Abstract
Periodontitis results from an ecological shift in the composition of subgingival biofilms. Subgingival community maturation is modulated by inter-organismal interactions and the relationship of communities with the host. In an effort to better understand this process, we evaluated biofilm formation, with oral commensal species, by three strains of the subgingivally prevalent microorganism Fusobacterium nucleatum and four strains of the periodontopathogen Porphyromonas gingivalis. We also tested the effect of serum, which resembles gingival exudates, on subgingival biofilms. Biofilms were allowed to develop in flow cells using salivary medium. We found that although not all strains of F. nucleatum were able to grow in mono-species biofilms, forming a community with health-associated partners Actinomyces oris and Veillonella parvula promoted biofilm growth of all F. nucleatum strains. Strains of P. gingivalis also showed variable ability to form mono-species biofilms. P. gingivalis W50 and W83 did not form biofilms, while ATCC 33277 and 381 formed biofilm structures, but only strain ATCC 33277 grew over time. Unlike the enhanced growth of F. nucleatum with the two health-associated species, no strain of P. gingivalis grew in three-species communities with A. oris and V. parvula. However, addition of F. nucleatum facilitated growth of P. gingivalis ATCC 33277 with health-associated partners. Importantly, serum negatively affected the adhesion of F. nucleatum, while it favored biofilm growth by P. gingivalis. This work highlights strain specificity in subgingival biofilm formation. Environmental factors such as serum alter the colonization patterns of oral microorganisms and could impact subgingival biofilms by selectively promoting pathogenic species.
Keywords: oral multi-species communities, subgingival, Fusobacterium nucleatum, Porphyromonas gingivalis, strain variability, serum, biofilms
Introduction
Oral microorganisms form complex biofilm communities in the subgingival environment, using the root surface as substratum for attachment and saliva and gingival crevicular fluid (GCF) as nutritional sources. Microorganisms in these communities interact physically, via adhesin-receptor mediated attachment, and metabolically, via cross-feeding, collective degradation of host macromolecules and exchange of metabolic signals [1–6]. Since it is nearly impossible to reconstruct in vitro the great complexity of subgingival microbial communities, the development of simplified models is fundamental to start dissecting the interactions that lead to community maturation and dysbiosis associated with periodontal disease.
Fusobacterium spp. are an important component of subgingival communities comprising ~10 to ~20% of the total flora, regardless of periodontal health status [7 and unpublished data from our laboratory]. Fusobacterium nucleatum appears to be the most abundant fusobacteria in the subgingival environment [8]. Due to its time of appearance during undisturbed plaque accumulation, abundance in plaque, coaggregation profile and ability to rapidly reduce the oxidative potential of the environment, F. nucleatum has been proposed to be important in the transition from health-associated communities to those associated with disease [9–11]. Modeling such transitions and dissecting the role of F. nucleatum in community maturation are thus important tasks if we are to gain a better understanding of the ecology of subgingival biofilm communities.
Among the bacterial species associated with periodontitis, Porphyromonas gingivalis plays a prominent role in the onset and progression of the disease [12, 13]. P. gingivalis virulence has been attributed to a great variety of factors involved in processes such as nutrient acquisition, immune activation and host evasion [14]. Some attention has also been paid to the process by which this microorganism establishes itself within subgingival biofilm communities. It seems that a pre-established community of other bacteria has an important role facilitating retention of P. gingivalis in vivo [15]. Furthermore, using a flow cell system with saliva as sole nutritional source, Periasamy and Kolenbrander [16] demonstrated that partnering with another oral species was in most cases beneficial and allowed P. gingivalis ATCC 33277 to increase its biofilm biomass over an 18 hour period. P. gingivalis has also been shown to form biofilms with the initial colonizer Streptococcus gordonii; however, in response to the presence of the streptococci, P. gingivalis decreases transcription of adhesion and signaling genes, a possible indication of switching to a “detachment” phenotype [6, 17]. It is thus likely that other community members modulate the establishment of P. gingivalis in the subgingival environment.
A less studied aspect of oral biofilm development is the variability among different strains from the same species in their colonization patterns. For example, strains of F. nucleatum appear to have heterogeneous coaggregation patterns, an indication of intra-species variability in their ability to recognize cell surfaces from other genera [18]. Variable cell surface characteristics are also likely to affect biofilm formation. Moreover, the population structure of P. gingivalis is highly heterogeneous from a genotypic perspective [19, 20]. Phenotypically, heterogeneity in P. gingivalis virulence has been observed with some strains considered “virulent” due to their ability to disseminate systemically, as shown using the subcutaneous mouse abscess model [21]. According to this assay, the type strains P. gingivalis W83 and W50 are considered “virulent”, while P. gingivalis 381 is considered “avirulent”, as it only produces localized abscesses [21]. Interestingly, genotypically similar strains to those classified as “avirulent” using the subcutaneous abscess model are more prevalent in subgingival plaque of periodontally healthy subjects, while the “virulent” genotypes occur more frequently in subjects with periodontitis. [20]. These differences have been attributed to the presence in “virulent” strains of a capsular polysaccharide structure, which decreases bacterial attachment, biofilm formation and the effectiveness of host cell responses, thus facilitating systemic spread [22–24]. A question that remains unanswered, however, is how encapsulated “virulent” strains colonize and persist in the oral environment, since an ability to form biofilms is a prime requirement for any bacterial species to maintain its population levels in the oral cavity, where saliva and GCF flow constantly.
Subgingival biofilm communities receive a constant influx of GCF, a serum-derived exudate that flows at a higher rate when local inflammation develops at the gingival margin [25]. Main components of GCF include albumin, transferrin, immunoglobulins and defensins, among many others [26, 27]. The role of serum, as an in vitro substitute for GCF, in the physiology of subgingival microorganisms has been studied in batch-wise enrichment cultures of subgingival plaque [28]. These studies demonstrated that several enrichment steps lead to the accumulation of bacteria classified, at the time, as Bacteroides spp., peptostreptococci and fusobacteria. Enriched natural consortia collectively degraded serum macromolecules, a pattern also observed with artificially assembled consortia [29]. These studies suggest that serum may promote the growth of certain subgingival species. However, the effect of serum on adhesion and biofilm formation of oral microorganisms is less clear. Human serum has been shown to inhibit the attachment of P. gingivalis to hydroxyapatite surfaces treated with type I collagen [30], however the effect of serum on adhesion and biofilm formation of subgingival consortia has not been evaluated.
Accordingly, the goal of this study was to characterize biofilm formation by different strains of F. nucleatum and P. gingivalis. We were also interested in testing whether the presence of other community members affected colonization patterns by these strains and in evaluating the role of serum on multi-species biofilms. In order to accomplish these goals we first tested three F. nucleatum strains for their ability to form mono-species or multi-species biofilms with health-associated partners Actinomyces oris and Veillonella parvula. Next, we evaluated the ability of four P. gingivalis strains to form mono-species biofilms and integrate into a 4-hour-old health-associated biofilm community in the presence and absence of F. nucleatum. We then evaluated the role of native serum as a potential environmental factor that modifies the characteristics of multi-species subgingival communities. These studies were aimed at increasing our understanding of the processes involved in subgingival biofilm development by the establishment of models representative of in vivo conditions, where transitions from a health-associated to a pathogenic biofilm could be investigated.
