Abstract
The present study establishes the effectiveness of natural drug delivery mechanisms and investigates the interactions between drug and its natural carrier. The binding between the isoflavone diadzein (DZN) and the natural carrier hemoglobin (HbA) was studied using optical spectroscopy and molecular dynamics simulations. The inherent fluorescence emission characteristics of DZN along with that of tryptophan (Trp) residues of the protein HbA were exploited to elucidate the binding location and other relevant parameters of the drug inside its delivery vehicle HbA. Stern-Volmer studies at different temperatures indicate that static along with collisional quenching mechanisms are responsible for the quenching of protein fluorescence by the drug. Molecular dynamics and docking studies supported the hydrophobic interactions between ligand and protein, as was observed from spectroscopy. DZN binds between the subunits of HbA, ~15 Å away from the closest heme group of chain α1, emphasizing the fact that the drug does not interfere with oxygen binding site of HbA.
Keywords: Diadzein, Natural drug carrier, Intrinsic Fluorescence, Molecular dynamics, docking
Introduction
Understanding and designing drug delivery systems is an increasingly promising discipline in pharmaceutical development, allowing sensible and reasonable handling of the pharmacological profiles of drugs and the beneficial roles associated with them. Recently, erythrocytes (red blood cells) have come into prominence as drug delivery systems, due to their biocompatibility, biodegradability, usability and easy loading capability [1–4]. But the factor that critically limits their use as drug carrier is their degradation in vivo by the reticuloendothelial system (RES) [5] and in some cases the toxic effects caused by erythrocytes under certain conditions [6, 7]. In order to design and improve an erythrocyte based drug delivery system, the binding interactions of drugs with the main component of erythrocytes, hemoglobin (HbA), have to be fully understood. Hemoglobin is the most abundant blood protein and consists of two α and two β subunits which are noncovalently associated within erythrocytes as a 64.5 kDa tetramer [8–10]. Delivery of oxygen from the lungs to respiring tissues is the main function of the heme group of hemoglobin. Recently, using radiolabeled HbA, the liver was confirmed as the main site for HbA uptake [7]. By using HbA as a drug carrier, the natural mechanisms of HbA uptake and decomposition, the latter of which leads to unloading of any bound compounds, can be employed to selectively transport therapeutic drugs to physiological sites of interest for the treatment of diseases.
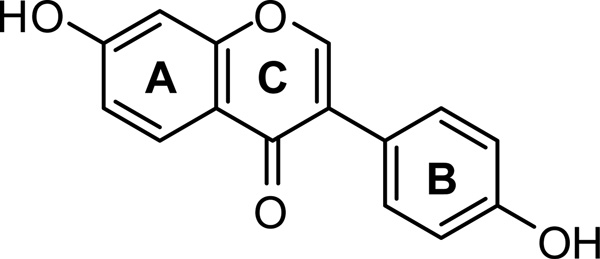
Isoflavones are naturally occurring polyphenolic compounds with antioxidant properties, and exert many health benefits such as lowering the risk of breast and prostate cancers, osteoporosis and cardiovascular diseases [11, 12]. Genistein and daidzein (DZN) are isoflavonoids commonly found in legumes. Both of these isoflavones have a wide spectrum of physiological and pharmacological functions including antiestrogenic [13, 14], anticancer [15, 16], anti-inflammatory [17], cardioprotective [18] and enzyme-inhibitory effects [13, 14]. The Dietary Reference Intakes, established by the National Academy of Sciences [19], highlight that plant based polyphenols are important dietary constituents. The objective of the present study is to examine the structural and binding interactions between HbA and diadzein (DZN, structure shown in Scheme 1) by absorption, fluorescence and circular dicroism spectroscopy, along with docking and molecular dynamics (MD) simulations. To study drug transport using a fluorescence based approach, the availability of a non-invasive and photo-stable fluorescent substrate is required [20]. The intrinsic fluorescence of DZN, which can probe the binding microenvironment, combined with its high therapeutic potency and low systemic toxicity fulfills these requirements. The results obtained for DZN in the protein matrix are useful for the identification of its location in HbA at the molecular level. The strong binding interactions between ligand and protein are evident from the high fluorescence anisotropy as well as the computational studies which show the presence of hydrophobic interactions between DZN and the surrounding amino acids in the HbA matrix.
Scheme 1.
Structure of the isoflavone Diadzein (7-hydroxy-3(-4’-hydroxy-phenyl)-chromen-4-one)
The significance of the present work lies in the fact that HbA is the natural drug carrier in physiological systems and its binding studies with the therapeutically potent fluorescent isoflavone DZN provide insights into the underlying mechanisms of interactions between the drug and its delivery vehicle. This approach can be well extrapolated to drugs of similar nature and helps in understanding the pharmacokinetics and biological activities of plant flavonoids.
Experimental
Materials
Lyophilized powder of human hemoglobin (molecular weight 64,500 D), diadzein, and phosphate buffer were purchased from Sigma Chemicals, USA, and used without further purification after confirming their purity by comparing their electronic absorption and emission spectra with published data [11]. Solvents used were of spectroscopic grade and obtained from Sigma. Purity of DZN was further confirmed by thin layer chromatography which showed only one spot under UV light. Absorption and fluorescence spectroscopic measurements were performed with DZN concentrations of 1 × 10−5 M. HbA was dissolved in pH 7.4 phosphate buffer solution (1 × 10−2 M) and the HbA stock solution (2 × 10−4 M) was kept in the dark at 277 K. The protein concentration was determined spectrophotometrically using the molar extinction coefficient of HbA at 276 nm (120,808 M−1cm−1) [21].
For fluorescence quenching studies, the HbA concentration was kept constant at 10−5 M. Varying aliquots of concentrated methanolic solution of DZN were added to obtain final concentrations ranging from 0 – 2.5 × 10−5 M. The concentrations of methanol were always kept <1% (by volume) in all samples.
Spectroscopic Measurements
Steady state absorption spectra were recorded with Shimadzu UV2550 spectrophotometers. Steady state fluorescence measurements were carried out with Shimadzu RF5301 (equipped with a Fisher temperature controlled accessory) spectrofluorometers. The fluorescence anisotropy (r) measurements were performed on a Fluoromax-3 (Jobin Yvon Horiba) spectrometer. The values were obtained using the expression r = (I VV − GI VH) / (I VV + 2GI VH), where I VV and I VH are the vertically and horizontally polarized components of probe emission with excitation by vertically polarized light at the respective wavelength and G is the sensitivity factor of the detection system [21]. Each intensity value used in this expression represents the computer-averaged values of five successive measurements. All spectral measurements were carried out at room temperature (298 K) with freshly prepared solutions. The emission spectra were taken by exciting the samples and measuring the emissions at relevant wavelengths; appropriate blanks were subtracted for respective measurements. The bandwidths were 3 nm for both the excitation and emission with integration times of 1 s. All spectra were collected using quartz cuvettes with 1 cm pathlengths.
Circular dichroism spectra were acquired with a J-710 spectropolarimeter (Jasco). The scan rate was 100 nm/min, and three consecutive spectra were averaged to produce the final spectrum.
