Abstract
Copy number variation (CNV) is becoming increasingly important as a feature of human variation in disease susceptibility studies. However, the consequences of copy number variation are not so well understood. Here we present data exploring the functional consequences of copy number variation of CCL3L1 in 55 independent UK samples with no known clinical phenotypes. Copy number of CCL3L1 was determined by the paralogue ratio test (PRT), and expression levels of MIP-1α and mRNA from stimulated monocytes were measured and analysed. The data show no statistically significant association of MIP-1α protein levels with copy number. However, there was a significant correlation between copy number and CCL3L1:CCL3 mRNA ratio. The data also provide evidence that expression of CCL3 predominates in both protein and mRNA, and therefore the observed variation of CCL3 is potentially more important biologically than that of copy number variation of CCL3L1.
Keywords: CCL3L1, MIP-1α, gene expression, copy number
Introduction
Copy number variation (CNV) is a frequent form of variation throughout the human genome. Over the last few years much research has been carried out to show the significant contribution of CNV to the variation observed between individuals and their influence upon disease susceptibility.
Whilst copy number variation has been shown to contribute to the heritable variation in human gene expression1, the full functional impact of CNVs is not completely recognized. At a genome-wide level copy number variation can directly alter the expression level of genes within the copy variable region1; 2. However, there is less progress in understanding the functional consequences of specific copy variable loci, particularly those CNVs that are multi-allelic.
The CCL3L1/CCL4L1 copy variable region is located on chromosome 17q123, and extends over 90kb. Each repeat unit contains a single copy of CCL3L1 and CCL4L1 and is flanked by the gene TBC1D3. The repeat region is situated adjacent to two very closely related, and commonly copy invariant genes CCL3 and CCL4, and is thought to have evolved by duplication of the invariant CCL3 and CCL4 region and successive divergence4. Consequently, members of each paralogous pair exhibit a high degree (96%) of nucleotide and protein similarity, with CCL3 and CCL3L1 being identical at 67/70 residues in the mature protein and 747/780 sites in the coding sequence, and CCL4 and CCL4L1 being identical at 68/69 resides in the mature protein and 644/667 sites in the coding sequence.
The genes CCL3 and CCL3L1 encode macrophage inflammatory protein (MIP)-1α, isoforms LD78α and LD78β respectively. MIP-1α is a low molecular weight β-chemokine. The chemokine acts as a pro-inflammatory cytokine, inducing a wide variety of immune cells, particularly CD8+ T cells and immature dendritic cells, and being inhibited by IL-4, IL-10 and IL-135; 6. It is secreted from most mature leucocytes, predominantly macrophages, in response to stimulus, and functions by attracting lymphocytes and macrophages to sites of infection and inflammation, with the isoform LD78β being 2-fold more efficient at chemoattracting human monocytes and lymphocytes than the LD78α isoform7. For this reason copy number variation of CCL3L1 has the potential to influence both immunological disorders and auto-immunity.
Furthermore MIP-1α is a natural ligand for CCR5, the co-receptor used by HIV-1 virus for cell entry8; 9. The isoform LD78β has been shown to be the most potent agonist for CCR57, with a superior antiviral activity10 and a 10-fold higher EC50 in inhibiting viral replication than LD78α7. Copy number variation of CCL3L1 has thus been suggested to limit HIV-1 entry into cells11, and is implicated in HIV-1 progression10; 12-18, although this is disputed19-22.
The aim of the study was to explore the functional consequences of the multi-allelic copy variable gene CCL3L1. Expression of both CCL3 and CCL3L1 mRNA was measured and compared and also the variation in expression level in relation to the diploid copy number was assessed. Additionally, the variation of MIP-1α protein levels relative to copy number was investigated.
Results
Variation in copy number
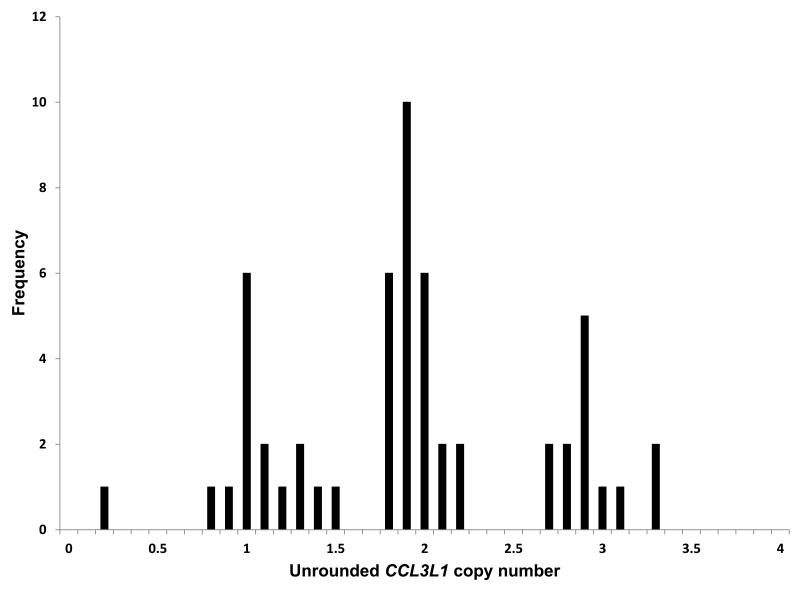
Copy number measurement of CCL3L1 by PRT was performed on all 55 independent UK samples. The 3 independent PRT systems assigned concordant copy numbers for 51/55 samples (to within 0.5 of the integer value for 75% of samples and to within 0.75 for 93%). The 4 samples that showed some discordance were typed with the two microsatellite assays which allowed confident integer copy number calling23. The calibrated unrounded copy number distribution is shown in figure 1 and shows that the samples all cluster around the predicted integer with discernable gaps. Variation in CCL3L1 copy number between 0-3 was observed (see table I), with a copy number of 2 being the most common. As the three different PRT measurement systems show a high level of agreement in all samples typed this provides no evidence for differences in the copy number between CCL3L1 and CCL4L1, nor of copy number variation in the reference genes CCL3 and CCL4.