Methods
Microorganisms and microbiological media
The microorganisms used in this study were Actinomyces oris T14V, Veillonella parvula PK1910, Fusobacterium nucleatum subsp. polymorphum ATCC 10953, Fusobacterium nucleatum subsp. polymorphum 12230, Fusobacterium nucleatum subsp. nucleatum ATCC 25586, Porphyromonas gingivalis W83, Porphyromonas gingivalis W50 (ATCC 53978), Porphyromonas gingivalis ATCC 33277 and Porphyromonas gingivalis 381. A. oris was grown in brain heart infusion (BHI) medium. V. parvula was grown in BHI medium supplemented with 0.17 M lactic acid, 0.5 g liter−1 cysteine and 5 mg liter−1 hemin. F. nucleatum strains were grown in BHI medium supplemented with 0.5 g liter−1 cysteine. P. gingivalis strains were grown in tryptic soy broth (TSB) supplemented with 0.5 g liter−1 cysteine, 5 mg liter−1 hemin and 1 mg liter−1 menadione. All microorganisms were grown at 37°C in an anaerobic chamber with an atmosphere of H2:CO2:N2 (5:5:90).
Saliva, used to supplement the biofilm growth medium was collected from 10 systemically healthy volunteers according to a protocol approved by the Institutional Review Board of the University of Connecticut Health Center (IRB #10-216-2). Methods for saliva processing and storage have been previously described [31].
Flow cell experiments
Biofilm growth of microorganisms was evaluated using flow cells harboring a 24 × 60 mm glass cover slip as the attachment substratum. General procedures for flow cell set up and sterilization have been described previously [31]. Briefly, prior to each experiment, flow cells were cleaned with 0.1 M HCl and rinsed with sterile distilled water. Subsequently, 10% (vol/vol) hypochlorite was pumped for 2 hours followed by a rinse with sterile distilled water overnight. The flow cells were then placed at 37°C in an anaerobic chamber and treated with saliva-supplemented growth medium (22.5% (vol/vol) sterile human saliva and 10% (vol/vol) BHI, supplemented with 0.05 g liter−1 cysteine, 0.5 mg liter−1 hemin and 0.1 mg liter−1 menadione, in PBS) for 15 min to allow formation of a salivary pellicle on the glass surface. Saliva diluted to a similar concentration has been shown to support the growth of oral biofilms [16, 32, 33]. When the effect of serum was evaluated, 10% (vol/vol) commercially available human native serum (Mediatech, Inc., Manassas, VA) was added to saliva-supplemented growth medium. This serum concentration is reported to support biofilm formation by P. gingivalis [34]. For experiments with serum, unless otherwise indicated, medium containing serum was used for resuspension of the bacterial inoculum and at all times during the flow cell experiment. For some experiments, saliva-supplemented growth medium without serum was used for inoculum resuspension, during the conditioning period and attachment of microorganisms and serum was only introduced to the system in the flow, after initial attachment had occurred.
For inoculation of flow cells, bacterial cultures were allowed to grow until late logarithmic phase, which was defined as an OD600nm equal to 1.3 for A. oris, V. parvula and F. nucleatum and an OD600 nm equal to 1.1 for P. gingivalis. Bacterial cultures were then normalized to an OD600 nm equal to 1.0 and microbial cells were washed in salivary growth medium, a step that has been shown to increase attachment of cells to the conditioned surface [32]. Flow cells were inoculated with mono-, dual- or multi-species inocula. The inoculum for each flow cell was prepared by mixing appropriate combinations of 1 × 107 cells of each species. After injecting the inoculum, flow cells were inverted and microorganisms were allowed to attach for 30 minutes under static conditions. Flow cells were then placed upright and the pump started with a flow rate for all experiments of 100 μL min−1. Biofilms were allowed to develop for 4 and 16 hours. For multi-species biofilms containing P. gingivalis, the microorganism was inoculated after 4 hours of biofilm development by previously inoculated species, unless otherwise indicated. Biofilms were allowed to develop for an additional 4 and 16 hours after P. gingivalis inoculation. All flow cell experiments were repeated at least three times.
Staining, imaging and quantification of biofilms
Biofilms were fixed in 4% paraformaldehyde at 4°C for 3 hours. Biofilms were then stained by fluorescence in-situ hybridization (FISH) with oligonucleotide probes labeled with Alexa dyes. Briefly, flow cells were washed with D-PBS and bacterial cells were permeabilized with lysozyme (70,000 U mL−1 in 100 mM Tris/HCl pH 7.5 and 5 mM EDTA) for 10 min, at 37°C. Biofilms were then dehydrated in a series of ethanol washes and exposed to hybridization buffer (0.9 M NaCl, 20 mM Tris/HCl pH 7.5, 0.01% sodium dodecyl sulfate and 25% formamide) containing 10 ng mL−1 of probe. Biofilms were then incubated at 46°C for 90 min and washed for 15 min at 48°C in washing buffer (20 mM Tris/HCl pH 7.5, 5 mM EDTA, 0.01% sodium dodecyl sulfate and 159 mM NaCl). The oligonucleotide probes JF201 [35], VEI488 [36], FUS664 [35] and Bg-8 [37] were used to detect A. oris, V. parvula., F. nucleatum and P. gingivalis, respectively. Biofilms were visualized with a Zeiss LSM 510 confocal scanning laser microscope equipped with an argon laser, using a water immersion C-Apochromat 40× objective (NA1.2). Stacks of z-plane images from at least 8 different fields of view per sample were acquired and later reconstructed into 3D images using IMARIS software (Bitplane Inc., Saint Paul, MN). Surface reconstructions using the surpass mode were used to calculate the biovolume (in μm3) of each microorganism. Significant growth from early to late time points was evaluated via student's t-test. Differences in biovolumes of species across conditions, at specific time points, were also evaluated via student's t-test.
Planktonic growth curves
Planktonic growth of F. nucleatum ATCC 10953 in mono-culture or in the presence of a community of A. oris and V. parvula was evaluated by inoculating 106 cells of each microorganism into triplicate test tubes containing 10 mL of pre-reduced saliva-supplemented growth medium in which the vol/vol of BHI was 30%. Cultures were incubated at 37°C under anaerobic conditions until stationary phase was reached. F. nucleatum was quantified by sampling microbial cultures at various time points. Culture samples were sonicated for 10 s, as previously described [31], to disrupt microbial aggregates. Samples were then serially diluted in PBS and plated on BHI-blood agar. F. nucleatum was identified in mixed-culture samples by colony morphology.
In order to assess the effect of serum on viability, microorganisms were grown planktonically in the absence or presence of native human serum. 106 bacterial cells were inoculated into pre-reduced microorganism-specific growth medium supplemented with either 10% human serum or D-PBS. Triplicate cultures for each condition were incubated anaerobically at 37 °C until stationary phase was reached. Growth was monitored by determining OD(600nm).
Coaggregation assays
Coaggregation among strains used in this study was evaluated visually according to the method of Cisar et al. [38]. Planktonic coaggregation was evaluated in saliva and also in coaggregation buffer. Scores for the degree of coaggregation ranged from 0 to 4 and were assigned by the following criteria: 0-no visible aggregates in the cell suspension; 1-small uniform coaggregates in suspension; 2-definite coaggregates easily seen but suspension remained turbid without immediate settling of coaggregates; 3-large coaggregates which settled rapidly leaving some turbidity in the supernatant fluid; 4-clear supernatant fluid and large coaggregates which settled immediately. Autoaggregation was also tested following these procedures and taken into account when interpreting coaggregation data.