Molecular docking
AutoDock4 [22] was employed to gain an insight into the DZN binding with HbA. 3-D atomic coordinates of HbA were obtained from the Brookhaven Protein Data Bank (PDB ID 2D60) and prepared for docking. Hemoglobin was considered as a tetramer. All hetero atoms were deleted and non-polar hydrogens were merged. The Kollman united-atom charge model was applied to the protein. Particular attention was given to the parameterization of the porphyrin rings. Partial atomic charges for the porphyrin ring were assigned using the Gasteiger-Marsili method while the state of the Iron (Fe) was added manually. Atomic solvation parameters and fragmental volumes were added to the protein. Grid maps used by the empirical free-energy scoring function in AutoDock were generated. A grid box of 100×100×100 grid points in size with a grid-point spacing of 0.375 Å was considered for docking. The map was centered such that it covered the entire protein including all possible binding sites.
The 3-D structure of DZN was built using the HYPERCHEM [23] molecular builder module and optimized using the AM1 semi-empirical method to an RMS convergence of 0.001 kcal/(Ǻ mol) with the Polak-Ribiere conjugate gradient algorithm implemented in the HYPERCHEM 7.5 package. Rotatable bonds were assigned for the ligand and partial atomic charges were calculated using the Gasteiger-Marsili method after merging non-polar hydrogens. 100 docking runs were performed and for each run, a maximum of 2,500,000 GA operations were performed on a single population of 150 individuals. The weights for crossover, mutation and elitism were default parameters of 0.8, 0.02 and 1 respectively.
Molecular Dynamics of the HbA-Diadzein complex
GROMACS molecular dynamics code [24] with OPLS all atom force-field [25] were employed to carry out the MD simulations on free hemoglobin and its complex with DZN. The parameters for the ligand (DZN) were developed from OPLS force-field [25] defined atomic groups. The partial atomic charges, which were adjusted to keep the charge neutrality of the atomic groups and make the ligand charge neutral, were tested by comparing the Gromacs optimized structure with i) the parameter-independent QM optimized structure of DZN in vacuum, and ii) the QM/MM optimized structures of DZN in explicit water. For QM code, we employed CPMD [26a] and for QM/MM, we employed GROMACS–CPMD [26b]. For QM/MM, the DZN molecule was considered in the QM sub-system and the water was considered in MM sub-system and their interphase interaction was described by the QM/MM Hamiltonian [26].
The crystal structure coordinates of HbA obtained from the protein data bank (PDB ID: 2D60) and used in the docking study, were considered for the simulation of free HbA. For HbA-DZN complex, the lowest energy docked complex obtained from the docking study was used for MD simulation. The OPLS parameters for the heme prosthetic group were taken from the previously published parameter set [27]. The HbA structure was primarily subjected to molecular dynamics simulation using GROMACS to check the planarity of the porphyrin ring and proper positioning of the distal histidine residue of hemoglobin that coordinates to the Iron (Fe) of the heme moiety, an essential structural feature for the biological functioning HbA. Both ligand bound/unbound structures of HbA were subjected to in vacuo minimization using steepest descent algorithm. Each structure was soaked in a water box containing SPC water molecules. All the protein atoms were at a distance equal or greater than 1 nm from the box edges. Then the system was further minimized using 500 steps of steepest descent algorithm in a water box, followed by 100 ps of position restrained dynamics where proteins were kept fixed by adding restraining forces, but water molecules were allowed to move. Final production simulations were performed in the isothermal-isobaric (NPT) ensemble at 300 K, using an external bath with a coupling constant of 0.1 ps. Pressure was kept constant (1 bar) by using the time-constant for pressure coupling set to 1 ps. The LINCS [28] algorithm was used to constrain bond lengths, allowing the use of 2 fs time steps. The van der Waals and Coulomb interactions were truncated at 1.2 nm. Conformations generated during MD simulation were stored at every 5 ps.
Results
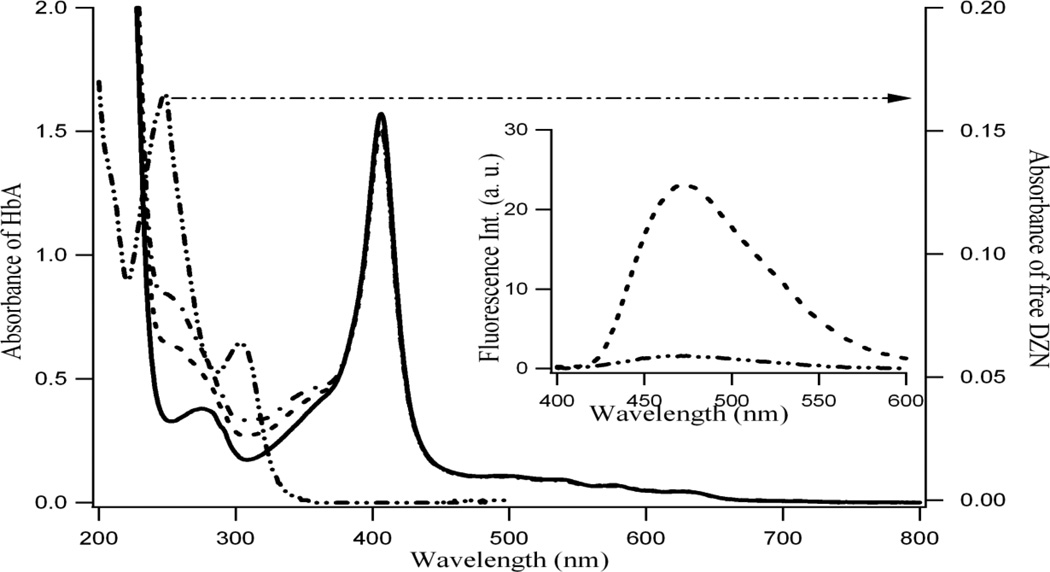
Absorption spectroscopy is a very basic but powerful tool to depict the binding interaction between ligand and protein. Figure 1 shows the absorption spectra of HbA with varying concentrations of DZN (chemical structure shown in Scheme 1). The absorption spectra of free HbA is shown in sold line in Figure 1 (using left axis) where as that for free DZN is indicated by -..-..- line (using right axis, shown by the arrow). In presence of DZN, the HbA absorption profiles are shown by - - - and -.-.- lines for two different concentrations of DZN. We observe that for the complex, two absorption bands gradually appear at 250 nm and 310 nm; these are the areas where although free HbA has no absorbance but free DZN has (shown by arrow in Figure 1, right axis). However, these two absorption bands in DZN bound HbA still exhibit significant differences in wavelengths from free DZN absorption profile indicating interactions and complex formation with HbA. It is pertinent to mention that the Soret band of HbA at 405 nm is unchanged in the presence of DZN. The inset of Figure 1 indicates the increase of fluorescence of DZN in a protein HbA matrix (---) compared to that in aqueous system (-..-) when λex = 340 nm where the protein does not absorb.
Figure 1.
Typical absorption spectra of hemoglobin (1 × 10−5M, solid line, free) and with 6 × 10−6 M (- - -); 1.5 × 10−5 M (-.-.) of diadzein (see left axis). Right axis presents the absorption of 1 × 10−5 M free diadzein in aqueous phase (-..-..). Inset: Fluorescence spectra of 1.0 × 10−5 M diadzein in aqueous phase (-..-..) and protein matrix (-----). λex=340 nm.
