Abstract
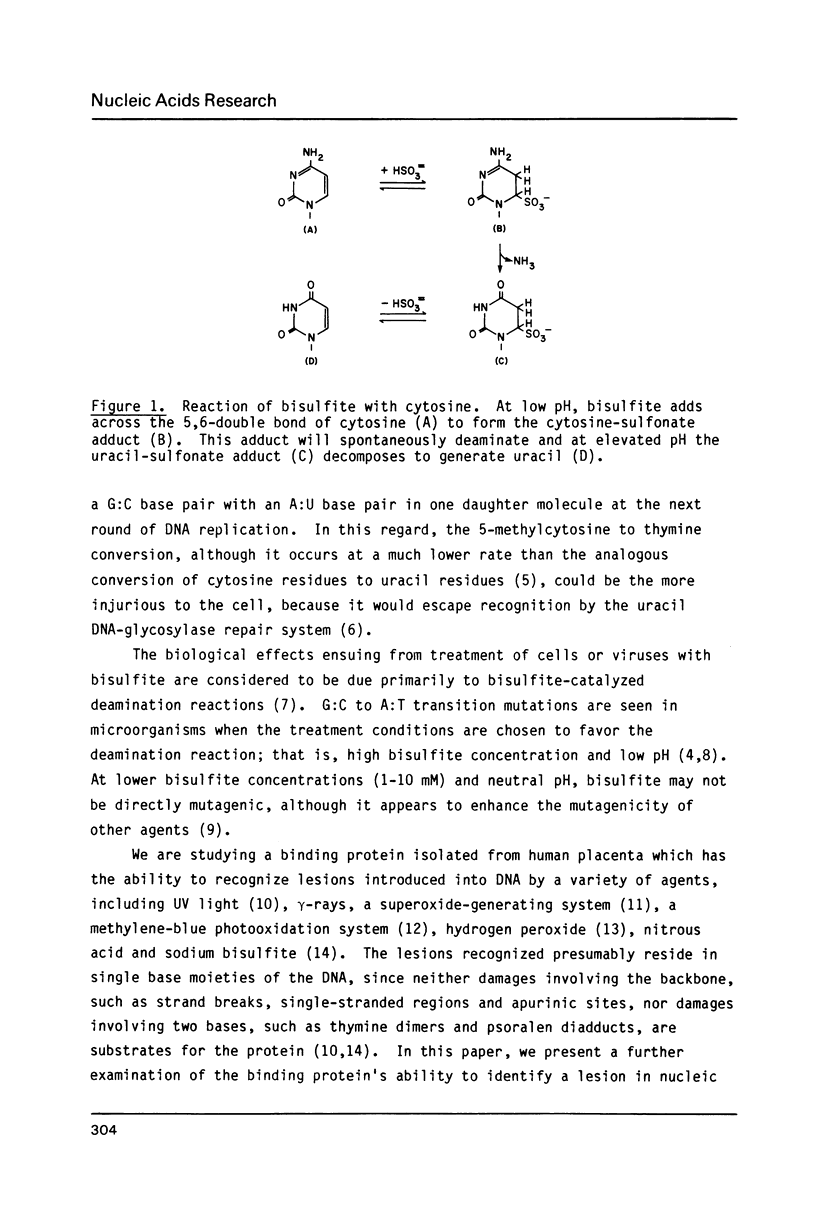
Sodium bisulfite reacts with cytosine and 5-methylcytosine, forming the 5,6-dihydrosulfonate adducts which deaminate to the uracil and thymine adducts, respectively. At alkaline pH, the sulfonate groups are then released, generating uracil and thymine. In DNA, the resulting G:U and G:T base mismatches generated are potential sites of mutagenesis. Using a human damage-specific DNA binding protein as a probe, we have found protein-recognizable lesions in bisulfite-treated DNA and poly d(I-C), but not in treated poly d(A-T) or poly d(A-U). Although this suggests that the lesion recognized is cytosine-derived, there was no correlation between the number of uracils induced and the number of binding sites, suggesting that the protein-bound damage is not a uracil-containing mismatch. Modification of the treatment protocol to reduce elimination of the bisulfite from the base adducts increased the level of binding, suggesting that the protein recognizes a base-sulfonate adduct.