Results
Strain-specificity in biofilm formation by F. nucleatum and its interaction with early colonizers A. oris and V. parvula
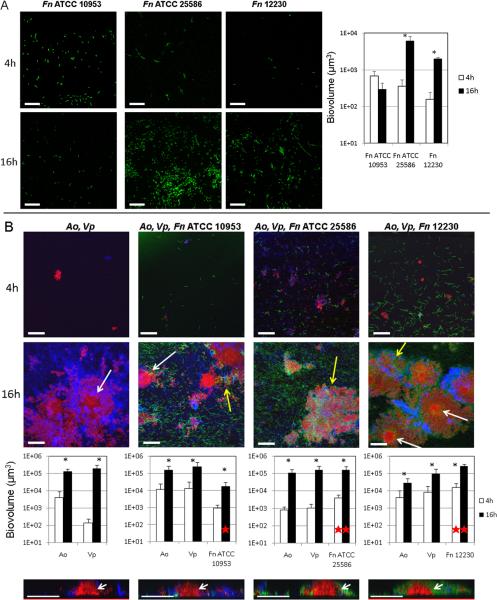
We first tested the ability of each of three strains of F. nucleatum to grow in a mono-species biofilm using flow cells and saliva-supplemented medium. As shown in Figure 1A, early biofilms (4h) did not differ among the three strains evaluated. However, mono-species biofilms of F. nucleatum ATCC 25586 and strain 12230 increased in biomass from 4h to 16h, while F. nucleatum ATCC 10953 was not able to grow as a mono-species biofilm between these time points. We then tested the effect of a community formed by A. oris and V. parvula on the biofilm growth of the three F. nucleatum strains. We confirmed results previously reported [39], in that A. oris and V. parvula formed dual-species biofilms in which both partners grew (Figure 1B). Figure 1B also shows that each strain of F. nucleatum grew as part of this community benefiting from the interaction with other community members. F. nucleatum ATCC 10953, unable to grow in mono-species biofilms, demonstrated a more than 10-fold increase in biovolume from 4h to 16h in the presence of A. oris and V. parvula. Although able to grow as a mono-species, F. nucleatum ATCC 25586 also benefited demonstrating an increased biovolume at both 4h and 16h time points as part of a community in comparison to mono-species biofilms (mono-species vs multi-species 4h P=0.017 and 16h P=0.007). Moreover, F. nucleatum 12230 showed an increase in biovolume of ~100 fold at both time points as part of a biofilm community in comparison to mono-species biofilms (mono-species vs multi-species 4h P=0.004 and 16h P=<0.001).
Figure 1.
Mono-species and multi-species biofilm formation by three strains of F. nucleatum. Biofilms were allowed to develop for 4 and 16h in flow cells using saliva-supplemented medium as nutritional source. Panel A depicts representative CLSM 3-D image projections and biovolume measurements (in μ3) for mono-species biofilms formed by three strains of F. nucleatum (green). Panel B depicts multi-species biofilm formation by F. nucleatum strains (green) with initial colonizers A. oris (red) and V. parvula (blue). White arrows in 16h CLSM 3-D image projections and 2-D x-z planes slices (bottom) indicate central columnar micro-colonies of A. oris. Yellow arrows indicate three-species inter-digitated structures. F. nucleatum = Fn, A. oris = Ao and V. parvula = Vp. * indicates a P value of less than 0.05 when biovolumes for each species were compared between 4h and 16h. A red star indicates a P value of less than 0.05 when biovolumes of F. nucleatum were compared in mono-species biofilms vs. multi-species biofilms. Bar in all images = 50 μm.
With respect to biofilm architecture of microbial communities, all strains of F. nucleatum formed three-species inter-digitated structures with A. oris and V. parvula (Figure 1B, yellow arrow). Another commonly observed biofilm architecture arrangement was central colonies of A. oris surrounded by V. parvula in dual-species biofilms or by F. nucleatum and V. parvula in three-species biofilms (Figure 1B, white arrows in 3-D image projections and 2-D slices of x-z planes). These results highlight the synergy among these three species and the robust ability to form biofilms displayed by the three strains of F. nucleatum tested when part of a multi-species community.
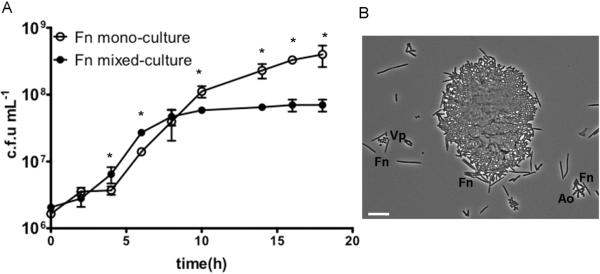
To investigate further the mechanisms mediating the interaction between F. nucleatum and an A. oris/ V. parvula community, we then tested the ability of V. parvula and A. oris to stimulate F. nucleatum ATCC 10953 in planktonic batch co-culture in a saliva-based medium. Figure 2A shows planktonic growth curves of F. nucleatum ATCC 10953 in the presence and absence of A. oris and V. parvula. Planktonic growth of F. nucleatum ATCC 10953 as part of a community in mixed-culture was slightly stimulated compared to mono-cultures. F. nucleatum entered logarithmic phase and reached stationary phase earlier in mixed-culture, the latter presumably because of nutrient exhaustion. Microscopic observation of planktonic mixed cultures revealed that the three microorganisms formed planktonic coaggregates of various sizes (Figure 2B). These findings demonstrate that the growth stimulatory effect of A. oris and V. parvula on F. nucleatum can also be detected in batch mixed cultures; however, the effect is more prominent in biofilms where microorganisms exist in close physical proximity under a flowing nutrient supply.
Figure 2.
Planktonic growth of F. nucleatum ATCC 10953 in salivary growth medium as a mono-culture or in co-culture with A. oris and V. parvula. Panel A shows planktonic growth curves. Data points represent measurements from three independent replicate experiments. Panel B shows a representative phase contrast micrograph of a mixed-species culture after 10 hours of growth in saliva-supplemented medium. Short rods are A. oris (Ao); coccoid cells are V. parvula (Vp) and thin long rods are F. nucleatum (Fn). Scale bar = 10 μm. * indicates a P value of less than 0.05 when colony forming units (c.f.u) mL−1 of Fn were compared between mono-culture and co-culture conditions at each time point.
Strain-specificity in mono-species biofilm formation by P. gingivalis and the effect of other community members on P. gingivalis colonization traits
Our next goal was to evaluate inter-strain variability in biofilm formation by four strains of the periodontal pathogen P. gingivalis. Additionally, we aimed to model a transition from a health-associated biofilm to a disease-associated biofilm by testing how different strains of P. gingivalis integrated into a community of health-associated early colonizers (A. oris-V. parvula), evaluating whether this community affected the colonization patterns of different strains. Since F. nucleatum is considered to play a central role in microbial succession, we tested its effect on P. gingivalis by including it in the community.