Quenching studies of protein tryptophan fluorescence in the presence of the ligand help to understand the microenvironment of the ligand inside the protein matrix and are also a measure of the proximity of the tryptophan residue to the quencher [29, 30]. With increasing DZN concentrations the emission of HbA gradually decreases (Figure 1S), indicating fluorescence quenching. Figure 2A presents the corresponding Stern-Volmer plots based on the equation (1)
| (1) |
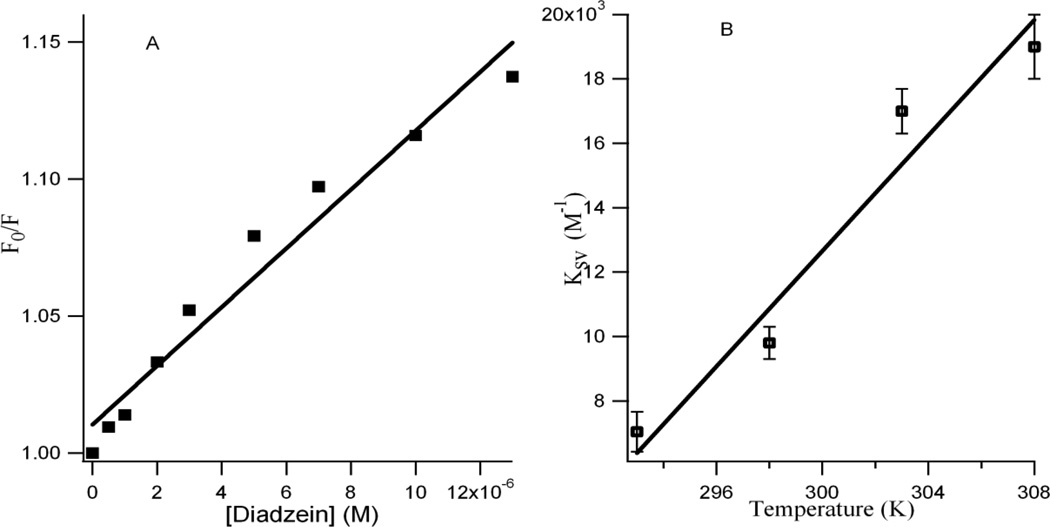
where KSV is the Stern-Volmer quenching constant for the quenching of HbA Trp fluorescence by the isoflavonoid [21]. The ratio F0/F for each addition of DZN are plotted against the DZN concentrations (see Figure 2A), where F0 and F are the fluorescence intensities of Trp in HbA in the absence and presence of DZN, respectively. It is observed that the ratio F0/F increases linearly with the DZN concentration and a linear regression equation following Stern-Volmer relation is obtained. The Stern-Volmer plots are essentially linear for flavonoid concentrations up to 1.2 × 10−5M (Figure 2A), after which the slope of the linear plot changes. This indicates that more than one type of quenching mechanism is involved [21, 30]. A KSV value of 0.98 × 104 M−1 is obtained for DZN which indicates that HbA tryptophan fluorescence is efficiently quenched by the flavonoid.
Figure 2.
A: Stern-Volmer plots of the HbA tryptophan fluorescence quenching with increasing diadzein concentration at ambient temperature. B: Variation of Ksv with temperatures. Each data point indicates the average of three experiments; error bars indicate standard deviations.
To understand the fluorescence quenching better, temperature dependent studies were performed. Figure 2B presents the variation of KSV with increasing temperature where the direct proportionality between the two is observed within the experimental error.
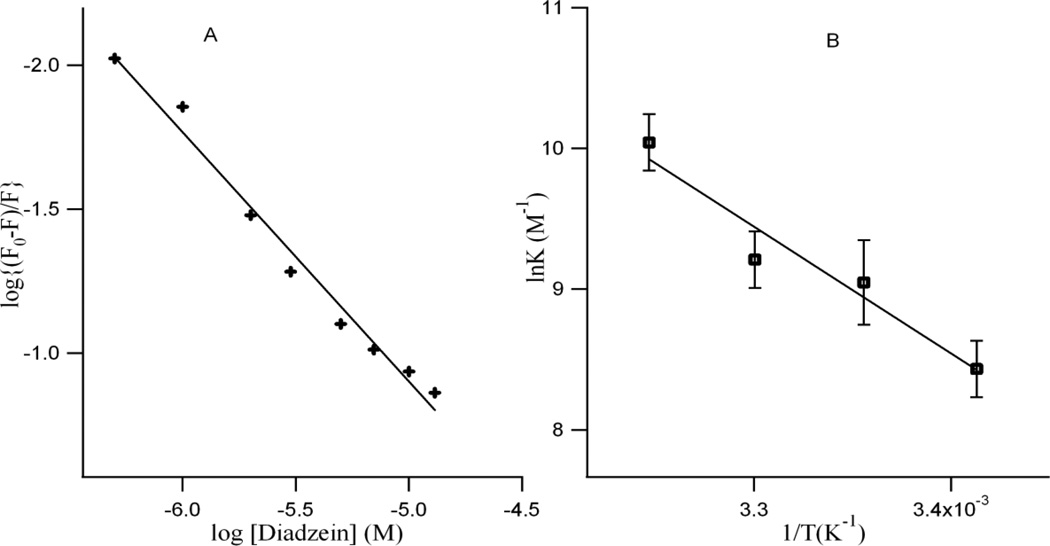
The apparent binding constant ‘K’ and the number of binding site(s) ‘n’ were estimated from fluorescence titration studies, using the plot of log(F0-F)/F vs. log[Q] [31] (Figure 3A) which is based on equation (2) [31, 32]
| (2) |
where F0 and F are the fluorescence intensity of HbA in absence and presence of isoflavonoid (DZN) respectively, Q is the DZN concentration, n is the number of binding sites and K is the binding constant. Table 1 shows the Stern-Volmer quenching (KSV) and binding constants ‘K’ for the binding of DZN to HbA within the studied concentration range of the flavonoids. The value of n approximates unity, indicating that there is one binding site in HbA for DZN.
Figure 3.
A: The plot of log(F0-F)/F vs. log[DZN] of the fluorescence quenching data at room temperature. The concentration of HbA= 1.0 × 10−5 M. B: van’t Hoff plot of the binding of DZN with 1.0 × 10−5 M HbA. Each data point is average of three independent measurements and error bars indicate standard deviations.
Table 1.
Stern Volmer Quenching (Ksv) and Binding (K) constants at different temperatures along with thermodynamic parameters of binding of diadzein with hemoglobin.
| Temperature | |||||
|---|---|---|---|---|---|
| 292 K | 298 K | 303 K | 308 K | ||
| Ksv (M−1) | 7.0 × 103 | 9.8 × 103 | 1.7 × 104 | 1.9 × 104 | |
| K (M−1) | 4.6 × 103 | 8.7 × 103 | 1.0 × 104 | 1.7 × 104 | |
| ΔG0 (kcal) | −4.74 | −5.27 | −5.51 | −6.16 | |
| ΔH0 | 17.9 kcal | ||||
| ΔS0 | 77.8 kcal | ||||
The thermodynamic parameters associated with temperature variation were analyzed in order to further characterize the acting forces between DZN and HbA in phosphate buffer of pH 7.4. The thermodynamic parameters such as enthalpy change (ΔH0) and entropy change (ΔS0) of the binding reaction are the main evidence for the confirmation of the binding modes. From a thermodynamic standpoint, ΔH > 0 and ΔS > 0 implies a hydrophobic interaction; ΔH < 0 and ΔS < 0 reflects van der Waals forces or hydrogen bond formation; and ΔH ≈ 0 and ΔS > 0 suggests an electrostatic force [31, 33]. The thermodynamic parameters are determined using the van’t Hoff equations:
| (3) |
| (4) |
where K and R are the binding constant and gas constant, respectively. The temperature-dependence of the binding constant was studied at four different temperatures (293, 298, 303, and 308 K) in which ΔH0 and ΔS0 of reaction could be determined from the linear relationship between lnK and the reciprocal of absolute temperature. The free energy (ΔG0) could be calculated by Eq. (4).The thermodynamic parameters were determined using a van’t Hoff plot (Figure 3B) and are presented in Table 1. The values for the standard enthalpy (ΔH0) and entropy changes (ΔS0) of the binding reaction between DZN and HbA are found to be positive.