Figure 1.
Distribution histogram of the unrounded CCL3L1 copy number in 55 unrelated UK samples generated using PRT. The distribution shows a range of 0-3 copies, with peaks centred on the integer values and gaps between clusters.
Table 1.
Integer copy number distribution of the 55 samples
| Copy number | Number of samples |
|---|---|
| 0 | 1 |
| 1 | 15 |
| 2 | 26 |
| 3 | 13 |
| TOTAL | 55 |
Correlation of copy number with protein production
In order to deduce a suitable time point to measure protein expression, monocytes were isolated from an initial cohort of 7 individuals, and supernatants collected at 2, 4, 8, 24 and 48 hours post LPS stimulation. Protein was measured by ELISA, with a lower detection limit of 0.1ng/ml, and despite a single outlier sample, the time course shows an initial increase in protein production over the first 4 hours, which levelled off by 8 hours and then declined gradually for all copy numbers (see supplementary figure 1). There was no detectable protein expression from the unstimulated cells. This is as expected as MIP-1α is not constitutively expressed but is induced upon stimulation. For measurement of protein levels all cell supernatants were subsequently harvested 4 hours post stimulation and MIP-1α production from macrophages was measured in triplicate for all samples.
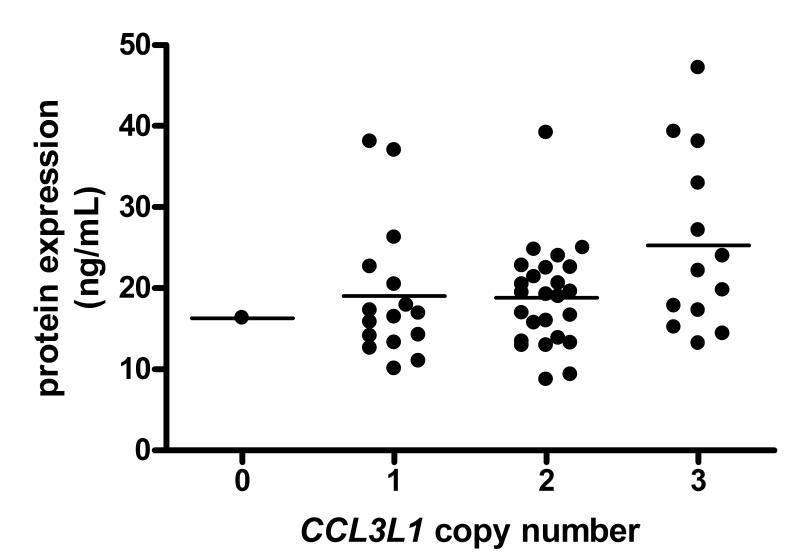
There is a high level of sequence similarity between the two isoforms of MIP-1α that are encoded by CCL3 and CCL3L1, and we are not aware of any commercially available antibodies that can reliably distinguish between them. As our dataset contained one individual who had a copy number of zero (zero copy individuals make up approximately 2% of the UK population) we were able to measure the specificity of a number of commercial antibodies (both monoclonal and polyclonal) (AbCam, Cambridge, UK; R&D systems, Abingdon, UK; Lifespan Biosciences, Seattle, WA) for the LD78α isoform. There were no antibodies tested that were completely specific to LD78β and thus all available antibodies measured both MIP-1α isoforms. Therefore the ELISA results presented in figure 2 are for total MIP-1α expression, as with other previously published reports that also only measure total MIP-1α expression24. Figure 2 shows MIP-1α expression grouped by gene copy number, together with the stratified means. Whilst a CCL3L1 copy number of 3 has a greater spread of expression variability (S.D.=10.93), there was no significant difference in mean MIP-1α expression level between copy numbers. There was, however suggestive evidence for a weak correlation of MIP-1α expression and copy number (r=0.2707; p=0.0309).
Figure 2.
MIP-1α protein expression levels (ng/mL) classified by integer CCL3L1 copy number.
Furthermore, considering that the zero copy sample corresponds to CCL3-encoded protein expression only, at least for this individual it shows the background level of expression of the copy constant CCL3 protein, LD78α, against which other samples can be compared. Therefore, in comparing the MIP-1α expression level for all copy number samples to that of the zero copy samples, the data actually suggests that the majority of the expressed MIP-1α protein is composed of the LD78α isoform. Additionally, within all 3 classes of copy number, at least some individuals have a level of expression for total MIP-1α less than that observed with the zero copy sample, suggesting that the expression of LD78α is also variable.
The sandwich ELISA was used to measure MIP-1α expression in sera at the time of extraction. There was no protein detected in any of the samples, with a lower detection limit of 0.1ng/ml.
To look at the variation in protein expression within each individual 10 volunteers from the 55 agreed to be re-tested. These 10 repeat samples comprise 4 one-copy individuals, 3 two-copy individuals and 3 three-copy individuals. A comparison of the original MIP-1α expression and the expression in the repeat samples is shown in supplementary figure 2a. The data show a single individual, sample F, to have a very high MIP-1α expression in the original experiment. However, for this individual there was also evidence for MIP-1α expression in the unstimulated cells, potentially due to clinical symptoms, although not symptomatic at the time of blood extraction. For this individual only the repeat sample data has been used in the further analysis. It should be noted that sample F was the only one that had any protein expression in the unstimulated cells. Analysis of repeat measures found that neither of the within subjects factors (time and measurement) were significant for measurement of MIP-1α expression.