Full text
PDF



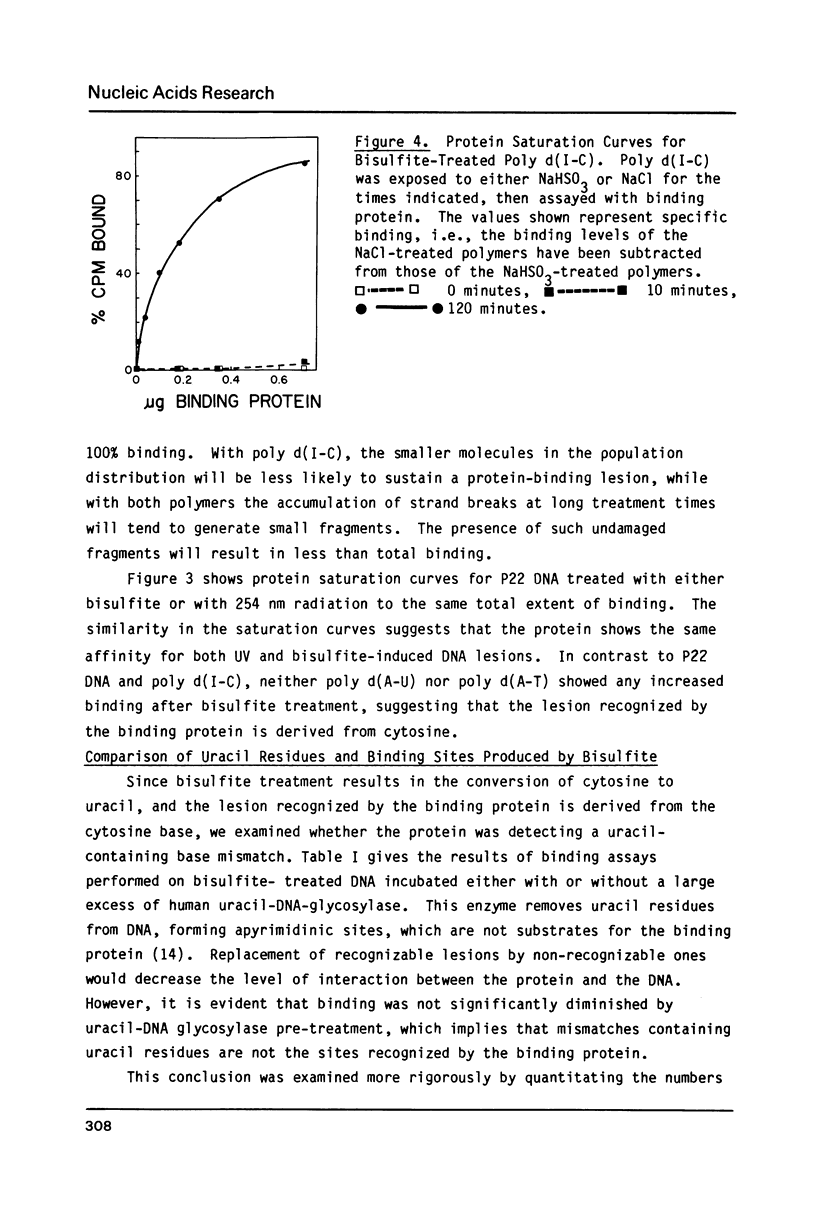
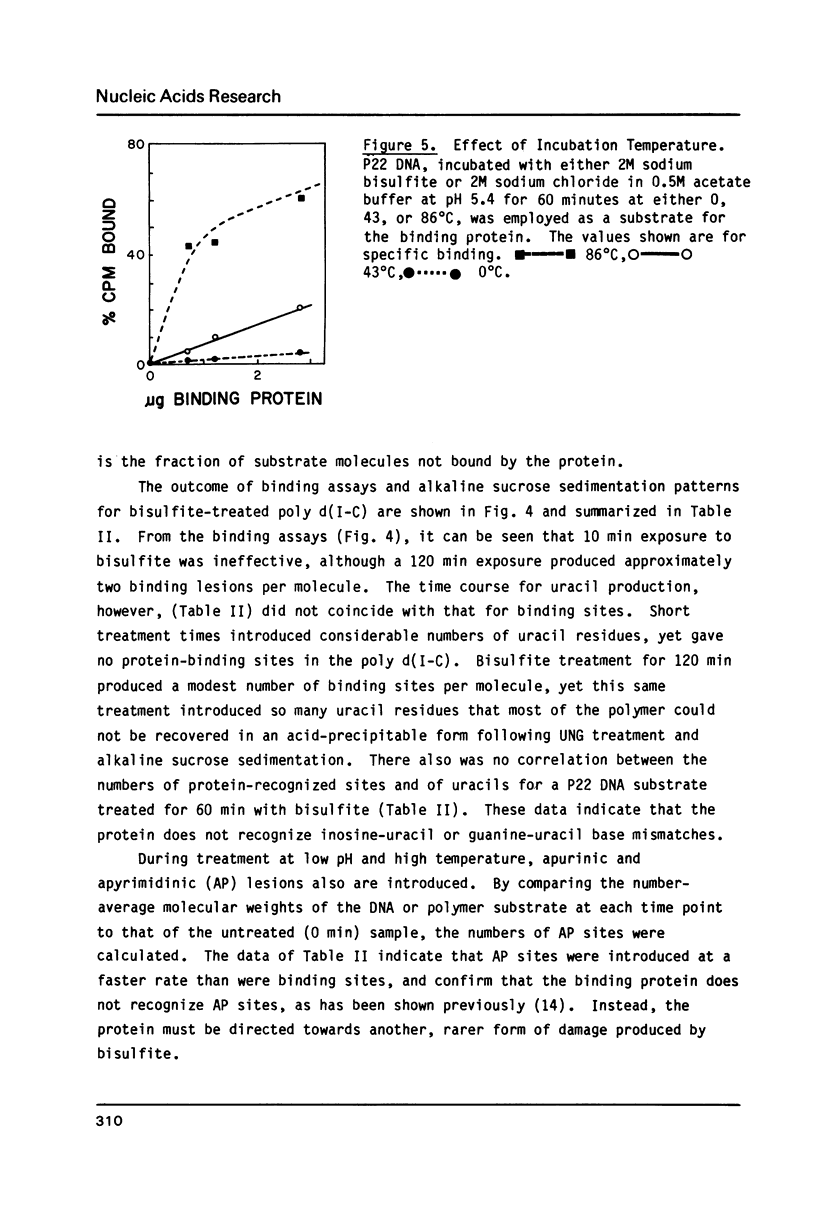




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Botstein D. Synthesis and maturation of phage P22 DNA. I. Identification of intermediates. J Mol Biol. 1968 Jun 28;34(3):621–641. doi: 10.1016/0022-2836(68)90185-x. [DOI] [PubMed] [Google Scholar]
- Brash D. E., Haseltine W. A. UV-induced mutation hotspots occur at DNA damage hotspots. Nature. 1982 Jul 8;298(5870):189–192. doi: 10.1038/298189a0. [DOI] [PubMed] [Google Scholar]
- Burd J. F., Wells R. D. Effect of incubation conditions on the nucleotide sequence of DNA products of unprimed DNA polymerase reactions. J Mol Biol. 1970 Nov 14;53(3):435–459. doi: 10.1016/0022-2836(70)90076-8. [DOI] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H., Farabaugh P. J., Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978 Aug 24;274(5673):775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- Duncan B. K., Weiss B. Specific mutator effects of ung (uracil-DNA glycosylase) mutations in Escherichia coli. J Bacteriol. 1982 Aug;151(2):750–755. doi: 10.1128/jb.151.2.750-755.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldberg R. S., Brown C., Carew J. A., Lucas J. L. Probing photodynamic damage in nucleic acids with a damage-specific DNA binding protein: a comparison of the B and Z DNA conformations. Photochem Photobiol. 1983 May;37(5):521–524. doi: 10.1111/j.1751-1097.1983.tb04511.x. [DOI] [PubMed] [Google Scholar]
- Feldberg R. S., Carew J. A. Water radiolysis products and nucleotide damage in gamma-irradiated DNA. Int J Radiat Biol Relat Stud Phys Chem Med. 1981 Jul;40(1):11–17. [PubMed] [Google Scholar]
