Abstract
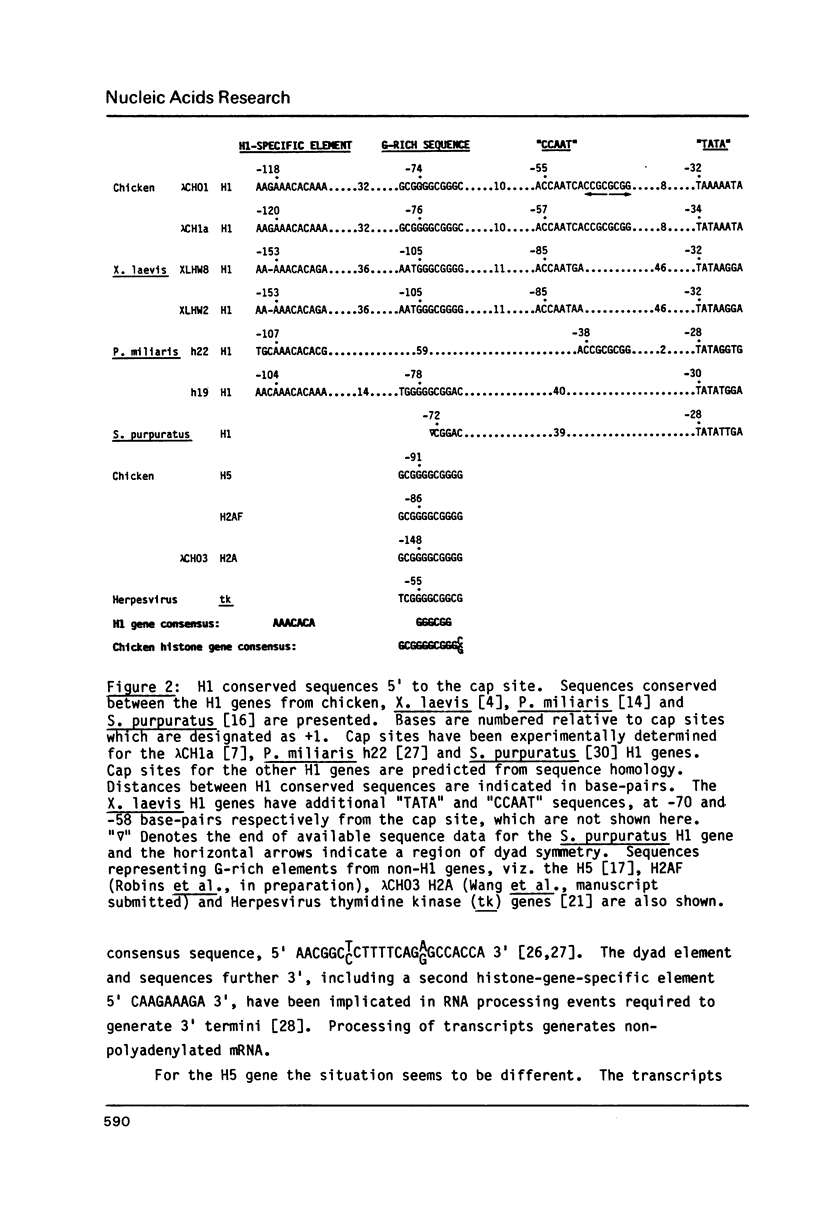
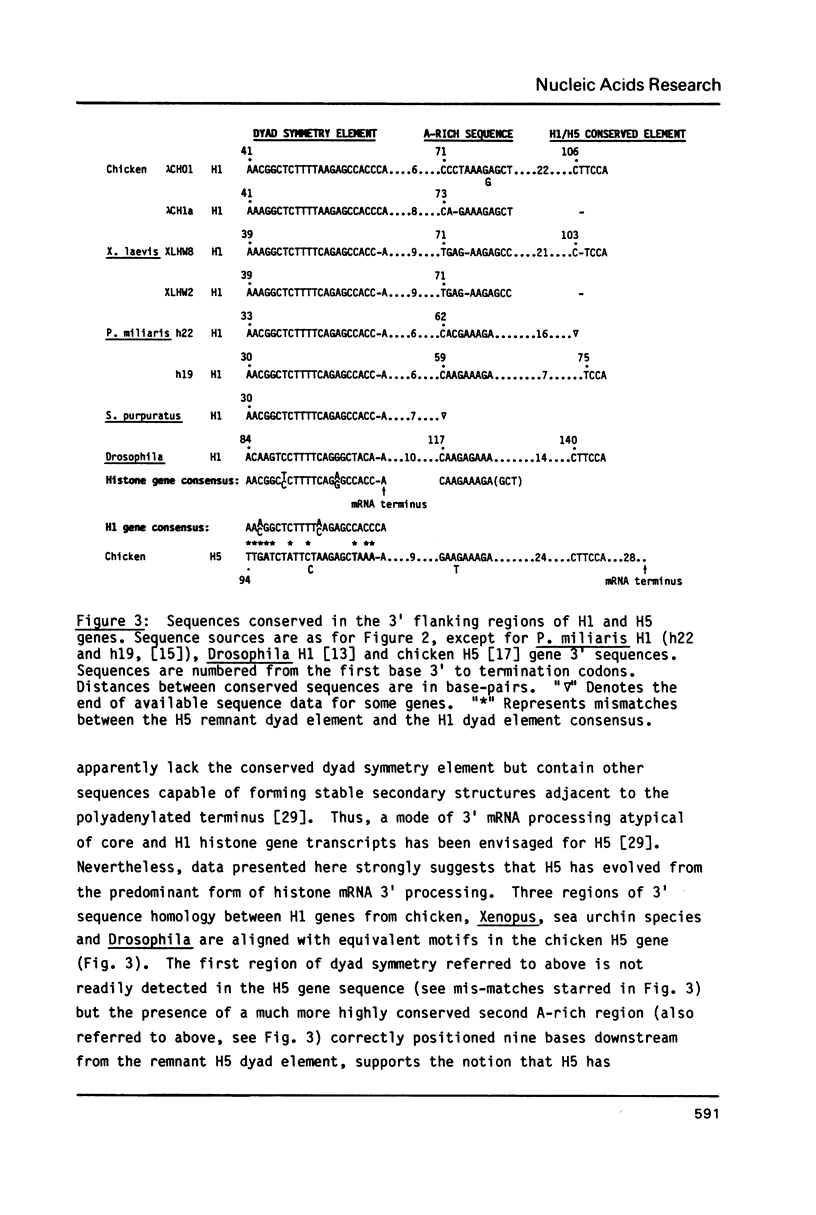
In previous studies we have shown that the H5 gene is not closely linked to the dispersed clusters of core and H1 histone genes. Here we emphasise features of H1 and H5 genes relevant to their expression in the chicken genome. Of particular note is an H1 gene-specific 5' element, 5' AAACACA 3' found upstream of all H1 genes studied to date. This "H1-box" is not found in the related H5 gene, which is expressed only in erythroid cells. A second aspect relates to generation of histone mRNA 3' termini. The H5 gene is shown to contain a remnant of the dyad symmetry element (as well as other conserved sequences) associated with core and H1-histone gene transcript 3' processing. However, it appears as if H5 has evolved a different mechanism in which the mRNA terminus (which is polyadenylated) is displaced downstream from the dyad element. The two clear differences noted here have the potential to affect transcriptional (H1-box) and post-transcriptional (3' terminus processing) regulation of H1 and H5 gene expression.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allan J., Hartman P. G., Crane-Robinson C., Aviles F. X. The structure of histone H1 and its location in chromatin. Nature. 1980 Dec 25;288(5792):675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- Birchmeier C., Folk W., Birnstiel M. L. The terminal RNA stem-loop structure and 80 bp of spacer DNA are required for the formation of 3' termini of sea urchin H2A mRNA. Cell. 1983 Dec;35(2 Pt 1):433–440. doi: 10.1016/0092-8674(83)90176-9. [DOI] [PubMed] [Google Scholar]
- Birchmeier C., Schümperli D., Sconzo G., Birnstiel M. L. 3' editing of mRNAs: sequence requirements and involvement of a 60-nucleotide RNA in maturation of histone mRNA precursors. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1057–1061. doi: 10.1073/pnas.81.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Busslinger M., Portmann R., Birnsteil M. L. A regulatory sequence near the 3' end of sea urchin histone genes. Nucleic Acids Res. 1979 Jul 11;6(9):2997–3008. doi: 10.1093/nar/6.9.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M., Cole R. D. Species and organ specificity in very lysine-rich histones. J Biol Chem. 1968 Sep 10;243(17):4500–4505. [PubMed] [Google Scholar]