Correlation of copy number with mRNA transcript
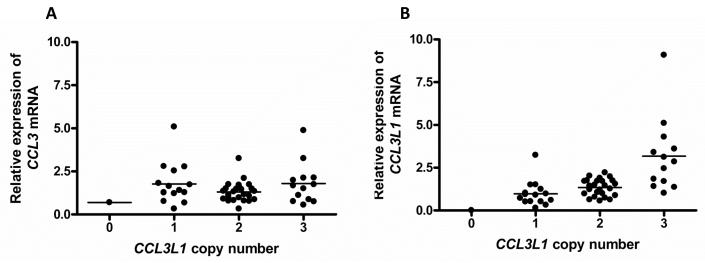
One major advantage of investigating mRNA transcripts is that it is possible to differentiate between the transcripts of CCL3 and CCL3L1 by use of sequence-specific primers. Again a time trial was performed on an initial 7 samples with total RNA being isolated from the lysed macrophages 2, 4, 8, 24 and 48 hours post stimulation with LPS, and subsequent measurement of CCL3L1 by real time PCR. There was an initial high expression of CCL3L1 specific mRNA at 2 hours post stimulation that declined sharply over time (supplementary figure 1b). Consequently all RNA was collected 2 hours post stimulation. Without stimulation there was no detectable mRNA production for either CCL3 or CCL3L1. It is also interesting to note that the one-copy sample that had elevated MIP-1α protein expression in the time trial (supplementary figure 1), does not have elevated CCL3L1 mRNA expression. Thus the high MIP-1α expression observed is potentially due to an increased expression of CCL3 mRNA, and indeed observations of CCL3 mRNA expression for these same samples show this particular sample to have a greater than expected level of expression for CCL3 (mean RQ for all 7 samples = 1.34; RQ for this sample = 2.14). Whilst this may suggest that genetic variation may play role through the presence of a promoter polymorphism or enhancer element, targeted upstream sequence analysis did not find any variants (data not shown).
As sequence-specific primers can be designed real-time PCR can be used to measure both CCL3 and CCL3L1 mRNA transcripts individually. Figure 3 shows mRNA expression for both CCL3 and CCL3L1 grouped by copy number with stratified means. Perhaps as expected, the CCL3 mRNA expression shows similar clusters of data for each copy number group and no evidence for a correlation (r=0.052) For each copy number the clusters of data show a spread of expression, levels of between 0 and 5 fold greater expression relative to GAPDH, suggesting that the inherent expression of CCL3 is variable. The data for the CCL3L1 specific transcripts show a statistically significant correlation of expression with copy number (r=0.633; p<0.0001), suggesting CCL3L1 mRNA transcript level increases with higher copy numbers. This also suggests the potential for post-transcriptional regulation of MIP-1α as the correlation with copy number is not preserved in the protein.
Figure 3.
mRNA expression of (A) CCL3 and (B) CCL3L1 by integer CCL3L1 copy number. The mRNA expression levels of both CCL3 and CCL3L1 were calculated using the ΔΔCt method and mRNA expression of GAPDH as the endogenous control.
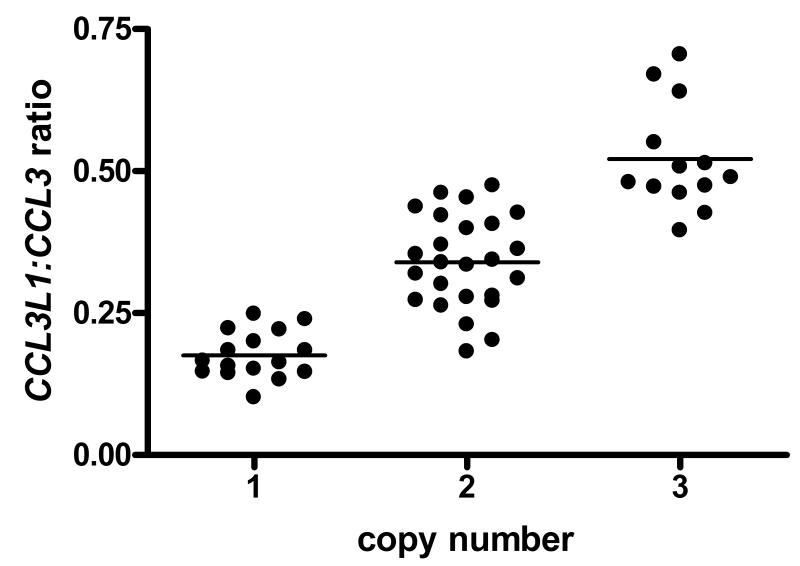
The ratio of CCL3L1:CCL3 mRNA transcript was measured using PRT for all samples at 2 hours post stimulation (see figure 4). The data show a statistically significant correlation (r=0.877, p=0.0001) between copy number and CCL3L1:CCL3 mRNA ratio, with clear clusters for each copy number. Additionally, the CCL3L1:CCL3 ratio data would suggest that there is a greater proportion of CCL3 mRNA transcript than CCL3L1; as the CCL3L1:CCL3 ratio would be predicted to approximate 1 for the 2-copy samples as equal quantities of transcripts would be expected but the data actually give a mean ratio of 0.34. The mean ratio of the one-copy samples is 0.18, and for the three-copy samples is 0.52, altogether suggesting that there are approximately 2-3 times more CCL3 transcripts than CCL3L1, for all copy numbers of CCL3L1 at 2 hours post stimulation.