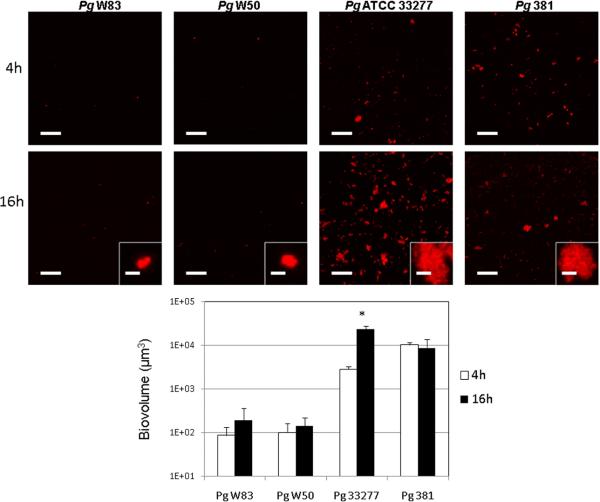
We found great inter-strain variability in P. gingivalis mono-species biofilm formation and biofilm growth capacity. As Figure 3 depicts, P. gingivalis W50 and P. gingivalis W83 displayed a poor ability to form biofilms, while P. gingivalis ATCC 33277 and 381 were able to attach to the saliva-conditioned surfaces and form biofilms. However, P. gingivalis ATCC 33277 grew between time points, while P. gingivalis 381 did not increase its biovolume between 4h and 16h. Insets on Figure 3 also depict differences in micro-colony size of adhered organisms with strains W50 and W83 showing micro-colonies of no more than 3 μm in diameter, while strains ATCC 33277 and 381 formed micro-colonies of various sizes, some reaching ~40 μm.
Figure 3.
Mono-species biofilm formation by four strains of P. gingivalis. Biofilms were allowed to develop for 4 and 16h in flow cells using saliva-supplemented medium as nutritional source. P. gingivalis (Pg) appears in red in CLSM 3-D image projections. * indicates a P value of less than 0.05 when biovolumes were compared between 4h and 16h. Left scale bar in all images = 50 μm. Scale bar in inset digital zoom images = 3 μm.
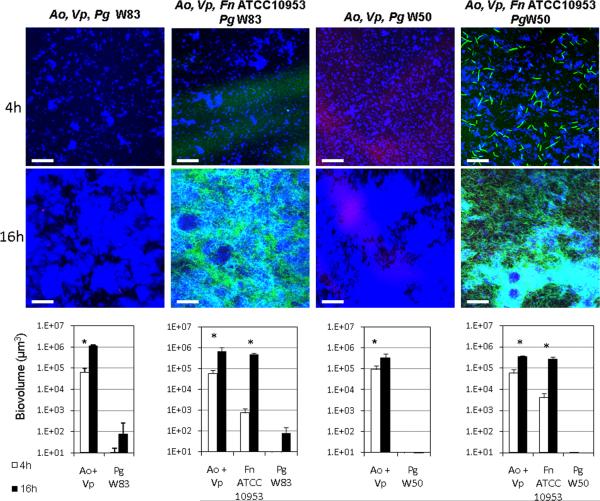
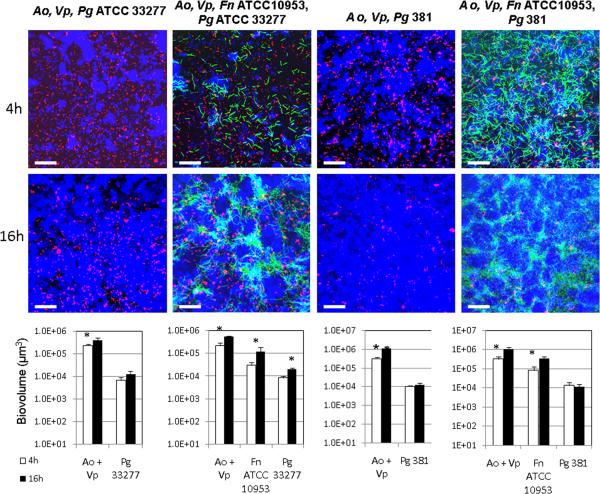
We then evaluated the ability of these four strains of P. gingivalis to integrate into a 4h-old biofilm of A. oris and V. parvula or a 4h-old biofilm of A. oris, V. parvula and F. nucleatum. For these experiments we first tested F. nucleatum ATCC 10953 since we previously identified a metabolic interaction occurring between this strain and P. gingivalis W50 [9]. As Figure 4 shows, P. gingivalis W50 and W83 did not improve in their ability to form biofilms in the presence of A. oris and V. parvula or when F. nucleatum ATCC 10953 formed part of the biofilm community (similar results observed for F. nucleatum 12230 and ATCC 25586; data not shown). Microscopic examination of planktonic coaggregation between F. nucleatum strains and P. gingivalis W83 or P. gingivalis W50 in the presence of saliva revealed minimal interaction of the two species. Furthermore, no coaggregation was observed between these two strains of P. gingivalis and A. oris or V. parvula, as tested by the visual coaggregation assay of cells suspended in saliva or buffer (see Methods; data not shown). These weak or absent cell-to-cell interactions might explain the lack of coadhesion of P. gingivalis W50 and W83 into preformed early biofilms.
Figure 4.
Ability of virulent P. gingivalis strains W50 and W83 to integrate into 4h-old biofilm communities of health-associated species. In panel A, biofilms of A. oris and V. parvula or A. oris, V. parvula and F. nucleatum were allowed to develop for 4h in flow cells using saliva-supplemented medium as nutritional source. After 4h, P. gingivalis was inoculated, and biofilms were allowed to develop for an additional 4h and 16h. In CLSM 3-D image projections A. oris (Ao) and V. parvula (Vp) appear in blue, F. nucleatum (Fn) appears in green and P. gingivalis (Pg) in red. A. oris and V. parvula were quantified as a combined biovolume (Ao + Vp). * indicates a P value of less than 0.05 when biovolumes were compared between 4h and 16h. Bar in all images = 50 μm.
The ability of P. gingivalis ATCC 33277 and 381 to integrate into a biofilm of A. oris and V. parvula (Figure 5, biovolume at 4h) was similar to their ability to form an early biofilm as mono-species (Figure 3, biovolume at 4h). However, no statistically significant increase in biovolume of P. gingivalis ATCC 33277 was observed between time points when forming part of a community with A. oris and V. parvula (Figure 5), in contrast to its growth in monospecies biofilms (Figure 3). As Figure 5 also shows, the presence of F. nucleatum ATCC 10953 showed a small but statistically significant stimulation of the ability of P. gingivalis ATCC 33277 to form biofilms as part of a community. Similar results were observed when F. nucleatum ATCC 10953 was replaced by either F. nucleatum 12230 or by ATCC 25586 (data not shown). Figure 5 also shows that P. gingivalis 381 was not affected by the presence of A. oris and V. parvula or A. oris, V. parvula and F. nucleatum ATCC 10953 and although it integrated into the biofilm community, it did not exhibit growth between time points, consistent with results of mono-species biofilms (Figure 3). Taken together, these results show that strains of P. gingivalis display a markedly different ability to form biofilms. Interaction with a community only affected strain ATCC 33277, while the other three strains tested were not influenced in their biofilm forming patterns.
Figure 5.
Ability of avirulent P. gingivalis strains ATCC 33277 and 381 to integrate into 4h-old biofilm communities of health-associated species. Biofilms of A. oris and V. parvula or A. oris, V. parvula and F. nucleatum were allowed to develop for 4h in flow cells using saliva-supplemented medium as nutritional source. After 4h, P. gingivalis was inoculated, and biofilms were allowed to develop for an additional 4h and 16h. In CLSM 3-D image projections A. oris (Ao) and V. parvula (Vp) appear in blue, F. nucleatum (Fn) appears in green and P. gingivalis (Pg) in red. A. oris and V. parvula were quantified as a combined biovolume (Ao + Vp). * indicates a P value of less than 0.05 when biovolumes were compared between 4h and 16h. Bar in all images = 50 μm.