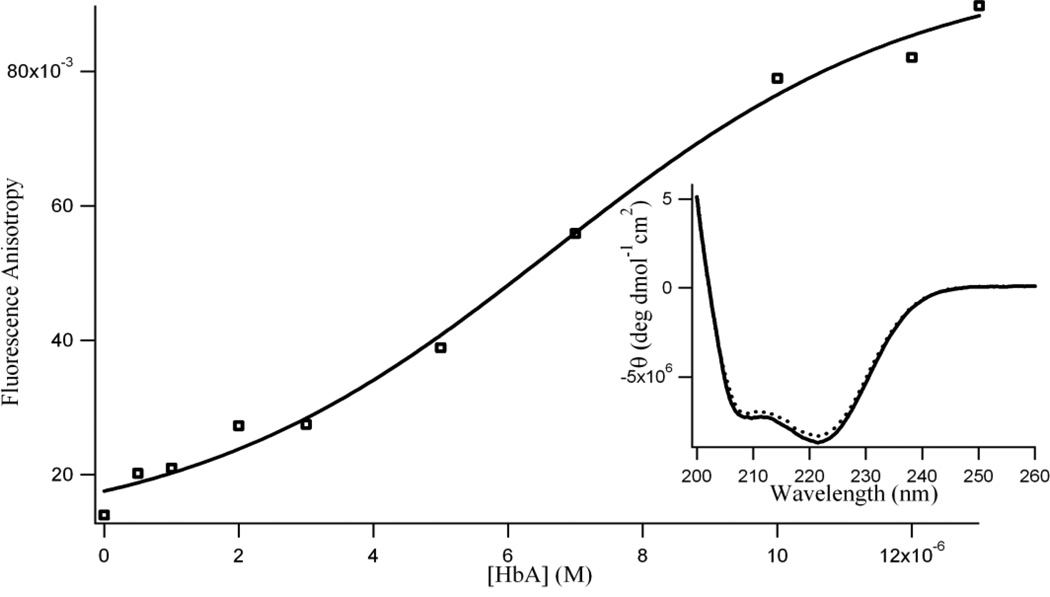
To ascertain the binding of DZN with HbA, we measured the fluorescence anisotropy of DZN in the presence of HbA. Fig. 4 presents the variation of the fluorescence anisotropy value for DZN with increasing HbA concentration. The plot shows the increase of the anisotropy (r) of DZN ([DZN] = 1.0 × 10−5 M) from 0.01 in aqueous buffer to 0.08 (at [HbA] = 1.0 × 10−5 M). The anisotropy gradually levels off at higher HbA concentrations (r = 0.09 at [DZN] = 1.3 × 10−5 M. The gradual increase in ‘r’ of DZN suggests that more and more DZN molecules bind with HbA, until HbA ~ 1.0 × 10−5 M.
Figure 4.
Variation in fluorescence anisotropy (r) of diadzein (1.0 × 10−5 M) with increasing protein (HbA) concentration (λex = 340 nm; λem = 470 nm, excitation and emission bandwidths were 5 nm). Each data point indicates the average of three determinations. Inset: Circular dichroism spectra of 3.0 × 10−6 M hemoglobin (HbA) in absence (solid line) and presence of 10.0 × 10−6 M diadzein (dotted line) in 0.01 M phosphate buffer, pH 7.4.
To investigate the possible effect of the isoflavonoid binding on the secondary structure of HbA, we carried out far-UV CD spectroscopy. The CD spectrum of HbA in aqueous buffer (in the absence of flavonoids) has two characteristic peaks of negative ellipticity at 208 nm and 222 nm indicating its predominantly α-helical secondary structure (Figure 4 inset). As shown in the figure, the CD spectrum of the HbA shows no significant change upon addition of 1 × 10−5 M of DZN (at constant hemoglobin concentration).
The HbA molecule contains three tryptophan residues in each α,β dimer, but only β-37 Trp is located at the interface between the two dimers. The intrinsic fluorescence signal of HbA arises from the indole group of tryptophan β-37. The fluorescence signal of this tryptophan residue plays an important role in depicting the changes that appear in the quaternary structure of HbA upon ligand binding [34, 35]. It is well known that the Trp amino acid exhibits extremely sensitive fluorescence emission properties [21]. In order to observe any structural perturbation in the protein matrix induced by DZN the intrinsic Trp fluorescence was exploited. Figure 2S displays the variation in the intrinsic emission of Trp residue with increasing [HbA] in absence (A) and presence (B) of DZN. Fluorescence intensity gradually increases with protein concentration until 5×10−5 M after which the intensity decreases for both cases. However the hypsochromic shift in the Trp [21, 34, 35] (right axis of the plots) with the increasing [HbA] is slightly different when DZN is present indicating that DZN is bound in close proximity to β-37 Trp residue, as is evident from the docking and molecular dynamics study, discussed below.
To compare the structural stability of free and DZN bound HbA, we have analyzed the dynamic structural properties (using root mean square deviation, (RMSD) and radius of gyration, (Rg)) of both systems, according to MD simulation which are summarized in Table 2. The first 2 ns of both simulations were considered to be the equilibration phase where slight structural re-organization takes place and properties were averaged over the last 3 ns of the simulations. RMSD and Rg are two important parameters that provide quantitative descriptions of changes in the tertiary structure of the simulated protein. Similar RMSD were observed for free and DZN bound HbA, which were 0.53 ± 0.034 nm and 0.56 ± 0.09 nm respectively. The calculated Rg value for free HbA was 2.43 ± 0.01 nm, while that of DZN bound HbA was 2.45 ± 0.035 nm, which again signifies no structural unfolding induced by the ligand, DZN. The binding energy components were analyzed using MD simulation during the last 2ns of the simulation and Lennard-Jones (LJ) is found to be the main determining factor of the total binding energy with an average value of −17.66.± 4.5 kcal/mole while Coulombic interaction is found to be −9.95 ± 3.85 kcal/mole. The high standard deviation of average Coulombic interactions indicates insignificance of the interaction to the total binding affinity, while the LJ interaction was stable throughout the simulation, as is presented in Figure 3S, which signifies its contribution to the total interaction energy. It is to be noted that no significant hydrogen bonding contacts are observed between DZN and HbA during the MD simulation. The last 3 ns of simulation contains 0–1 hydrogen bonding interactions between HbA and DZN.
Table 2.
