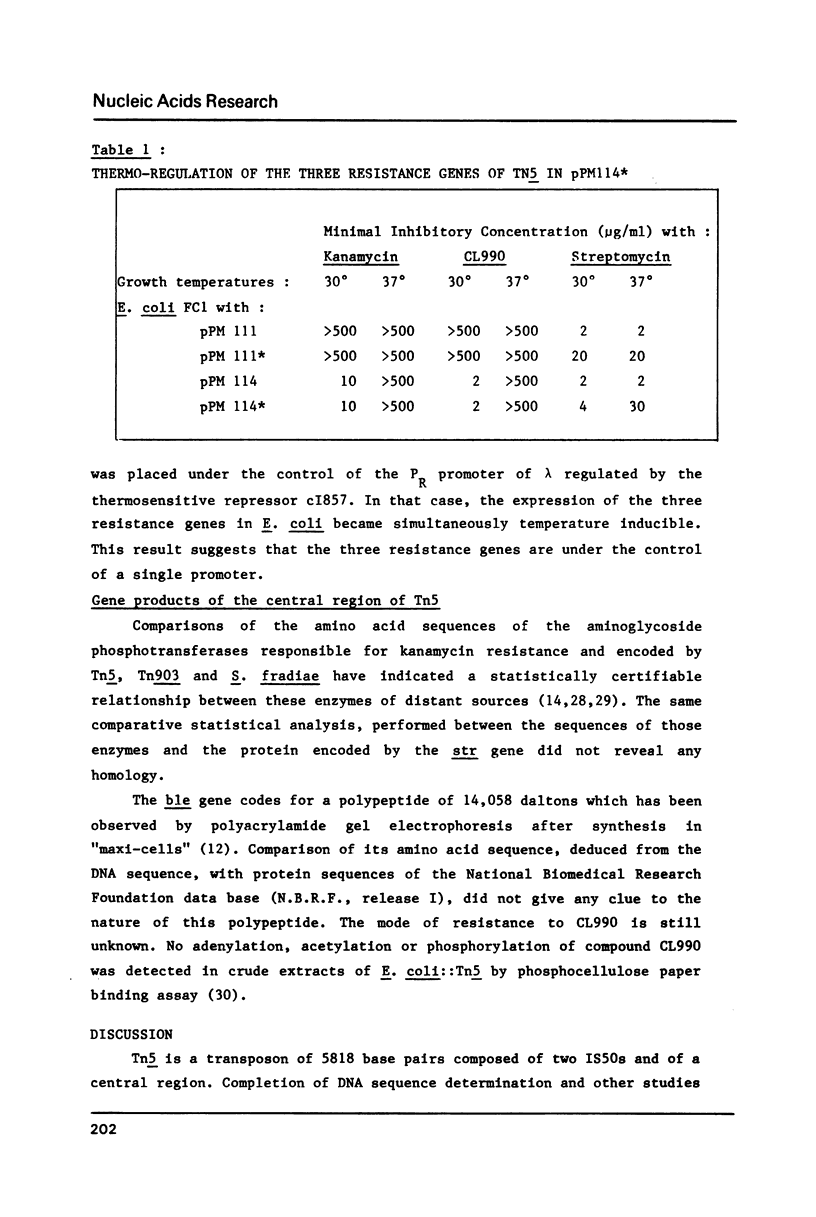
The DNA sequence of the region located downstream from the kanamycin resistance gene of Tn5 up to the right inverted repeat IS50R has been determined. This completes the determination of the sequence of Tn5 which is 5818 bp long. The 2.7 Kb central region contains three resistance genes: the kanamycin-neomycin resistance gene, a gene coding for resistance to CL990 an antimitotic-antibiotic compound of the bleomycin family and a third gene that confers streptomycin resistance in some bacterial species but is cryptic in E. coli. A Tn5* mutant able to express streptomycin resistance in E. coli was isolated. With this mutant, it was demonstrated that in E. coli the expression of the three resistance genes is coordinated in a single operon.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auerswald E. A., Ludwig G., Schaller H. Structural analysis of Tn5. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):107–113. doi: 10.1101/sqb.1981.045.01.019. [DOI] [PubMed] [Google Scholar]
- Beck E., Ludwig G., Auerswald E. A., Reiss B., Schaller H. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982 Oct;19(3):327–336. doi: 10.1016/0378-1119(82)90023-3. [DOI] [PubMed] [Google Scholar]
- Berg D. E., Davies J., Allet B., Rochaix J. D. Transposition of R factor genes to bacteriophage lambda. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3628–3632. doi: 10.1073/pnas.72.9.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Davies J., Brzezinska M., Benveniste R. The problems of drug-resistant pathogenic bacteria. R factors: biochemical mechanisms of resistance to aminoglycoside antibiotics. Ann N Y Acad Sci. 1971 Jun 11;182:226–233. doi: 10.1111/j.1749-6632.1971.tb30659.x. [DOI] [PubMed] [Google Scholar]
- Davies J., Smith D. I. Plasmid-determined resistance to antimicrobial agents. Annu Rev Microbiol. 1978;32:469–518. doi: 10.1146/annurev.mi.32.100178.002345. [DOI] [PubMed] [Google Scholar]
- De Vos G. F., Finan T. M., Signer E. R., Walker G. C. Host-dependent transposon Tn5-mediated streptomycin resistance. J Bacteriol. 1984 Jul;159(1):395–399. doi: 10.1128/jb.159.1.395-399.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garoff H., Ansorge W. Improvements of DNA sequencing gels. Anal Biochem. 1981 Aug;115(2):450–457. doi: 10.1016/0003-2697(81)90031-2. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., Ames G. F., Barnes W. M., Clement J. M., Hofnung M. A novel intercistronic regulatory element of prokaryotic operons. Nature. 1982 Aug 19;298(5876):760–762. doi: 10.1038/298760a0. [DOI] [PubMed] [Google Scholar]
- Humphreys G. O., Willshaw G. A., Anderson E. S. A simple method for the preparation of large quantities of pure plasmid DNA. Biochim Biophys Acta. 1975 Apr 2;383(4):457–463. doi: 10.1016/0005-2787(75)90318-4. [DOI] [PubMed] [Google Scholar]