- Carozzi N., Marashi F., Plumb M., Zimmerman S., Zimmerman A., Coles L. S., Wells J. R., Stein G., Stein J. Clustering of human H1 and core histone genes. Science. 1984 Jun 8;224(4653):1115–1117. doi: 10.1126/science.6719136. [DOI] [PubMed] [Google Scholar]
- Clerc R. G., Bucher P., Strub K., Birnstiel M. L. Transcription of a cloned Xenopus laevis H4 histone gene in the homologous frog oocyte system depends on an evolutionary conserved sequence motif in the -50 region. Nucleic Acids Res. 1983 Dec 20;11(24):8641–8657. doi: 10.1093/nar/11.24.8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierks P., van Ooyen A., Cochran M. D., Dobkin C., Reiser J., Weissmann C. Three regions upstream from the cap site are required for efficient and accurate transcription of the rabbit beta-globin gene in mouse 3T6 cells. Cell. 1983 Mar;32(3):695–706. doi: 10.1016/0092-8674(83)90055-7. [DOI] [PubMed] [Google Scholar]
- Gjerset R., Gorka C., Hasthorpe S., Lawrence J. J., Eisen H. Developmental and hormonal regulation of protein H1 degrees in rodents. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2333–2337. doi: 10.1073/pnas.79.7.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R., Mächler M., Rohrer U., Birnstiel M. L. A functional component of the sea urchin H2A gene modulator contains an extended sequence homology to a viral enhancer. Nucleic Acids Res. 1983 Dec 10;11(23):8123–8136. doi: 10.1093/nar/11.23.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R., Wasylyk B., Chambon P., Birnstiel M. L. Point mutation in the TATA box curtails expression of sea urchin H2A histone gene in vivo. Nature. 1981 Nov 12;294(5837):178–180. doi: 10.1038/294178a0. [DOI] [PubMed] [Google Scholar]
- Harvey R. P., Robins A. J., Wells J. R. Independently evolving chicken histone H2B genes: identification of a ubiquitous H2B-specific 5' element. Nucleic Acids Res. 1982 Dec 11;10(23):7851–7863. doi: 10.1093/nar/10.23.7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C., Irminger J. C., Bucher P., Birnstiel M. L. Sea urchin histone mRNA termini are located in gene regions downstream from putative regulatory sequences. Nature. 1980 May 15;285(5761):147–151. doi: 10.1038/285147a0. [DOI] [PubMed] [Google Scholar]
- Kinkade J. M., Jr Qualitative species differences and quantitative tissue differences in the distribution of lysine-rich histones. J Biol Chem. 1969 Jun 25;244(12):3375–3386. [PubMed] [Google Scholar]