List of dynamic parameters and interaction energies observed in free and DZN bound HbA averaged over the last 3ns of the simulation. RMSD and Rg are averaged over the last 3ns of the simulation while the interaction energies are averaged over last 2ns of the simulation.
| RMSD (nm) | Rg (nm) | Coulombic Interaction (kcal/mole) |
LJ Interaction (kcal/mole) |
|
|---|---|---|---|---|
| HbA | 0.53 ± 0.034 | 2.43 ± 0.01 | ||
| HbA-DZN complex | 0.56 ± 0.09 | 2.45 ± 0.035 | −9.95 ± 3.85 | −17.66.± 4.5 |
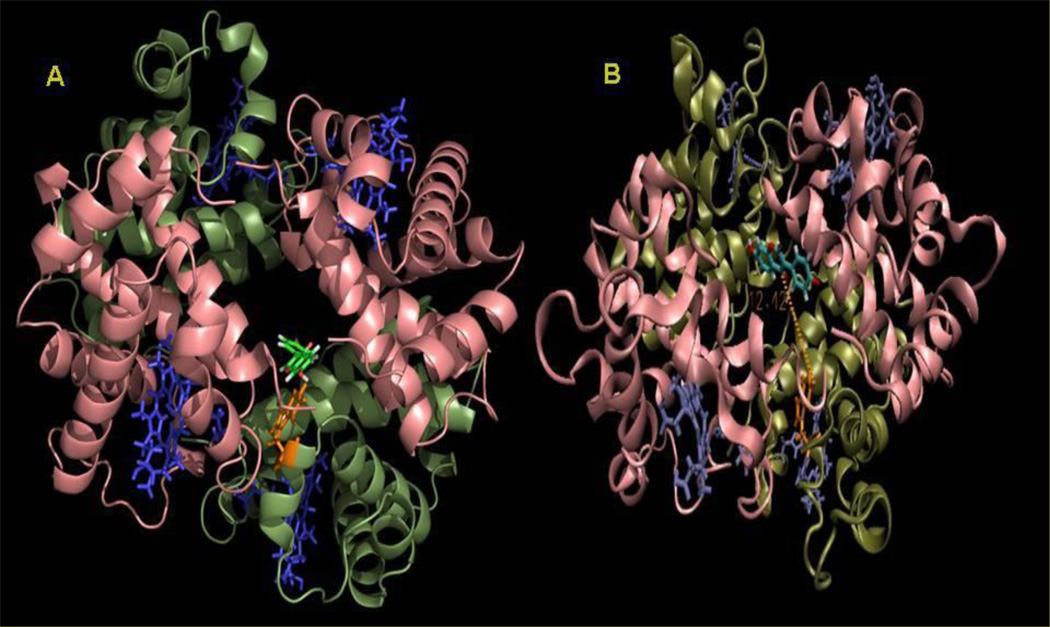
Molecular docking and dynamics was employed to understand the binding of DZN with HbA in atomistic details as well as to identify key interactions involved in the complex formation. The lowest energy docked conformation and the MD average structure starting from the lowest energy docked complex is shown in Figure 5 A & B respectively.
Figure 5.
A. Lowest energy docked conformation of diadzein (shown in green) inside hemoglobin. B. MD average structure of HbA-daidzein complex starting from the lowest energy docked complex obtained from MD simulation. Two α and β subunits are colored pink and dark green respectively. Trp-37 and daidzein are rendered as sticks and colored as orange and light green respectively in B. Porphyrine rings are represented as blue sticks.
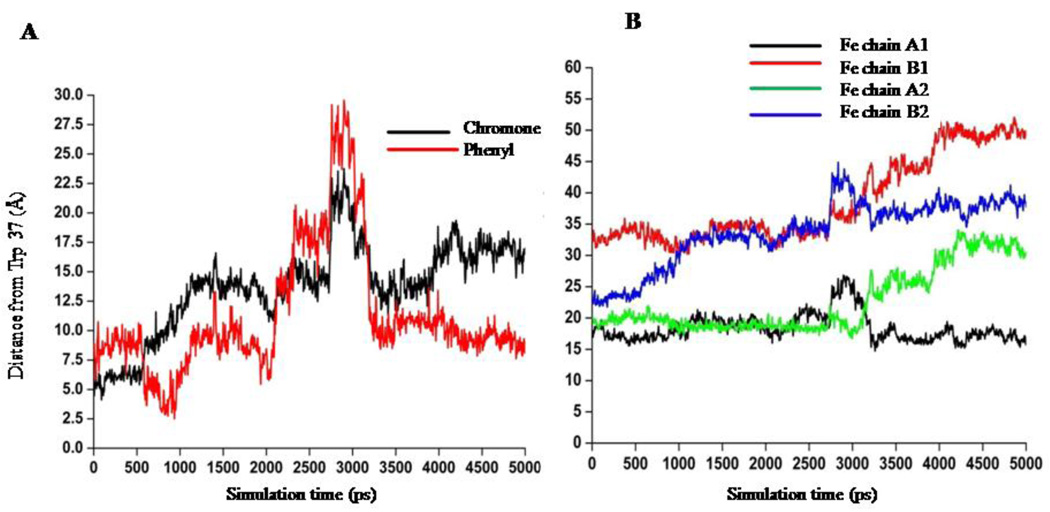
It is clearly evident from Fig. 5 that DZN readily enters the central cavity formed by the four subunits of HbA (α1, α2, β1, β2, shown in pink and dark green, respectively). It is pertinent to mention that this binding site is similar to the binding site of benzimidazole-biphenyl derivatives evident from docking and MD simulation [5, 26] and also the binding site for the effector RSR-13 which is further confirmed by high-resolution crystallography [36]. Initially, the DZN binds very close to the Trp-37, evident from the docking study (Figure 5A), moves a little apart during the MD simulation and stabilizes ~12 Å away from the Trp-37 residue, as is shown in Figure 5B. During the MD simulation DZN moves towards the central cavity of the HbA formed by the four subunits. The phenyl and the chromone ring of DZN becomes more planar compared to the conformation of the bound DZN obtained from docking study. We further analyzed the orientational dynamics of the DZN with respect to Trp 37 as shown in Figure 6A. Here the 4’-OH group has been taken as representative of the ‘phenyl’ moiety of DZN while the C(4)=O group has been considered as a representative of the chromone moiety of DZN. In the lowest energy docked complex, DZN docked very close to the Trp-37 with the chromone moiety of the DZN orienting towards Trp-37 that flipped during the MD simulation and the ‘phenyl’ ring approaches Trp-37. During MD simulation, DZN positioned itself at the center of the HbA central cavity and stabilizes with its ‘phenyl’ ring ~ 8 Å and the chromone ring ~12–15 Å away from the Trp-37 residue. In fact, in the MD averaged structure, the chromone ring of daidzein is 12.42 Å away from the Trp-37. This further confirms that the nearby Trp-37 serves as good optical probe for monitoring DZN binding to HbA.
Figure 6.
A: Orientation dynamics of daidzein with respect to Trp 37 (β1) obtained from MD simulation. 4’-OH group has been taken as representative of the ‘phenyl’ moiety of DZN while the C(4)=O group has been considered as a representative of the chromone moiety of DZN. B: Minimum distance between DZN and the four iron atoms of the heme groups of HbA obtained from MD simulation.
Another important aspect we observed from our MD simulation is the distance between DZN and the iron atoms of the heme moieties. We calculated the minimum distances between DZN and the Fe atom of the four heme groups during MD simulation and results are shown in Figure 6 B. DZN binds between the subunits of HbA, which is sufficiently away from the heme. The closest distance of DZN to any of the four heme group is 15–17 Å away (heme group of α1), emphasizing the fact that the drug does not interfere with oxygen binding of HbA, which has already been concluded from the unaltered Soret band in the absorption spectra of diadzein bound HbA in Figure 1.