- Isberg R. R., Lazaar A. L., Syvanen M. Regulation of Tn5 by the right-repeat proteins: control at the level of the transposition reaction? Cell. 1982 Oct;30(3):883–892. doi: 10.1016/0092-8674(82)90293-8. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Yin J. C., Reznikoff W. S. Control of Tn5 transposition in Escherichia coli is mediated by protein from the right repeat. Cell. 1982 Oct;30(3):873–882. doi: 10.1016/0092-8674(82)90292-6. [DOI] [PubMed] [Google Scholar]
- Jorgensen R. A., Rothstein S. J., Reznikoff W. S. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177(1):65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- Klenow H., Overgaard-Hansen K., Patkar S. A. Proteolytic cleavage fo native DNA polymerase into two different catalytic fragments. Influence of assay condtions on the change of exonuclease activity and polymerase activity accompanying cleavage. Eur J Biochem. 1971 Oct 14;22(3):371–381. doi: 10.1111/j.1432-1033.1971.tb01554.x. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- O'Neill E. A., Kiely G. M., Bender R. A. Transposon Tn5 encodes streptomycin resistance in nonenteric bacteria. J Bacteriol. 1984 Jul;159(1):388–389. doi: 10.1128/jb.159.1.388-389.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnoky P., Kiss G. B., Ott I., Kondorosi A. Tn5 carries a streptomycin resistance determinant downstream from the kanamycin resistance gene. Mol Gen Genet. 1983;191(2):288–294. doi: 10.1007/BF00334828. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Rothstein S. J., Jorgensen R. A., Postle K., Reznikoff W. S. The inverted repeats of Tn5 are functionally different. Cell. 1980 Mar;19(3):795–805. doi: 10.1016/s0092-8674(80)80055-9. [DOI] [PubMed] [Google Scholar]
- Rothstein S. J., Reznikoff W. S. The functional differences in the inverted repeats of Tn5 are caused by a single base pair nonhomology. Cell. 1981 Jan;23(1):191–199. doi: 10.1016/0092-8674(81)90284-1. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Selvaraj G., Iyer V. N. Transposon Tn5 specifies streptomycin resistance in Rhizobium spp. J Bacteriol. 1984 May;158(2):580–589. doi: 10.1128/jb.158.2.580-589.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. F., Waterman M. S., Fitch W. M. Comparative biosequence metrics. J Mol Evol. 1981;18(1):38–46. doi: 10.1007/BF01733210. [DOI] [PubMed] [Google Scholar]
- Thompson C. J., Gray G. S. Nucleotide sequence of a streptomycete aminoglycoside phosphotransferase gene and its relationship to phosphotransferases encoded by resistance plasmids. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5190–5194. doi: 10.1073/pnas.80.17.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3'5"-aminoglycoside phosphotransferase type III. Gene. 1983 Sep;23(3):331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]