Serum as an ecological determinant of adhesion and biofilm architecture
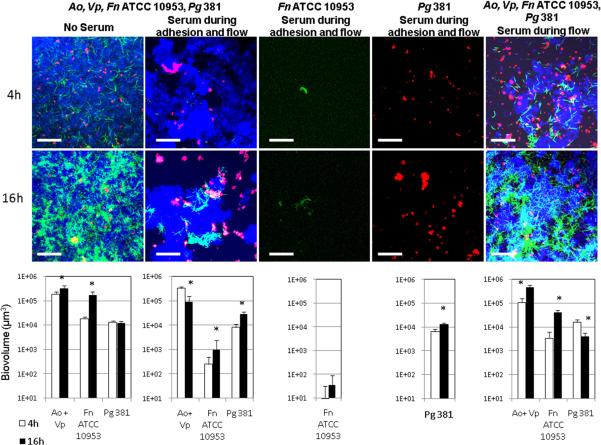
We then evaluated whether the model biofilms developed were affected by the presence of human native serum, a nutritional source and possibly a limiting factor for the growth of microorganisms in the subgingival environment. We chose to use native serum, as opposed to heat-inactivated serum, to more closely simulate in vivo conditions. For these experiments, we inoculated flow cells simultaneously with A. oris, V. parvula, F. nucleatum ATCC 10953 and P. gingivalis 381. Figure 6 shows the effect of serum on multi-species biofilms and also on mono-species biofilms of F. nucleatum ATCC 10953 and P. gingivalis 381. The addition of serum to multi-species communities, during flow cell conditioning, attachment period and in the flow, resulted in a marked decrease in the biovolumes of F. nucleatum at both time points evaluated compared to the non-serum condition (4h P=0.0002; 16h P=0.005; Figure 6, two left panels), although F. nucleatum was still able to grow between time points in the presence of serum. A similar result was observed for F. nucleatum mono-species biofilms with a dramatic decrease in biovolume in the presence of serum (Figure 6, middle panel) compared to mono-species biofilms formed in the absence of serum (Figure 1A). On the contrary, in multi-species biofilms, biovolumes of A. oris/V. parvula or P. gingivalis 381 were not affected by the presence of serum at 4h when compared to the control condition. However, between time point comparisons showed that in the presence of serum, A. oris/V. parvula decreased in their biofilm biomass from 4h to 16h, while we observed for the first time an increase in P. gingivalis 381 biovolume between time points. Figure 6 also shows that serum also allowed P. gingivalis 381 to grow as a mono-species biofilm, in sharp contrast to results obtained in the absence of serum (Figure 3). These results suggest that serum has a selective effect on members of microbial communities favoring biofilm growth by some species such as P. gingivalis and limiting attachment and biofilm formation by others such as F. nucleatum.
Figure 6.
Effect of serum on mono-species and multi-species biofilm communities. Left image and biovolume data represent the control condition in which no serum was added to the salivary growth medium. In the three central images and graphs serum was present in the salivary medium during inoculum resuspension, substratum conditioning, attachment and in the medium flow. In right image and biovolume data graph, serum was only added to the salivary medium during flow. * indicates a P value of less than 0.05 when biovolumes were compared between 4h and 16h. Bar in all images = 65 μm. A. oris (Ao) and V. parvula (Vp) appear in blue, F. nucleatum ATCC 10953 (Fn) appears in green and P. gingivalis 381 (Pg) in red.
We then investigated if the observed decrease in biovolumes of F. nucleatum in the presence of serum was the result of an effect on adhesion, as serum was present during conditioning of flow cells and attachment, or alternatively serum affected viability of F. nucleatum. To test the role of serum in inhibition of bacterial attachment, we performed an experiment in which we conditioned flow cells and inoculated microorganisms in the presence of serum-free salivary medium, thus allowing the formation of an acquired pellicle over the substratum devoid of serum components. We introduced serum in the flow only after the attachment period of microorganisms had elapsed. As Figure 6 shows, when serum was introduced only in the flow, F. nucleatum biovolumes were still significantly lower than in the absence of serum (no serum vs serum during flow 4h P=0.0004, 16h P=0.01), but higher than when serum was present during adhesion (serum during adhesion and flow vs serum during flow 4h P=0.06; 16h P=0.0005). These results suggest that the effect of serum on F. nucleatum is mostly due to an inhibition of its adhesion to the initially-formed pellicle. However, to rule out an effect on viability, we grew microorganisms planktonically in the presence and absence of serum. As Figure 7 demonstrates, serum did not affect viability of F. nucleatum.
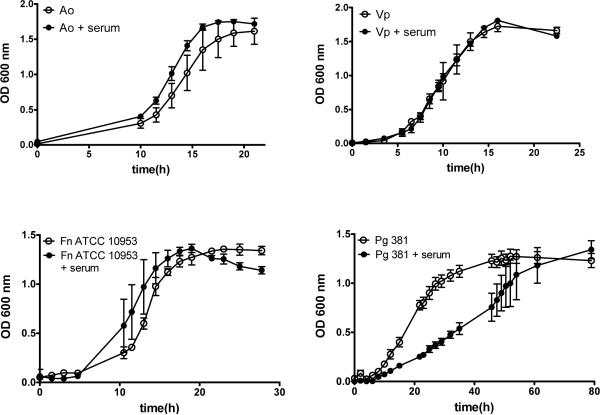
Figure 7.
Effect of serum on viability of A. oris (Ao), V. parvula (Vp), F. nucleatum ATCC 10953 (Fn) and P. gingivalis 381 (Pg). Planktonic growth of microorganisms was evaluated in their appropriate nutrient-rich medium supplemented with 10% native human serum or with PBS as control. Data represent measurements from three independent replicate experiments.
Figure 7 also shows that in planktonic culture serum did not affect the viability of A. oris and V. parvula. However, serum decreased the growth rate of P. gingivalis 381, without completely inhibiting its growth as it reached a final OD similar to the no-serum condition. The negative effect of serum on P. gingivalis in planktonic culture contrasts with its positive effect on P. gingivalis 381 biofilm growth over time, particularly when serum was part of the initial pellicle (Figure 6). However, when microorganisms were inoculated as a multi-species biofilm under serum-free conditions, and serum was introduced only in the flow, P. gingivalis 381 failed to grow. These results confirm that the main effect serum exerts on biofilm microorganisms is mediated by its integration into the acquired pellicle covering the surface where microorganisms attach and develop biofilms. Collectively, these results suggest human native serum is a strong ecological determinant of subgingival biofilm formation.
Discussion
Microorganisms in the oral cavity survive by attaching to oral surfaces. This obligatory biofilm lifestyle differs from a planktonic existence in that microorganisms must be able to display surface characteristics that allow them to attach to the substratum or to neighboring cells. Moreover, in biofilms, microorganisms encounter competition for resources from other cells living in their vicinity and must adapt to the beneficial or deleterious effects of metabolites released from other community members. These factors may explain the non-random association patterns seen in vivo in undisrupted dental plaque, where a remarkably intricate architectural organization is observed with individual species occupying specific physical niches [1, 40, 41]. Spatial organization of oral communities has only been studied at the species level; however, it is likely that because of intra-species population heterogeneity, strain variability exists in biofilm formation and inter-species association patterns. Here, we use an in vitro model of oral biofilm communities to study colonization patterns displayed by different strains of F. nucleatum and P. gingivalis, either as mono-species or as part of a community. We show that both F. nucleatum and P. gingivalis display intra-species variability in biofilm formation. A genome sequence comparison of three F. nucleatum subspecies has shown that the population of F. nucleatum is highly variable with the three subspecies compared only sharing ~62% of genes [42]. It is thus difficult to point to specific candidate genes responsible for the variable phenotype observed. Moreover, in our study, strains ATCC 10953 and 12330 showed a different biofilm phenotype, although they both belong to the polymorphum subspecies. On the contrary, the genomes of the “virulent” strain P. gingivalis W83 and that of the “avirulent” strain P. gingivalis ATCC 33277 only differ by 7% [43]. Among the variable genes are those encoding for enzymes involved in capsular polysaccharide biosynthesis, which Davey and Duncan [23] later showed to be responsible for the “poor biofilm former” phenotype of P. gingivalis W83. It is thus likely that the inability to form biofilms of P. gingivalis W50, a strain genotypically very similar to W83 [20] is also due to the presence of a capsule polysaccharide.