- Feldberg R. S., Grossman L. A DNA binding protein from human placenta specific for ultraviolet damaged DNA. Biochemistry. 1976 Jun 1;15(11):2402–2408. doi: 10.1021/bi00656a024. [DOI] [PubMed] [Google Scholar]
- Feldberg R. S., Lucas J. L., Dannenberg A. A damage-specific DNA binding protein. Large scale purification from human placenta and characterization. J Biol Chem. 1982 Jun 10;257(11):6394–6401. [PubMed] [Google Scholar]
- Feldberg R. S. On the substrate specificity of a damage-specific DNA binding protein from human cells. Nucleic Acids Res. 1980 Mar 11;8(5):1133–1143. doi: 10.1093/nar/8.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa H., Sekiguchi M. Repair of deaminated cytosine residues of DNA: biological significance of the absence of uracil from DNA. Biochem Biophys Res Commun. 1978 Aug 29;83(4):1312–1318. doi: 10.1016/0006-291x(78)91364-5. [DOI] [PubMed] [Google Scholar]
- Hayakawa H., Sekiguchi M. Repair of deaminated cytosine residues of DNA: biological significance of the absence of uracil from DNA. Biochem Biophys Res Commun. 1978 Aug 29;83(4):1312–1318. doi: 10.1016/0006-291x(78)91364-5. [DOI] [PubMed] [Google Scholar]
- Hayatsu H. Bisulfite modification of nucleic acids and their constituents. Prog Nucleic Acid Res Mol Biol. 1976;16:75–124. doi: 10.1016/s0079-6603(08)60756-4. [DOI] [PubMed] [Google Scholar]
- Hayatsu H., Miura A. The mutagenic action of sodium bisulfite. Biochem Biophys Res Commun. 1970 Apr 8;39(1):156–160. doi: 10.1016/0006-291x(70)90771-0. [DOI] [PubMed] [Google Scholar]
- Hayatsu H., Wataya Y., Kai K., Iida S. Reaction of sodium bisulfite with uracil, cytosine, and their derivatives. Biochemistry. 1970 Jul 7;9(14):2858–2865. doi: 10.1021/bi00816a016. [DOI] [PubMed] [Google Scholar]