Figure 4.
Comparison of the ratio of CCL3L1:CCL3 mRNA expression measured by PRT between integer CCL3L1 copy numbers. The bold black bar represents the mean ratio for each copy number.
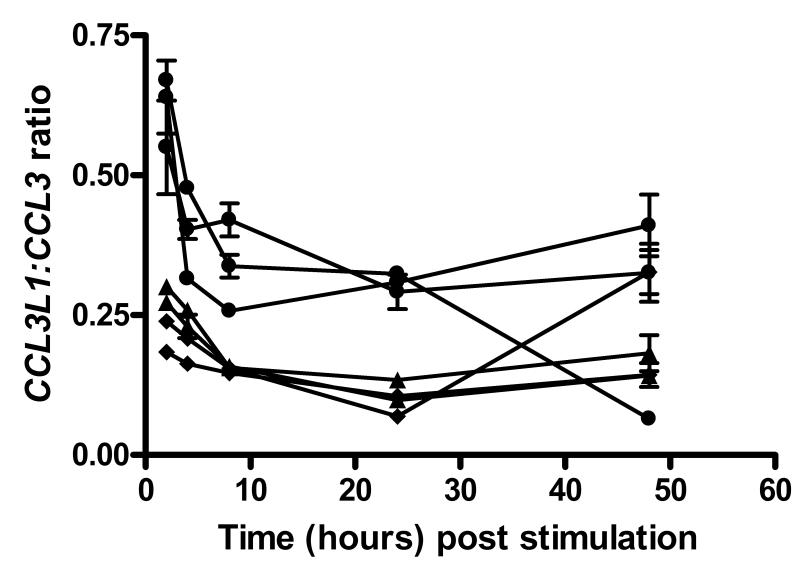
Furthermore, the ratio of CCL3L1:CCL3 was measured by PRT for the 7 time trial samples (see figure 5). The time trial data give evidence for a drop in the ratio of CCL3L1:CCL3 for these samples over the first 12 hours, followed by a relative plateauing of the ratio. Such a drop in the ratio would suggest that the decay rate of the transcripts of CCL3 and CCL3L1 is not equal. The ratio measurements would suggest that it is the decay of CCL3L1 that is faster than that for CCL3, suggesting that over time the CCL3 transcripts can predominate. If we assume that the relative mRNA levels are preserved at the protein level, then as the CCL3 mRNA transcripts predominate, it is most likely that the vast majority of MIP-1α expressed is of the isoform LD78α (figure 6), corroborating our observations with MIP-1α protein. Confirmation by a specific antibody is required, however.
Figure 5.
Time trial graph of CCL3L1:CCL3 mRNA expression, measured by PRT, from isolated macrophages post LPS stimulation for 7 initial samples. These samples comprised 2 one-copy samples ( ), 2 two-copy samples (
), 2 two-copy samples ( ) and 3 three-copy samples (●). There was no expression detected from unstimulated cells.
) and 3 three-copy samples (●). There was no expression detected from unstimulated cells.
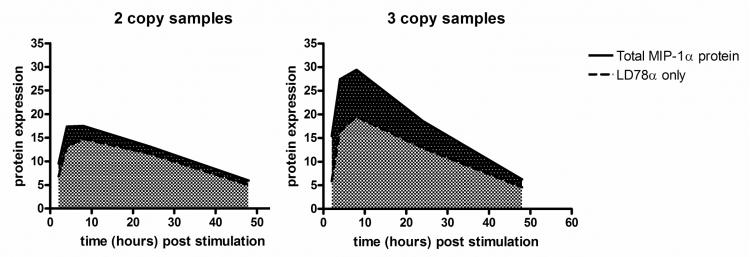
Figure 6.
Graphs for the time trial samples illustrating the proportion of all MIP-1α protein expression that is accounted for by isoform LD78α expression two-copy and three-copy samples, assuming that mRNA levels are preserved in the protein. The area under the curve calculations show that for two copy samples LD78α accounts for 85% of the total expressed protein, and for three copies 67%.
We also observed two samples with elevated mRNA expression for both CCL3 and CCL3L1, one for a one-copy samples and the other for a three-copy sample. However both had a CCL3:CCL3L1 ratio that was within range for the copy number. This could suggest that a potentially shared enhancer or promoter element causes raised expression of both CCL3 and CCL3L1. Further sequence work may identify some variant at the nucleotide level that could influence expression of both mRNAs in these samples.
To look at the consistency of mRNA expression within each individual the 10 repeat samples were evaluated. A comparison of the original CCL3L1 mRNA expression and the expression in the repeat samples is shown in supplementary figure 2b. The repeat measure analysis found that neither of the within subjects factors (time and measurement) were significant for the measurement of CCL3L1 mRNA expression, thought there is some disparity for the 3 three copy samples (H, I, J). However, comparing this data with the ratio data for the repeat samples (supplementary figure 2c), it can be seen that the ratio data do not differ significantly within individuals, and thus the disparity observed in the CCL3L1 mRNA expression for the higher copies is mirrored in the CCL3 expression also.