The inability of F. nucleatum ATCC 10953 to grow as a mono-species biofilm in saliva-supplemented flow cells has been documented before [33]. The Kolenbrander laboratory also reported that a lack of biofilm growth by F. nucleatum ATCC 10953 could be relieved by the independent presence of either A. oris [33] or V. parvula [44], both of which are abundant components of subgingival plaque, particularly during periodontal health [7, 8, 45]. Our study shows that despite variability among F. nucleatum strains in their biofilm forming ability as mono-species, the presence of community members A. oris and V. parvula was beneficial, not only relieving the inability of F. nucleatum ATCC 10953 to grow in biofilms, but also facilitating biofilm formation by the two other strains tested, a suggestion of a potentially species-wide synergistic interaction. This synergistic community correlates with the in vivo abundance of these three genera in subgingival plaque during health [7]. Moreover, the positive interaction observed between A. oris, V. parvula and F. nucleatum coincided with a display of specific physical association patterns among strains in these three-species communities, in which A. oris formed central chimney-like micro-colonies surrounded by V. parvula and F. nucleatum. This architectural arrangement suggests A. oris occupies a central role in community dynamics. Interestingly, in vivo observations have revealed similar architecture in 48h-old biofilms developing on glass slabs carried in the mouth of volunteers, in which Actinomyces sp. were observed to form columnar micro-colonies surrounded by streptococci and other unidentified bacteria [46]. A defined spatial structure has been experimentally shown to be important in maintaining the stability of microbial communities, as it allows for the creation of chemical gradients were maximum productivity of specific strains within a syntropic context is achieved [47]. In this respect, it was also interesting to observe that the synergistic effect of A. oris and V. parvula on F. nucleatum was more pronounced in multi-species biofilm communities than in planktonic batch cultures. Furthermore, it is also possible that a close physical association with A. oris and V. parvula increases the biofilm forming ability of F. nucleatum and thus, its increased biomass in a flow cell community is not only a consequence of an augmented growth capacity due to effective resource utilization but rather a change in gene expression of adhesion-related genes. The nature of this synergistic interaction is currently under investigation.
Different strains of P. gingivalis also displayed marked variability in their biofilm forming capacity. These differences are in agreement with previous observations of P. gingivalis mono-species biofilms formed on plastic surfaces in a closed environment [23]. The presence of a community did not markedly benefit the “poor biofilm former” strains of P. gingivalis W83 and W50, which were almost undetectable in biofilms under all conditions tested. Although P. gingivalis can fulfill its role as a pathogenic species even at low levels of oral colonization [48], persistence in the oral cavity would still require a minimal capacity to form biofilms on oral surfaces. Thus, the mechanisms by which P. gingivalis “virulent” strains colonize the tooth surfaces remain unclear. It is possible that biofilm formation by these strains requires synergy with specific partners or alternatively, environmental conditions different to the ones tested in this study modulate in vivo colonization by P. gingivalis encapsulated strains.
Contrary to the lack of multi-species biofilm formation by P. gingivalis W50 and W83, the “avirulent” but “good biofilm-formers” P. gingivalis ATCC 33277 and 381 were able to integrate into a community of early health-associated colonizers, achieving similar biovolumes at the 4h time point as those achieved in early mono-species biofilms. However, the presence of A. oris/V. parvula impeded biofilm growth by P. gingivalis ATCC 33277, unless F. nucleatum was part of the community. On the contrary, the presence of the middle colonizer F. nucleatum did not markedly change the biofilm growth of P. gingivalis 381. F. nucleatum has been proposed to have a fundamental role in the establishment of periodontal pathogens during microbial succession events, based on in vivo and in vitro observations [9, 12, 49, 50]. The lack of observation in this study of a species-wide effect of F. nucleatum on the growth of P. gingivalis, highlights the specificity of inter-organismal interactions. Moreover, it is possible that different environmental conditions to those used in this study could enhance the ability of F. nucleatum to metabolically support P. gingivalis. For example, the positive interaction we previously reported in planktonic continuous cultures between these two species, where F. nucleatum supplied CO2 to P. gingivalis and reduced the environment, allowing the latter to grow under more oxygenated conditions that those it tolerates as a mono-culture [9], could not have occurred under the CO2-rich anaerobic conditions used in the present study. Furthermore, the previously reported ability of F. nucleatum to allow the biofilm growth of P. gingivalis ATCC 33277 in flow cells was tested in a system with saliva as the sole nutrient supply [16], in contrast with the current experiments in which the salivary medium was supplemented with BHI, hemin and cysteine, a condition that supported mono-species biofilm growth by P. gingivalis ATCC 33277. It is also possible that the health-associated community of A. oris and V. parvula changed P. gingivalis phenotype towards one compatible with detachment, as occurs when P. gingivalis forms biofilms over a surface previously colonized by S. gordonii [6]. A situation consistent with a switch towards detachment is possibly indicated by the decreased micro-colony size displayed by P. gingivalis ATCC 33277 and 381 when forming part of a community in comparison to mono-species biofilms (Figures 4 and 6).
In this study we also show that the presence of serum alters colonization patterns by oral microorganisms. Bovine serum and bovine serum albumin have been shown to inhibit the adhesion to abiotic surfaces and subsequent biofilm development of non-oral species Pseudomonas aeruginosa [51] and Helicobacter pylori [52]. Similarly, in this study, we found that when human serum was part of the acquired pellicle formed over the glass substratum, it had a marked negative effect on the adhesion of F. nucleatum, while it did not affect the initial adhesion of P. gingivalis or A. oris//V. parvula. This is thus an indication that the main role of the gingival crevicular exudate flowing from the adjacent gingival tissues and constantly bathing the tooth surfaces is to serve as an innate immunity defense mechanism limiting adhesion of certain species. Interestingly, serum as part of the substratum allowed P. gingivalis 381 to form a commensal interaction with the three-species community, growing as part of the community and displaying micro-colonies of greater size, similar to those observed in mono-species biofilms. Importantly, the presence of serum only in the flowed medium did not replicate this result, an indication that the surface characteristics of the initially formed pellicle determine the type of biofilm that develops on the surface. This may explain why eradication of periodontal pathogens from tooth surfaces is a challenge, as after removal of these biofilms via dental procedures such as scaling and root planning, the bleeding tissues and inflammatory fluids immediately form a pellicle over the debrided tooth surface that favors the reestablishment of a pathogenic biofilm. It is thus important that future studies address the role of serum on biofilm formation by other periodontal pathogens.
In conclusion, these studies demonstrate that strain specificity in biofilm formation exists and should be considered when establishing models of subgingival plaque. Importantly, this variability in biofilm formation can be modulated by species-specific interactions with other community members, as demonstrated by the synergistic effect of A. oris and V. parvula on F. nucleatum that allowed all strains to grow as a biofilm. In contrast, P. gingivalis variability in biofilm formation remained when part of a multi-species community. Our experiments also suggest that environmental factors, such as the presence of a serum-like gingival exudate could have profound effects on microbial adhesion to surfaces and in the ability of certain microorganisms to flourish in the biofilm community.