- Kai K., Tsuruo T., Hayatsu H. The effect of bisulfite modification on the template activity of DNA for DNA polymerase I. Nucleic Acids Res. 1974 Jul;1(7):889–899. doi: 10.1093/nar/1.7.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallon R. G., Rossman T. G. Bisulfite (sulfur dioxide) is a comutagen in E. coli and in Chinese hamster cells. Mutat Res. 1981 Feb;88(2):125–133. doi: 10.1016/0165-1218(81)90011-2. [DOI] [PubMed] [Google Scholar]
- SCHACHMAN H. K., ADLER J., RADDING C. M., LEHMAN I. R., KORNBERG A. Enzymatic synthesis of deoxyribonucleic acid. VII. Synthesis of a polymer of deoxyadenylate and deoxythymidylate. J Biol Chem. 1960 Nov;235:3242–3249. [PubMed] [Google Scholar]
- Shapiro R., Braverman B., Louis J. B., Servis R. E. Nucleic acid reactivity and conformation. II. Reaction of cytosine and uracil with sodium bisulfite. J Biol Chem. 1973 Jun 10;248(11):4060–4064. [PubMed] [Google Scholar]
- Simmons R. R., Friedberg E. C. Enzymatic degradation of uracil-containing deoxyribonucleic acid. V. Survival of Escherichia coli and coliphages treated with sodium bisulfite. J Bacteriol. 1979 Mar;137(3):1243–1252. doi: 10.1128/jb.137.3.1243-1252.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklyadneva V. B., Chekanovskaya L. A., Nikolaeva I. A., Tikchonenko T. I. The secondary structure of bacteriophage DNA in situ. VIII. The reaction of sodium bisulphite with intraphage cytosine as a probe for studying the DNA-protein interaction. Biochim Biophys Acta. 1979 Nov 22;565(1):51–66. doi: 10.1016/0005-2787(79)90082-0. [DOI] [PubMed] [Google Scholar]
- Wang R. Y., Gehrke C. W., Ehrlich M. Comparison of bisulfite modification of 5-methyldeoxycytidine and deoxycytidine residues. Nucleic Acids Res. 1980 Oct 24;8(20):4777–4790. doi: 10.1093/nar/8.20.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]