Discussion
UV-visible absorption spectroscopy is a simple method used for the exploration of structural changes and the understanding of complex formation (binding). The absorption spectrum of HbA in presence of DZN in Figure 1 shows the appearance of bands in the region where free DZN exhibits absorption bands (right axis), suggesting that DZN is interacting (binding) with hemoglobin in the ground state. It is noteworthy that the Soret band at 406 nm, which is the heme group absorption of HbA, is unaffected by the complex formation. This is in accordance with the results obtained from the MD study where the DZN binding site is sufficiently away from the heme groups of HbA (Figure 6). The closest distance of diadzein to any of the four heme group is 15–17 Å away (heme group of α1), emphasizing the fact that the DZN does not interfere with oxygen binding of HbA which is the essential biological functioning of the protein. The significant increase in fluorescence (Figure 1, inset) of DZN in the protein matrix compared to that in the aqueous phase, supports the binding interaction between HbA and DZN, as was observed in Figure 1. With increasing DZN concentration, a gradual decrease in protein fluorescence without a shift in emission wavelength was observed (refer to Figure 1S). This suggests that DZN is binding in close proximity to the β-Trp-37 residue inside the HbA matrix and its binding does not change the microenvironment. In fact computational analysis further depicts that DZN binds within the central cavity formed by four subunits of HbA and the chromone moiety of DZN is only 12.42 Å away from the β-Trp 37 residue. Initially, the chromone moiety of the DZN orients towards Trp-37 residue as was observed in the docking study, which gradually moves away during the MD simulation and stabilizes at a distance of ~12–15 Å from the Trp-37 residue. Thus Trp-37 serves as good optical probe for monitoring DZN binding to HbA. The quenching plot in Figure 2A illustrates that the fluorescence quenching in the DZN-HbA complex is in good agreement with the linear Stern-Volmer equation until ~ 1.3 × 10−5 M. However, at higher flavonoid concentrations (> 13 µM), the Stern Volmer plots become non-linear which may be attributed to two complicating factors: (i) existence of more than one type of quenching mechanisms at higher flavonoids concentrations, (ii) attenuation of tryptophan fluorescence due to an inner filter effect arising from significant absorption of flavonoids in the tryptophan emission region with these (> 13 µM) quencher (flavonoid) concentrations. Hence, further analysis has been restricted beyond this flavonoid concentration range. The typical fluorescence emission spectra of protein HbA in presence of different concentrations of diadzein is presented in Figure 1S where the λex = 280 nm. It is evident from the figure that with increasing diadzein concentrations the protein emission decreases, simultaneously diadzein emission (although low at λex= 280 nm) increases, which supports the occurrence of non-linear Stern Volmer plots at diadzein concentration > 13 × 10 −6 M. The inset of Figure 1S provides the excitation profiles of HbA (λem= 340 nm, dotted line) as well as diadzein (λem= 470 nm, solid line) in the diadzein bound protein solution. Here the excitation wavelength of HbA was found to be ~ 280 nm (the normal region where Trp absorbs) which again supports the fact that DZN is not altering the protein’s inherent structure as is depicted in the CD spectrum (Figure 4, inset).
The KSV value obtained is ca. 0.98 × 104 M−1 at 25°C for DZN, which indicates that HbA tryptophan fluorescence is efficiently quenched by the flavonoid. Two possible mechanisms are responsible for this observed quenching, static and dynamic quenching. Both mechanisms may appear in a quenching process, but usually one of them has a larger contribution to the total quenching effect. Static quenching occurs by ground state interactions between fluorophore and quencher, resulting in the formation of a non-fluorescent complex. This type of quenching does not rely on diffusion or molecular collisions [21]. The difference in the absorption spectra of free (solid line) and DZN bound HbA (- - -, -.-.-) in Figure 1 clearly provides the evidence of ground state complex formation of DZN with the protein. This gives rise to static quenching of Trp fluorescence in presence of DZN. On the other hand dynamic quenching process is due to the collisions that appear between fluorophore and quencher molecules in the excited state. Since higher temperatures lead to larger diffusion coefficients, dynamic quenching constants are expected to increase with growing temperature. At the same time, at high temperatures, dissociation of weakly bound molecules takes place, decreasing the contribution of static quenching [31, 37]. Figure 2B shows the temperature dependence of KSV where a positive correlation was observed, suggesting dynamic quenching mechanism also to participate. This is because higher temperatures result in faster diffusion and hence larger amounts of collisional quenching. The latter is the experimental evidence that the tryptophan fluorescence quenching by DZN is also governed by dynamic collisions rather than solely by ground state complex formation.
The negative value of free energy change (ΔG0, Table 1) obtained from the binding constant in Figure 2 (following eqn. 4) indicates the thermodynamic favorability of the binding process. The positive values for both enthalpy and entropy change are associated with hydrophobic interactions [33]. A positive value for ΔS0 characterizes hydrophobic interactions, due to the tendency of water to form a more ordered structure near non-polar hydrocarbon groups. Positive values of both enthalpy change and entropy change indicate that hydrogen bond formation is less dominant than hydrophobic interactions [33]. This hydrophobically driven reaction is rather an exception than a rule for the majority of associations involving proteins [33]. Interaction energy analysis by molecular docking and dynamics also reveals a similar trend. Van der Waals interactions are found to be the main determining factor for the complex formation evident from both docking and dynamics studies. The average Lennard-Jones interactions are stable throughout the simulation with a value of −17.66.± 4.5 kcal/mole averaged over the last 2 ns of the simulation. Hydrogen bonding contacts were not observed between DZN and HbA during the MD simulation and an insignificant contribution of Coulombic interaction (with average value of −9.95 ± 3.85 kcal/mole) was predicted from the molecular dynamics study which never stabilized over the simulation period of the interaction energy analyses, further supporting the experimental observation that the binding process was mainly driven by hydrophobic interactions.
The high values of fluorescence anisotropy ‘r’ of DZN at high concentrations of HbA in Figure 4 suggest that the drug molecules are bound in a motionally constrained environment in the protein matrix [38]. The observed hydrophobic interaction between the ligand DZN and the protein supported the high ‘r’ value of DZN. Trp fluorescence spectroscopy provides a simple and reliable means to characterize the Trp microenvironment in a protein which is related to the tertiary conformation of proteins in solution [39]. The increasing fluorescence intensity in Figure 2S indicates a more rigid environment around the Trp residue [40, 41] with increase in protein concentration. Intrinsic Trp fluorescence is a function of the polarity of the microenvironment around the Trp and the change in the polarity affects the of the Trp fluorescence spectrum. A blue shift indicates the presence of a hydrophobic environment around the Trp or in other words the Trp residue being deeply embedded in the protein tertiary structure [10, 37–39]. We have postulated only tetramers to be present in appreciable abundance at the studied concentration (1 × 10−5 M) of HbA [10, 42] and analyzed the data in terms of the linkage scheme for such a system using computational approaches as discussed below.
The DZN molecule particularly binds to the interface cavity formed by α1, α2 and β1 chains and closely interacts with the Trp-37 residue in the β-chains of HbA, which is the fluorophoric moiety of HbA, as is depicted in Figure 5. Figure 5A also indicates that DZN is docked inside HbA in such a way that its B-ring is slightly tilted over the remaining chromone plane formed by the (A+C) rings of polyphenols (see Scheme 1). Docking interaction energy reveals that the main binding forces of this complexation are van der Waals interactions while a negligible portion of interactions is contributed by electrostatic interaction.