Discussion
In this study we sought to explore the functional consequences of copy number variation at CCL3L1 through analysis of variation in expression of CCL3L1 mRNA and protein. We found no significant association between copy number and MIP-1α protein expression. Whilst the data presented here are characteristic of a European population, with a narrow range and mean copy number of 2, a population with a higher mean copy number, for example Africans, may be useful to explore protein expression of higher copy numbers. Furthermore as our study suggests that the majority of MIP-1α products are of the isoform LD78α, then it is possible that any significant association between copy number and LD78β expression is being masked by the higher levels of LD78α. However, until a specific antibody can be developed this cannot be proven.
The mature proteins, LB78α and LD78β, differ at only 3 amino acids situated at position 3, 39 and 47. The dispersed distribution of these amino acid substitutions throughout the mature protein does potentially make the generation of a specific antibody to one or other of the isoforms more difficult, though these sites are not evolutionary conserved. Nevertheless the substitution of a serine by proline at position 3 could alter the protein configuration and thus theoretically support the generation of a specific monoclonal antibody.
To the best of our knowledge this is the first study to directly quantify both CCL3 and CCL3L1 mRNA transcripts separately. Whilst the ratio of CCL3L1:CCL3 significantly correlates with copy number, the quantity of the individual mRNA transcripts is not equal, with CCL3 mRNA transcripts predominating. As the protein data also suggest that the CCL3 isoform LD78α predominates, this observation with mRNA is potentially preserved in the protein.
The predominance of LD78α is interesting as it suggests that the variation observed with CCL3 is potentially more consequential biologically than the CCL3L1 copy number variation. Indeed, functionally, whilst LD78β is a more potent agonist than LD78α, LD78α nevertheless has agonist activity, and therefore, even if less potent it can outnumber LD78β 3 to 1. Whilst a prior study has established that LD78β is two-fold more chemoattractive than LD78α7, if the expression of LD78β in a two-copy individual is only 15% of the total MIP-1α expression, then the LD78α isoform will still account for most biological activity. It is thus possible that the variation in expression observed for CCL3 is as likely to influence susceptibility to infectious disease and HIV progression as the copy number variation of LD78β.
Our data does not support a previous study24 which found a significant association of copy number with MIP-1α expression. However, unfortunately it is not possible to directly compare the data as MIP-1α was collected 4 hours post stimulation in this study, whereas it was collected 48 hours post stimulation previously24, a time point which we observed to be poorly correlated with overall effect (see supplementary figure 1). Furthermore, whilst their ELISA system was the same as ours, the prior study did not contain any zero copy samples with which to compare their background levels of CCL3 encoded protein. Whilst the previous study did not specifically look at the separate mRNA transcripts, the ratio of CCL3L1:CCL3 was assessed and found to be significantly correlated with copy number, which is in agreement with our observations, though we cannot assess whether, like our data, there is a predominance of CCL3 transcript or not.
There have been a number of studies that have found associations between copy number variation of CCL3L1 and HIV-1 progression10; 13-16: the biological rationale is that there is strong competition between the MIP-1α isoform LD78β and HIV-1 for the receptor CCR5; thus the more LD78β there is, the greater the competition. However there are other studies that dispute this reported association20; 21, and in particular a series of highly powered studies published recently failed to replicate the association19; 22, whilst another study demonstrated the problems with the measurement of CCL3L1 copy number and how this may confound associations with copy number25. As our data suggest that the product of CCL3L1 is appreciably less abundant than that of CCL3, this would call into question the biological premise behind the associations observed between HIV-1 progression and increased CCL3L1 copy number.
To conclude, in exploring the functional consequences of copy number variation of CCL3L1 we find no statistically significant association of MIP-1α protein levels with copy number. In contrast we find evidence that it is CCL3 that predominates at both the protein and mRNA level and therefore variation of CCL3 expression has potentially more impact biologically than the copy number variation of CCL3L1.
Materials and methods
Study population
This study utilises 55 independent volunteers from the University of Nottingham staff and student body, with 10 randomly selected repeat volunteers (see supplementary figure 2), and were taken with full consent from individuals and under local ethical approval. All samples were of UK origin with no known clinical phenotype.
Sample preparation
20mL of whole blood was taken from each volunteer at approximately 9.30am (+/-10 minutes), from which sera, genomic DNA and monocytes (a primary source of MIP-1α) were isolated. 2mL of the whole blood was removed and spun down at 13000rpm for 2 minutes to generate 1mL of serum per sample, which was stored at −80°C.
Peripheral blood mononuclear cells were isolated from the remaining 18mL of whole blood via density gradient centrifugation over Ficoll-paque (Sigma, Gillingham, UK). Monocytes were isolated using positive selection by means of CD14 MicroBeads (Miltenyi Biotech, Bergisch Gladbach, Germany), and re-suspended in cell culture media, supplemented with fetal bovine serum, antibiotics and glutamine. Monocytes were transferred into 96-well plates to a concentration of 0.5×106 cells per well and left overnight to mature to macrophages. As only macrophages adhere to the base of the cell culture plate confirmation that all the cells isolated were macrophages was possible. Cells were then either left untreated or stimulated with 10ng/mL of LPS (Sigma); varying LPS concentrations (1μg/mL-10ng/mL) were tested and 10ng/mL was found to be the most appropriate (data not shown).
Genomic DNA was prepared from the remaining mononuclear cells using reagents from Qiagen (Crawley, UK).