Highlights
We studied the role of strain variability and serum on oral biofilm formation.
Strains of Fusobacterium nucleatum and Porphyromonas gingivalis differ in their ability to form monospecies biofilms.
Actinomyces oris and Veillonella parvula allow all strains of F. nucleatum tested to grow as biofilms.
Growth as part of a community does not markedly change inter-strain variability in P. gingivalis biofilm formation.
Serum limits biofilm formation by F. nucleatum, while it enhances P. gingivalis biofilm growth.
Acknowledgments
This work was supported by grant R21DE020166 from the National Institute of Dental and Craniofacial Research, National Institutes of Health. We would like to thank colleagues (PE Kolenbrander, YW Han, ME Davey and BY Wang), who shared with our laboratory the bacterial strains used in these studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Zijnge V, van Leeuwen MB, Degener JE, Abbas F, Thurnheer T, Gmur R, Harmsen HJ. Oral biofilm architecture on natural teeth. PLoS One. 2010;5:e9321. doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kolenbrander PE. Surface recognition among oral bacteria: multigeneric coaggregations and their mediators. Crit Rev Microbiol. 1989;17:137–59. doi: 10.3109/10408418909105746. [DOI] [PubMed] [Google Scholar]
- [3].Bradshaw DJ, Homer KA, Marsh PD, Beighton D. Metabolic cooperation in oral microbial communities during growth on mucin. Microbiology. 1994;140(Pt 12):3407–12. doi: 10.1099/13500872-140-12-3407. [DOI] [PubMed] [Google Scholar]
- [4].Jakubovics NS, Gill SR, Vickerman MM, Kolenbrander PE. Role of hydrogen peroxide in competition and cooperation between Streptococcus gordonii and Actinomyces naeslundii. FEMS Microbiol Ecol. 2008;66:637–44. doi: 10.1111/j.1574-6941.2008.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jakubovics NS, Gill SR, Iobst SE, Vickerman MM, Kolenbrander PE. Regulation of gene expression in a mixed-genus community: stabilized arginine biosynthesis in Streptococcus gordonii by coaggregation with Actinomyces naeslundii. J Bacteriol. 2008;190:3646–57. doi: 10.1128/JB.00088-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chawla A, Hirano T, Bainbridge BW, Demuth DR, Xie H, Lamont RJ. Community signalling between Streptococcus gordonii and Porphyromonas gingivalis is controlled by the transcriptional regulator CdhR. Mol Microbiol. 2010;78:1510–22. doi: 10.1111/j.1365-2958.2010.07420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6:1176–85. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol 2000. 2005;38:135–87. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- [9].Diaz PI, Zilm PS, Rogers AH. Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbon-dioxide-depleted environments. Microbiology. 2002;148:467–72. doi: 10.1099/00221287-148-2-467. [DOI] [PubMed] [Google Scholar]
- [10].Loe H, Theilade E, Jensen SB. Experimental gingivitis in man. J Periodontol. 1965;36:177–87. doi: 10.1902/jop.1965.36.3.177. [DOI] [PubMed] [Google Scholar]
- [11].Kolenbrander PE, Palmer RJ, Jr, Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. Bacterial interactions and successions during plaque development. Periodontol 2000. 2006;42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- [12].Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- [13].Craig RG, Boylan R, Yip J, Mijares D, Imam M, Socransky SS, Taubman MA, Haffajee AD. Serum IgG antibody response to periodontal pathogens in minority populations: relationship to periodontal disease status and progression. J Periodontal Res. 2002;37:132–46. doi: 10.1034/j.1600-0765.2002.00031.x. [DOI] [PubMed] [Google Scholar]
- [14].Curtis MA, Slaney JM, Aduse-Opoku J. Critical pathways in microbial virulence. J Clin Periodontol. 2005;32(Suppl 6):28–38. doi: 10.1111/j.1600-051X.2005.00817.x. [DOI] [PubMed] [Google Scholar]
- [15].Slots J, Gibbons RJ. Attachment of Bacteroides melaninogenicus subsp. asaccharolyticus to oral surfaces and its possible role in colonization of the mouth and of periodontal pockets. Infect Immun. 1978;19:254–64. doi: 10.1128/iai.19.1.254-264.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Periasamy S, Kolenbrander PE. Mutualistic biofilm communities develop with Porphyromonas gingivalis and initial, early, and late colonizers of enamel. J Bacteriol. 2009;191:6804–11. doi: 10.1128/JB.01006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Maeda K, Tribble GD, Tucker CM, Anaya C, Shizukuishi S, Lewis JP, Demuth DR, Lamont RJ. A Porphyromonas gingivalis tyrosine phosphatase is a multifunctional regulator of virulence attributes. Mol Microbiol. 2008;69:1153–64. doi: 10.1111/j.1365-2958.2008.06338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kolenbrander PE, Andersen RN, Moore LV. Coaggregation of Fusobacterium nucleatum, Selenomonas flueggei, Selenomonas infelix, Selenomonas noxia, and Selenomonas sputigena with strains from 11 genera of oral bacteria. Infect Immun. 1989;57:3194–203. doi: 10.1128/iai.57.10.3194-3203.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Frandsen EV, Poulsen K, Curtis MA, Kilian M. Evidence of recombination in Porphyromonas gingivalis and random distribution of putative virulence markers. Infect Immun. 2001;69:4479–85. doi: 10.1128/IAI.69.7.4479-4485.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Igboin CO, Griffen AL, Leys EJ. Porphyromonas gingivalis strain diversity. J Clin Microbiol. 2009;47:3073–81. doi: 10.1128/JCM.00569-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Neiders ME, Chen PB, Suido H, Reynolds HS, Zambon JJ, Shlossman M, Genco RJ. Heterogeneity of virulence among strains of Bacteroides gingivalis. J Periodontal Res. 1989;24:192–8. doi: 10.1111/j.1600-0765.1989.tb02005.x. [DOI] [PubMed] [Google Scholar]
- [22].Genco CA, Cutler CW, Kapczynski D, Maloney K, Arnold RR. A novel mouse model to study the virulence of and host response to Porphyromonas (Bacteroides) gingivalis. Infect Immun. 1991;59:1255–63. doi: 10.1128/iai.59.4.1255-1263.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Davey ME, Duncan MJ. Enhanced biofilm formation and loss of capsule synthesis: deletion of a putative glycosyltransferase in Porphyromonas gingivalis. J Bacteriol. 2006;188:5510–23. doi: 10.1128/JB.01685-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Singh A, Wyant T, Anaya-Bergman C, Aduse-Opoku J, Brunner J, Laine ML, Curtis MA, Lewis JP. The capsule of Porphyromonas gingivalis leads to a reduction in the host inflammatory response, evasion of phagocytosis, and increase in virulence. Infect Immun. 2011;79:4533–42. doi: 10.1128/IAI.05016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Holm-Pedersen P, Agerbaek N, Theilade E. Experimental gingivitis in young and elderly individuals. J Clin Periodontol. 1975;2:14–24. doi: 10.1111/j.1600-051x.1975.tb01722.x. [DOI] [PubMed] [Google Scholar]