We further explored the effect of DZN binding on the secondary and tertiary structural profile of HbA. Far-UV CD spectroscopy in combination with molecular dynamics study reveals that DZN binding to HbA induces no significant change in the secondary structure of HbA. Both free and DZN bound HbA exist in a predominately α-helical structure. Effect of DZN binding on the tertiary structure and size-shape distribution of HbA have been carried out by molecular dynamics simulation. RMSD and radius of gyration (Rg) are two very important parameters that provide a quantitative description of tertiary structure of the simulated system. Observed RMSD for free and DZN bound HbA are highly similar, 0.53 ± 0.034 nm and 0.56 ± 0.09 nm while the calculated Rg value for free and DZN bound HbA are 2.43 ± 0.01 nm and 2.45 ± 0.035 nm, respectively. Essentially unchanged values of RMSD, Rg along with CD profiles of free and ligand bound HbA signify no significant structural change of the carrier protein HbA in the presence of DZN.
Concluding remarks
The results clearly show the usefulness of the present study in exploiting the natural drug carrier hemoglobin for binding with the therapeutically important isoflavonoid diadzein, while utilizing the sensitive and discriminating emission properties of the drug. Since diadzein is neutral in nature, ionic interactions between protein and ligand can be ruled out. Instead, the results indicate that the binding interactions can be attributed to hydrophobic interactions. The molecular docking and dynamics studies are valuable in providing two important evidences: 1. β-Trp-37 serves as good indicator for the microenvironment of diadzein inside the protein matrix. 2. The drug diadzein does not interfere with the functionality of the heme group of porphyrin of hemoglobin, especially in oxygen binding. Furthermore, the far UV-CD study reveals that the overall secondary structure of the protein remains identical in the presence of the drug, validating the fact that binding of diadzein essentially does not produce any harm to the protein. Hence, hemoglobin, a natural carrier molecule of drugs, can be successfully utilized in medicinal biology for delivering flavonoid based therapeutics to the physiological targets.
Supplementary Material
Research Highlights.
-
►
Binding of isoflavonoid diadzein with human hemoglobin was investigated.
-
►
Spectroscopic, molecular docking and dynamics studies were carried out.
-
►
β-Trp-37 serves as good indicator for the microenvironment of diadzein inside the protein.
-
►
Diadzein does not interfere with the functionality of the heme of hemoglobin.
-
►
Overall secondary structure of the protein remains unchanged in the presence of the drug.
Acknowledgements
BSG and PKB like to thank the financial support from NIH/ NCMHHD/RIMI grant # 1P20MD002725. BSG thanks the research support from HBCU-UP Grant, NSF ID: 0811638 and JHS program with the contract # N01-HC-95172 at Tougaloo College. We thank Prof. P. K. Sengupta, SINP, Kolkata, India, for giving access to HYPERCHEM software. We thank Dr. Jeff Petty of Chemistry Department, Furman University for allowing us to use the fluorimeter for the anisotropy studies. PKB thanks the financial support from MS-INBRE (#USM-GR04015-05-9; NIH/NCRR #P20RR016476).WHK acknowledges financial support by Grant Number P20RR016476 from the National Institute of General Medical Sciences, National Institutes of Health (NIH), through the Mississippi INBRE at The University of Southern Mississippi (USM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental data available
Figure 1S, 2S and 3S are provided in the supplemental document.
References
- 1.Patel PD, Dand N, Hirlekar RS, Kadam VJ. Drug loaded erythrocytes as novel drug delivery system. Curr. Pharm. Des. 2008;14:63–70. doi: 10.2174/138161208783330772. [DOI] [PubMed] [Google Scholar]
- 2.Alanzi FK, Harisa GE-DI, Maqboul A, Hamid MA, Neau SH, Alsarra IA. Biochemically Altered Human Erythrocytes as a carrier for Targeted Delivery of Primaquine: an in vitro study. Ach. Pharm. Res. 2011;34:563–571. doi: 10.1007/s12272-011-0406-7. [DOI] [PubMed] [Google Scholar]
- 3.Hamidi M, Tajerzadeh H, Dehpour AR, Rouini MR, Ejtemaee-Mehr S. In vitro characterization of intact human erythrocytes loaded by enalaprilat. Drug. Delivery. 2001;8:223–230. doi: 10.1080/107175401317245903. [DOI] [PubMed] [Google Scholar]
- 4.Adams T, Alanzi F, Lu DR. Safety and utilization of blood components as therapeutic delivery systems. Curr. Pharm. Biotechnol. 2003;4:275–282. doi: 10.2174/1389201033489720. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Shi R, Yang C, Zhu X. Exploration of the binding of benzimidazole-biphenyl derivatives to hemoglobin using docking and molecular dynamics simulation. Int. J. Biol. Macromol. 2011;48:20–26. doi: 10.1016/j.ijbiomac.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Prokop A, Davidson JM. Nanovehicular Intracellular Delivery Systems. J. of Pharm. Sc. 2008;97:3518–3595. doi: 10.1002/jps.21270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brookes S, Biessels P, Ng NFL, Woods C, Bell DN, Adamson G. Synthesis and Characterization of a hemoglobin-Ribavarin conjugate for targeted drug delivery. Bioconjugate Chem. 2006;17:530–537. doi: 10.1021/bc0503317. [DOI] [PubMed] [Google Scholar]
- 8.Ackers GK, Johnson ML, Mills FC, Ip SHC. Energetics of oxygenation linked subunit interactions in human hemoglobin. Biochem. Biophys. Res. Commun. 1976;69:135–142. doi: 10.1016/s0006-291x(76)80283-5. [DOI] [PubMed] [Google Scholar]
- 9.Doyle ML, Holt JM, Ackers GK. Effects of NaCl on the linkages between O2 binding and subunit assembly in human hemoglobin: titration of the quaternary enhancement effect. Biophysical Chemistry. 1997;64:271–287. doi: 10.1016/s0301-4622(96)02235-1. [DOI] [PubMed] [Google Scholar]; Ackers GK. Energetics of subunit assembly and ligand binding in human hemoglobin. Biophys. J. 1980;32:331–343. doi: 10.1016/S0006-3495(80)84960-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang X, Chou J, Sun G, Yang H, Lu T. Synchronous Fluorescence Spectra of Hemoglobin: A Study of Aggregation States in Aqueous Solutions. Microchem. J. 1998;60:210–216. [Google Scholar]
- 11.Dwiecki K, Neunert G, Polewski P, Polewski K. Antioxidant activity of daidzein, a natural antioxidant, and its spectroscopic properties in organic solvents and phosphatidylcholine liposomes. J. Photochem. Photobiol. B: Biology. 2009;96:242–248. doi: 10.1016/j.jphotobiol.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Fengpei D, Peng B, Lu R, Gao H, Zhou Z. Structure, electronic properties, and radical scavenging mechanisms of daidzein, genistein, formononetin, and biochanin A: A density functional study. J. Mol. Struct.: Theochem. 2010;955:1–6. [Google Scholar]