Measurement of gene copy number by the paralogue ratio test
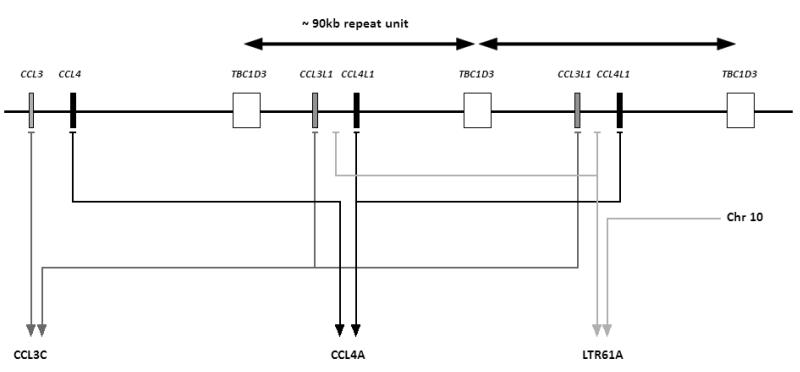
The copy number of CCL3L1 was measured from genomic DNA using the paralogue ratio test (PRT) previously described23; 26; 27. Briefly, the PRT method is essentially a PCR based assay using a single pair of primers to simultaneously amplify two specific products in a single reaction, one from a single-copy reference locus and the other from a copy variable test locus of interest. The copy number of the test locus is then estimated from the ratio of test to reference PCR products. In this study a single tube triplex assay was performed using three independent PRT assays which span the copy variable region27, with test loci in CCL3L1 (assay termed CCL3C), CCL4L1 (assay termed CCL4A) and within a long repeat sequence (assay termed LTR61A) to give three measures of copy number, which are then averaged into a single unrounded copy number value (see Figure 7). Fragment analysis of the test and reference loci were carried out by electrophoresis on an ABI3100 36 cm capillary using POP-4 polymer with an injection time of 30s. Products from the single PCR reactions were mixed with 10μl HiDi formamide with ROX-500 marker (Applied Biosystems, Warrington, UK). GeneMapper software (Applied Biosystems) was used to extract the peak areas for the three PRT systems and calculate the ratio of test to reference. Copy number values were calculated by calibrating the ratios from each experiment with 4 ECACC HRC-1 samples (http://www.hpacultures.org.uk/) of known copy number [C0075 with a copy number (CN)=1; C0150 with CN=2; C0007 with CN=3; and C0877 with CN=4], which were included in every experiment in duplicate. All experimental samples were repeated in a separate PCR to confirm copy number value.
Figure 7.
A schematic diagram showing the chromosomal region (17q12) containing the CCL3L1/CCL4L1 copy variable region on a chromosome with 2 copies of this region. The approximate locations of the primers are identified; the CCL3C system has a set of primers that simultaneously amplify sequences in a reference locus (CCL3) and a test locus (CCL3L1); the CCL4A system has a set of primers that simultaneously amplify sequences in a reference locus (CCL4) and a test locus (CCL4L1); the LTR61A system has a set of primers that simultaneously amplify sequences in a reference locus (an LTR sequence on chromosome 10) and a test locus (an LTR sequence within the copy variable region). This figure is modified from 23
For further confirmation of gene copy number two microsatellite PCRs were performed for each sample, as described previously23.
Measurement of MIP-1α protein
The extent of MIP-1α secreted by monocytes post LPS (S. minnesota) stimulation and the amount of MIP-1α present in the serum at time of extraction was measured using a sandwich MIP-1α ELISA system (R&D systems), according to manufacturer’s instructions. Assays were performed in duplicate on serial dilutions of recombinant MIP-1α LD78β protein of known concentration (R&D systems) to generate a standard curve. Assays were performed in duplicate for each individual sample and measured in triplicate.
Measurement of CCL3 and CCL3L1 mRNA transcripts
Total mRNA was prepared from lysed LPS-stimulated macrophages using a mini-prep (Qiagen) kit and reverse-transcription performed. RNA preparation was performed in duplicate for each individual sample.
Real time PCR was performed using SYBR green chemistry on an ABI7500 machine (Applied Biosystems) and the well established relative quantification ΔΔCt method. Primers (Invitrogen, Paisley, UK) were designed using Primer Express (Applied Biosystems). CCL3 specific mRNA transcripts were amplified using a final concentration of 0.1μM CCL3QF (5′-TGG CTC TCT GCA ACC AGT TC-3′) and CCL3QR (5′-CAC TGG CTG CTC GTC TCA AA-3′), CCL3L1 specific mRNA transcripts were amplified using a final concentration of 0.1μM CCL3L1QF (5′-GCT CTC TGC AAC CAG GTC C-3′) and CCL3QR (5′-CAC TGG CTG CTC GTC TCA AA-3′), and the constitutively expressed endogenous control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was amplified using a final concentration of 0.1μM GAPDHF (5′-ATC ATC AGC AAT GCC TCC TG-3′) and GAPDHR (5′-AGT CTT CTG GGT GGC AGT GA-3′). Primer concentrations were adjusted to improve yield according to the manufacturer’s instructions and all had efficiency greater than 95%. For every individual the mRNA expression level of CCL3 and CCL3L1 relative to GAPDH was calculated, Real time PCR assays were performed in triplicate for each individual sample and for all primer pairs.