- [26].Curtis MA, Sterne JA, Price SJ, Griffiths GS, Coulthurst SK, Wilton JM, Johnson NW. The protein composition of gingival crevicular fluid sampled from male adolescents with no destructive periodontitis: baseline data of a longitudinal study. J Periodontal Res. 1990;25:6–16. doi: 10.1111/j.1600-0765.1990.tb01202.x. [DOI] [PubMed] [Google Scholar]
- [27].Bostanci N, Heywood W, Mills K, Parkar M, Nibali L, Donos N. Application of label-free absolute quantitative proteomics in human gingival crevicular fluid by LC/MS E (gingival exudatome) J Proteome Res. 2010;9:2191–9. doi: 10.1021/pr900941z. [DOI] [PubMed] [Google Scholar]
- [28].ter Steeg PF, Van der Hoeven JS, de Jong MH, van Munster PJ, Jansen MJ. Enrichment of subgingival microflora on human serum leading to accumulation of Bacteroides species, Peptostreptococci and Fusobacteria. Antonie Van Leeuwenhoek. 1987;53:261–72. doi: 10.1007/BF00393933. [DOI] [PubMed] [Google Scholar]
- [29].Jansen HJ, van der Hoeven JS. Protein degradation by Prevotella intermedia and Actinomyces meyeri supports the growth of non-protein-cleaving oral bacteria in serum. J Clin Periodontol. 1997;24:346–53. doi: 10.1111/j.1600-051x.1997.tb00768.x. [DOI] [PubMed] [Google Scholar]
- [30].Naito Y, Gibbons RJ. Attachment of Bacteroides gingivalis to collagenous substrata. J Dent Res. 1988;67:1075–80. doi: 10.1177/00220345880670080301. [DOI] [PubMed] [Google Scholar]
- [31].Diaz PI, Xie Z, Sobue T, Thompson A, Biyikoglu B, Ricker A, Ikonomou L, Dongari-Bagtzoglou A. Synergistic Interaction between Candida albicans and Commensal Oral Streptococci in a Novel In Vitro Mucosal Model. Infect Immun. 2012;80:620–32. doi: 10.1128/IAI.05896-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Palmer RJ, Jr, Kazmerzak K, Hansen MC, Kolenbrander PE. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect Immun. 2001;69:5794–804. doi: 10.1128/IAI.69.9.5794-5804.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Periasamy S, Chalmers NI, Du-Thumm L, Kolenbrander PE. Fusobacterium nucleatum ATCC 10953 requires Actinomyces naeslundii ATCC 43146 for growth on saliva in a three-species community that includes Streptococcus oralis 34. Appl Environ Microbiol. 2009;75:3250–7. doi: 10.1128/AEM.02901-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Davey ME. Techniques for the growth of Porphyromonas gingivalis biofilms. Periodontol 2000. 2006;42:27–35. doi: 10.1111/j.1600-0757.2006.00183.x. [DOI] [PubMed] [Google Scholar]
- [35].Foster JS, Kolenbrander PE. Development of a multispecies oral bacterial community in a saliva-conditioned flow cell. Appl Environ Microbiol. 2004;70:4340–8. doi: 10.1128/AEM.70.7.4340-4348.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chalmers NI, Palmer RJ, Jr, Cisar JO, Kolenbrander PE. Characterization of a Streptococcus sp.-Veillonella sp. community micromanipulated from dental plaque. J Bacteriol. 2008;190:8145–54. doi: 10.1128/JB.00983-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Dix K, Watanabe SM, McArdle S, Lee DI, Randolph C, Moncla B, Schwartz DE. Species-specific oligodeoxynucleotide probes for the identification of periodontal bacteria. J Clin Microbiol. 1990;28:319–23. doi: 10.1128/jcm.28.2.319-323.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cisar JO, Kolenbrander PE, Mcintire FC. Specificity of Coaggregation Reactions between Human Oral Streptococci and Strains of Actinomyces-Viscosus or Actinomyces-Naeslundii. Infection and Immunity. 1979;24:742–52. doi: 10.1128/iai.24.3.742-752.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Periasamy S, Kolenbrander PE. Central role of the early colonizer Veillonella sp. in establishing multispecies biofilm communities with initial, middle, and late colonizers of enamel. J Bacteriol. 2010;192:2965–72. doi: 10.1128/JB.01631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Marsh PD, Moter A, Devine DA. Dental plaque biofilms: communities, conflict and control. Periodontol 2000. 2011;55:16–35. doi: 10.1111/j.1600-0757.2009.00339.x. [DOI] [PubMed] [Google Scholar]
- [41].Listgarten MA. Structure of the microbial flora associated with periodontal health and disease in man. A light and electron microscopic study. J Periodontol. 1976;47:1–18. doi: 10.1902/jop.1976.47.1.1. [DOI] [PubMed] [Google Scholar]
- [42].Karpathy SE, Qin X, Gioia J, Jiang H, Liu Y, Petrosino JF, Yerrapragada S, Fox GE, Haake SK, Weinstock GM, Highlander SK. Genome sequence of Fusobacterium nucleatum subspecies polymorphum - a genetically tractable fusobacterium. PLoS One. 2007;2:e659. doi: 10.1371/journal.pone.0000659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chen T, Hosogi Y, Nishikawa K, Abbey K, Fleischmann RD, Walling J, Duncan MJ. Comparative whole-genome analysis of virulent and avirulent strains of Porphyromonas gingivalis. J Bacteriol. 2004;186:5473–9. doi: 10.1128/JB.186.16.5473-5479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Periasamy S, Kolenbrander PE. Aggregatibacter actinomycetemcomitans builds mutualistic biofilm communities with Fusobacterium nucleatum and Veillonella species in saliva. Infect Immun. 2009;77:3542–51. doi: 10.1128/IAI.00345-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Diaz PI. Microbial Diversity and Interactions in Subgingival Communities. In: Kinane D, Mombelli A, editors. Periodontal Disease. Karger; 2012. pp. 17–40. [DOI] [PubMed] [Google Scholar]
- [46].Dige I, Raarup MK, Nyengaard JR, Kilian M, Nyvad B. Actinomyces naeslundii in initial dental biofilm formation. Microbiology. 2009;155:2116–26. doi: 10.1099/mic.0.027706-0. [DOI] [PubMed] [Google Scholar]
- [47].Kim HJ, Boedicker JQ, Choi JW, Ismagilov RF. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc Natl Acad Sci U S A. 2008;105:18188–93. doi: 10.1073/pnas.0807935105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, McIntosh ML, Alsam A, Kirkwood KL, Lambris JD, Darveau RP, Curtis MA. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bradshaw DJ, Marsh PD, Watson GK, Allison C. Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infect Immun. 1998;66:4729–32. doi: 10.1128/iai.66.10.4729-4732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kolenbrander PE, Parrish KD, Andersen RN, Greenberg EP. Intergeneric coaggregation of oral Treponema spp. with Fusobacterium spp. and intrageneric coaggregation among Fusobacterium spp. Infect Immun. 1995;63:4584–8. doi: 10.1128/iai.63.12.4584-4588.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hammond A, Dertien J, Colmer-Hamood JA, Griswold JA, Hamood AN. Serum inhibits P. aeruginosa biofilm formation on plastic surfaces and intravenous catheters. J Surg Res. 2010;159:735–46. doi: 10.1016/j.jss.2008.09.003. [DOI] [PubMed] [Google Scholar]
- [52].Williams JC, McInnis KA, Testerman TL. Adherence of Helicobacter pylori to abiotic surfaces is influenced by serum. Appl Environ Microbiol. 2008;74:1255–8. doi: 10.1128/AEM.01958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]