- Krieg P. A., Robins A. J., Colman A., Wells J. R. Chicken histone H5 mRNA: the polyadenylated RNA lacks the conserved histone 3' terminator sequence. Nucleic Acids Res. 1982 Nov 11;10(21):6777–6785. doi: 10.1093/nar/10.21.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg P. A., Robins A. J., D'Andrea R., Wells J. R. The chicken H5 gene is unlinked to core and H1 histone genes. Nucleic Acids Res. 1983 Feb 11;11(3):619–627. doi: 10.1093/nar/11.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S., Sures I., Kedes L. The nucleotide and amino acid coding sequence of a gene for H1 histone that interacts with euchromatin. The early embryonic H1 gene of the sea urchin Strongylocentrotus purpuratus. J Biol Chem. 1982 Aug 25;257(16):9438–9443. [PubMed] [Google Scholar]
- McKnight S. L. Functional relationships between transcriptional control signals of the thymidine kinase gene of herpes simplex virus. Cell. 1982 Dec;31(2 Pt 1):355–365. doi: 10.1016/0092-8674(82)90129-5. [DOI] [PubMed] [Google Scholar]
- Panyim S., Bilek D., Chalkley R. An electrophoretic comparison of vertebrate histones. J Biol Chem. 1971 Jul 10;246(13):4206–4215. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson E. C., Erba H. P., Gall J. G. Characterization of a cloned histone gene cluster of the newt Notophthalamus viridescens. Nucleic Acids Res. 1981 May 25;9(10):2281–2295. doi: 10.1093/nar/9.10.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugarman B. J., Dodgson J. B., Engel J. D. Genomic organization, DNA sequence, and expression of chicken embryonic histone genes. J Biol Chem. 1983 Jul 25;258(14):9005–9016. [PubMed] [Google Scholar]
- Sures I., Levy S., Kedes L. H. Leader sequences of Strongylocentrotus purpuratus histone mRNAs start at a unique heptanucleotide common to all five histone genes. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1265–1269. doi: 10.1073/pnas.77.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner P. C., Aldridge T. C., Woodland H. R., Old R. W. Nucleotide sequences of H1 histone genes from Xenopus laevis. A recently diverged pair of H1 genes and an unusual H1 pseudogene. Nucleic Acids Res. 1983 Jun 25;11(12):4093–4107. doi: 10.1093/nar/11.12.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]