The ratio of CCL3L1:CCL3 mRNA was also calculated using a novel cDNA PRT system. For each sample a single PCR was carried out using 5ng cDNA and 0.5U Taq DNA polymerase (NEB, Hitchin, UK) in a buffer with final concentrations of 50mM Tris–HCl pH8.8, 12.5mM ammonium sulphate, 1.4mM magnesium chloride, 7.5mM 2-mercaptoethanol, 125μg/ml BSA and 200μM each dNTP. Products were amplified with 1μM each of primers FAM-labelled CCL3CRNAF (TGC TCG TCT CAA AGT AGT CAG) with CCL3CR (AAT CAT GCA GGT CTC CAC T), for 22 cycles of 95°C for 30 seconds, 55°C for 30 seconds, 70°C for 1 minute followed by a final hold at 70°C for 40 minutes. CCL3 products were 167bp and CCL3L1 products were 170bp and can be readily distinguished by fragment analysis carried out by electrophoresis on an ABI3100. Products from the single PCR reactions were mixed with 10μl HiDi formamide with ROX-500 marker (Applied Biosystems), and run on an ABI3100 36cm capillary using POP-4 polymer with an injection time of 10s. GeneMapper software (Applied Biosystems) was used to extract the peak areas for each sample and the ratio was calculated by comparing the peak areas of the specific CCL3L1 transcript peak to the specific CCL3 transcript peak. All experimental samples were repeated in a separate PCR to confirm CCL3L1:CCL3 mRNA ratio.
Statistical analysis
Correlations between groups of copy number data and either protein expression or mRNA expression was assessed using a Spearman correlation in SPSS V16, and figures were drawn with the software GraphPadPrism. Repeat measures were analysed in SPSS using a general linear model with repeated measures, with the within subject factors defined as time and measurement and the between subject factor defined as copy number.
Supplementary Material
Supplementary Figure 1: Time trial graph of MIP-1α protein expression (a) and CCL3L1 mRNA expression (b) from isolated macrophages post LPS stimulation for 7 initial samples. Means and standard deviations are shown for these samples which comprised 2 one-copy samples ( ), 2 two-copy samples (
), 2 two-copy samples ( ) and 3 three-copy samples (●). There was no expression detected from the unstimulated cells.
) and 3 three-copy samples (●). There was no expression detected from the unstimulated cells.
Supplementary Figure 2: Comparison of MIP-1α expression (a), CCL3L1 mRNA expression (b) and CCL3L1:CCL3 ratio data (c) in 10 repeat samples, labelled A-J, with the initial expression measurement in black and the repeat in white. Samples A-D are all one-copy, samples E-G are all two-copies and samples H-J are three-copy samples.
Acknowledgements
This work and DC was supported by a Wellcome Trust grant (number 083929) awarded to JALA and RJP.
Footnotes
Conflict of interest The authors declare no conflict of interest.
Supplementary information is available at Genes and Immunity’s website
References
- 1.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, et al. Relative Impact of Nucleotide and Copy Number Variation on Gene Expression Phenotypes. Science. 2007;315(5813):848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henrichsen CN, Vinckenbosch N, Zollner S, Chaignat E, Pradervand S, Schutz F, et al. Segmental copy number variation shapes tissue transcriptomes. Nat Genet. 2009;41(4):424–429. doi: 10.1038/ng.345. [DOI] [PubMed] [Google Scholar]
- 3.Modi WS. CCL3L1 and CCL4L1 chemokine genes are located in a segmental duplication at chromosome 17q12. Genomics. 2004;83(4):735–738. doi: 10.1016/j.ygeno.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 4.Irving SG, Zipfel PF, Balke J, McBride OW, Morton CC, Burd PR, et al. Two inflammatory mediator cytokine genes are closely linked and variably amplified on chromosome 17q. Nucl Acids Res. 1990;18(11):3261–3270. doi: 10.1093/nar/18.11.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkman N, John M, Roesems G, Jose PJ, Barnes PJ, Chung KF. Inhibition of macrophage inflammatory protein-1 alpha expression by IL- 10. Differential sensitivities in human blood monocytes and alveolar macrophages. J Immunol. 1995;155(9):4412–4418. [PubMed] [Google Scholar]
- 6.Standiford T, Kunkel SL, Liebler JM, Burdick MD, Gilbert AR, Strieter RM. Gene Expression of macrophage inflammatory protein-1 alpha from human blood monocytes and alveolar macrophages is inhibited by interleukin-4. American Journal of Respiratory Cell and Molecular Biology. 1993;9:192–198. doi: 10.1165/ajrcmb/9.2.192. [DOI] [PubMed] [Google Scholar]
- 7.Menten P, Struyf S, Schutyser E, Wuyts A, De Clercq E, Schols D, et al. The LD78b isoform of MIP-1a is the most potent CCR5 agonist and HIV-1-inhibiting chemokine. The Journal of Clinical Investigation. 1999;104(4):R1–R5. doi: 10.1172/JCI7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, et al. The [beta]-Chemokine Receptors CCR3 and CCR5 Facilitate Infection by Primary HIV-1 Isolates. Cell. 1996;85(7):1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 9.Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, et al. CC CKR5: A RANTES, MIP-1alpha, MIP-1beta Receptor as a Fusion Cofactor for Macrophage-Tropic HIV-1. Science. 1996;272(5270):1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 10.Aquaro S, Menten P, Struyf S, Proost P, Van Damme J, De Clercq E, et al. The LD78b Isoform of MIP-1a Is the Most Potent CC-Chemokine in Inhibiting CCR5-Dependent Human Immunodeficiency Virus Type 1 Replication in Human Macrophages. J Virol. 2001;75(9):4402–4406. doi: 10.1128/JVI.75.9.4402-4406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xin X, Shioda T, Kato A, Liu H, Sakai Y, Nagai Y. Enhanced anti-HIV-1 activity of CC-chemokine LD78[beta], a non-allelic variant of MIP-1[alpha]/LD78[alpha] FEBS Letters. 1999;457(2):219–222. doi: 10.1016/s0014-5793(99)01035-2. [DOI] [PubMed] [Google Scholar]