- 13.Setchell KDR, Cassidy A. Dietary isoflavones: biological effects and relevance to human health. J. Nutr. 1999;129:758–767. doi: 10.1093/jn/129.3.758S. [DOI] [PubMed] [Google Scholar]
- 14.Jacobsen BK, Knutsen SF, Fraser GE. Does high soy milk intake reduce prostate cancer incidence? The Adventist Health Study (United States) Cancer Causes Control. 1998;9:553–557. doi: 10.1023/a:1008819500080. [DOI] [PubMed] [Google Scholar]
- 15.Esselen M, Boettler U, Teller N, Bachler S, Hutter M, Rufer CE, Skrbek S, Marko D. Anthocyanin-rich blackberry extract suppresses the DNA-damaging properties of topoisomerase I and II poisons in colon carcinoma cells. J. Agric. Food Chem. 2011;59:6966–6973. doi: 10.1021/jf200379c. [DOI] [PubMed] [Google Scholar]
- 16.Setchell KDR, Brown NM, Desai PB, Zimmer-Nechimias L, Wolfe B, Jakate AS, Creutzinger V, Heubi JE. Bioavailability, Disposition, and Dose-Response effects of Soy Isoflavones when consumed by healthy women at physiologically typical dietary intakes. J. Nutr. 2003;133:1027–1035. doi: 10.1093/jn/133.4.1027. [DOI] [PubMed] [Google Scholar]
- 17.Prochazkova D, Bousova I, Wilhelmova N. Antooxidant and prooxidant properties of flavonoids. Fitoterapia. 2011;82:513–523. doi: 10.1016/j.fitote.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 18.Manach C, Mazur A, Scalbert A. Polyphenols and prevention of cardiovascular diseases. Curr Opin Lipidol. 2005;16:77–84. doi: 10.1097/00041433-200502000-00013. [DOI] [PubMed] [Google Scholar]
- 19.Williamson G, Holst B. Dietary reference intake (DRI) value for dietary polyphenols: are we heading in the right direction? Br J Nutr. 2008;99(Suppl 3):S55–S58. doi: 10.1017/S0007114508006867. [DOI] [PubMed] [Google Scholar]
- 20.Mason JN, Farmer H, Tomlinson ID, Schwartz JW, Savchenko V, DeFelice LJ, Rosenthal SJ, Blakely RD. Novel fluorescence-based approaches for the study of biogenic amine transporter localization, activity, and regulation. J. Neurosc. Methods. 2005;143:3–25. doi: 10.1016/j.jneumeth.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 21.a Lakowicz JR. Principles of fluorescence spectroscopy. New York: Springer; 2006. [Google Scholar]; b Gratzer WB. Med. Res. Holly Hill, London: Council Labs; [Google Scholar]
- 22.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarkian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- 23.Hyperchem. USA: Hypercube Inc.; 2002. [Google Scholar]
- 24.Lindahl E, Hess B B, Spoel DVD. GROMACS 3.0 a package for molecular simulation and trajectory analysis. J. Mol. Model. 2001;7:306–317. [Google Scholar]
- 25.Jorgensen WL, Rives T. Development and testing of the OPLS all-atom force field on conformational energetic and properties of organic liquids. J. Am Chem Soc. 1988;110:1657–1666. [Google Scholar]
- 26.(a) Biswas PK, Gogonea VA. Journal of Chemical Physics. 2005;123:164114–164122. doi: 10.1063/1.2064907. [DOI] [PubMed] [Google Scholar]; (b) Car R, Parrinello M. Physical Review Letter. 1985;55:2471–2474. doi: 10.1103/PhysRevLett.55.2471. [DOI] [PubMed] [Google Scholar]
- 27.Gogonea V, Shy JM, II, Biswas PK. Electronic Structure, Ionization Potential, and Electron Affinity of the Enzyme Cofactor (6, R)-5,6,7,8-Tetrahydrobiopterin in the Gas Phase, Solution, and Protein Environments. J. Phys. Chem. B. 2006;110:22861–22871. doi: 10.1021/jp061653q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. J. Comp. Chem. 1997;18:1463–1472. [Google Scholar]
- 29.Kang J, Liu Y, Xie M, Li S, Jiang M, Wang Y. Interactions of human serum albumin with chlorogenic acid and ferulic acid. Biochim. Biophys. Acta. 2004;1674:205–214. doi: 10.1016/j.bbagen.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 30.Xie MX, Xu XY, Wang YD. Interaction between hesperetin and human serum albumin revealed by spectroscopic methods. Biochim Biophys Acta. 2005;1724:215–224. doi: 10.1016/j.bbagen.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 31.Yu X, Liu R, Yi R, Yang F, Huang H, Chen J, Ji D, Yang Y, Li X, Yi P. Study of the interaction between N-confused porphyrin and bovine serum albumin by fluorescence spectroscopy. Spectrochim. Acta Part A. 2011;78:1329–1335. doi: 10.1016/j.saa.2011.01.024. [DOI] [PubMed] [Google Scholar]
- 32.Hegde AH, Sandhya B, Seetharamappa J. Evaluation of binding and thermodynamic characteristics of interactions between a citrus flavonoid hesperitin with protein and effects of metal ions on binding. Mol. Biol. Rep. 2011;38:4921–4929. doi: 10.1007/s11033-010-0634-9. [DOI] [PubMed] [Google Scholar]
- 33.Ross DP, Subramanian S. Thermodynamics of protein association reactions: forces contributing to stability. Biochemistry. 1981;20:3096–3102. doi: 10.1021/bi00514a017. [DOI] [PubMed] [Google Scholar]
- 34.Hirsch RE, Nagel RL. Conformational studies of hemoglobins using intrinsic fluorescence measurements. J. Biol. Chem. 1981;256:1080–1083. [PubMed] [Google Scholar]
- 35.Chen Q, Hirsch RE. Quantification of effector binding to the hemoglobin central cavity by intrinsic and extrinsic steady-state fluorescence. J. of Fluorescence. 2003;13:25–31. [Google Scholar]
- 36.Abraham DJ, Wireko FC, Randad RS. Biochemistry. 1992;31:9141–9149. doi: 10.1021/bi00153a005. [DOI] [PubMed] [Google Scholar]
- 37.Gentili PL, Ortica F, Favaro G. Static and dynamic interaction of a naturally occurring photochromic molecule with bovine serum albumin studied by UV-visible absorption and fluorescence spectroscopy. J. Phys. Chem. B. 2008;112:16793–16801. doi: 10.1021/jp805922g. [DOI] [PubMed] [Google Scholar]
- 38.Samanta A, Paul BK, Guchhait N. Spectroscopic probe analysis for exploring probe–protein interaction: A mapping of native, unfolding and refolding of protein bovine serum albumin by extrinsic fluorescence probe. Biophys. Chem. 2011;156:128–139. doi: 10.1016/j.bpc.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 39.Sharma VK, Kalonia DS. Steady-state tryptophan fluorescence spectroscopy study to probe tertiary structure of proteins in solid powders. J. of Pharm. Sc. 2003;92:890–899. doi: 10.1002/jps.10354. [DOI] [PubMed] [Google Scholar]
- 40.Wright WW, Guffanti GT, Vanderkooi JM. Protein in Sugar Films and in Glycerol/Water as Examined by Infrared Spectroscopy and by the Fluorescence and Phosphorescence of Tryptophan. Biophysics J. 2003;85:1980–1995. doi: 10.1016/S0006-3495(03)74626-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eftink MR. The use of fluorescence methods to monitor unfolding transitions in proteins. Biophys J. 1994;66:482–501. doi: 10.1016/s0006-3495(94)80799-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mills FC, Johnson ML, Ackers GK. Oxygenation-Linked Subunit Interactions in Human Hemoglobin: Experimental Studies on the Concentration Dependence of Oxygenation Curves. Biochemistry. 1976;15:5350–5362. doi: 10.1021/bi00669a023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.