- 12.Shostakovich-Koretskaya L, Catano G, Chykarenko ZA, He W, Gornalusse G, Mummidi S, et al. Combinatorial content of CCL3L and CCL4L gene copy numbers influence HIV-AIDS susceptibility in Ukrainian children. AIDS. 2009;23(6):679–688. doi: 10.1097/QAD.0b013e3283270b3f. 610.1097/QAD.1090b1013e3283270b3283273f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, et al. The Influence of CCL3L1 Gene-Containing Segmental Duplications on HIV-1/AIDS Susceptibility. Science. 2005;307(5714):1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 14.Dolan MJ, Kulkarni H, Camargo JF, He W, Smith A, Anaya J-M, et al. CCL3L1 and CCR5 influence cell-mediated immunity and affect HIV-AIDS pathogenesis via viral entry-independent mechanisms. Nat Immunol. 2007;8(12):1324–1336. doi: 10.1038/ni1521. [DOI] [PubMed] [Google Scholar]
- 15.Kulkarni H, Marconi VC, Agan BK, McArthur C, Crawford G, Clark RA, et al. Role of CCL3L1-CCR5 Genotypes in the Epidemic Spread of HIV-1 and Evaluation of Vaccine Efficacy. PLoS ONE. 2008;3(11):e3671. doi: 10.1371/journal.pone.0003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahuja SK, Kulkarni H, Catano G, Agan BK, Camargo JF, He W, et al. CCL3L1-CCR5 genotype influences durability of immune recovery during antiretroviral therapy of HIV-1-infected individuals. Nat Med. 2008;14(4):413–420. doi: 10.1038/nm1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He W, Kulkarni H, Castiblanco J, Shimizu C, Aluyen U, Maldonado R, et al. Reply to: “CCL3L1 and HIV/AIDS susceptibility” and “Experimental aspects of copy number variant assays at CCL3L1”. Nat Med. 2009;15(10):1117–1120. doi: 10.1038/nm1009-1117. [DOI] [PubMed] [Google Scholar]
- 18.Huik K, Sadam M, Karki T, Avi R, Krispin T, Paap P, et al. CCL3L1 Copy Number Is a Strong Genetic Determinant of HIV Seropositivity in Caucasian Intravenous Drug Users. Journal of Infectious Diseases. 2010;201(5):730–739. doi: 10.1086/650491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urban TJ, Weintrob AC, Fellay J, Colombo S, Shianna KV, Gumbs C, et al. CCL3L1 and HIV/AIDS susceptibility. Nat Med. 2009;15(10):1110–1112. doi: 10.1038/nm1009-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fellay J, Ge D, Shianna KV, Colombo S, Ledergerber B, Cirulli ET, et al. Common Genetic Variation and the Control of HIV-1 in Humans. PLoS Genet. 2009;5(12):e1000791. doi: 10.1371/journal.pgen.1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shao W, Tang J, Song W, Wang C, Li Y, Wilson CM, et al. CCL3L1 and CCL4L1: variable gene copy number in adolescents with and without human immunodeficiency virus type 1 (HIV-1) infection. Genes Immun. 2007;8(3):224–231. doi: 10.1038/sj.gene.6364378. [DOI] [PubMed] [Google Scholar]
- 22.Bhattacharya T, Stanton J, Kim E-Y, Kunstman KJ, Phair JP, Jacobson LP, et al. CCL3L1 and HIV/AIDS susceptibility. Nat Med. 2009;15(10):1112–1115. doi: 10.1038/nm1009-1112. [DOI] [PubMed] [Google Scholar]
- 23.Walker S, Janyakhantikul S, Armour JAL. Multiplex Paralogue Ratio Tests for accurate measurement of multiallelic CNVs. Genomics. 2009;93(1):98–103. doi: 10.1016/j.ygeno.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 24.Townson JR, Barcellos LF, Nibbs RJ. Gene copy number regulates the production of the human chemokine CCL3-L1. European Journal of Immunology. 2002;32(10):3016–3026. doi: 10.1002/1521-4141(2002010)32:10<3016::AID-IMMU3016>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 25.Field SF, Howson JMM, Maier LM, Walker S, Walker NM, Smyth DJ, et al. Experimental aspects of copy number variant assays at CCL3L1. Nat Med. 2009;15(10):1115–1117. doi: 10.1038/nm1009-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Armour JAL, Palla R, Zeeuwen PLJM, den Heijer M, Schalkwijk J, Hollox EJ. Accurate, high-throughput typing of copy number variation using paralogue ratios from dispersed repeats. Nucl Acids Res. 2007;35(3):e19. doi: 10.1093/nar/gkl1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carpenter D, Walker S, Prescott N, Schwalkwijk J, Armour JAL. Accuracy and differential bias in copy number measurement of CCL3L1. BMC Genomics. 2011;12:418. doi: 10.1186/1471-2164-12-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Time trial graph of MIP-1α protein expression (a) and CCL3L1 mRNA expression (b) from isolated macrophages post LPS stimulation for 7 initial samples. Means and standard deviations are shown for these samples which comprised 2 one-copy samples ( ), 2 two-copy samples (
), 2 two-copy samples ( ) and 3 three-copy samples (●). There was no expression detected from the unstimulated cells.
) and 3 three-copy samples (●). There was no expression detected from the unstimulated cells.
Supplementary Figure 2: Comparison of MIP-1α expression (a), CCL3L1 mRNA expression (b) and CCL3L1:CCL3 ratio data (c) in 10 repeat samples, labelled A-J, with the initial expression measurement in black and the repeat in white. Samples A-D are all one-copy, samples E-G are all two-copies and samples H-J are three-copy samples.