Abstract
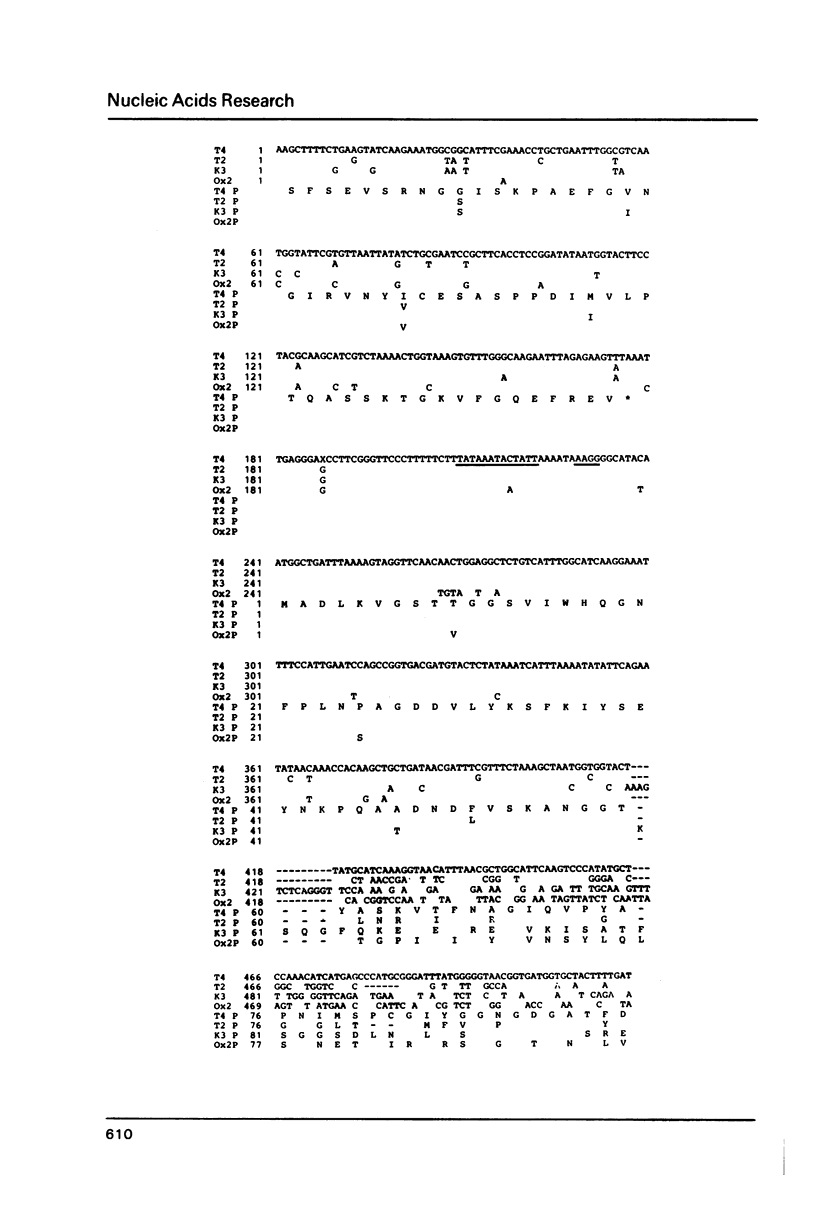
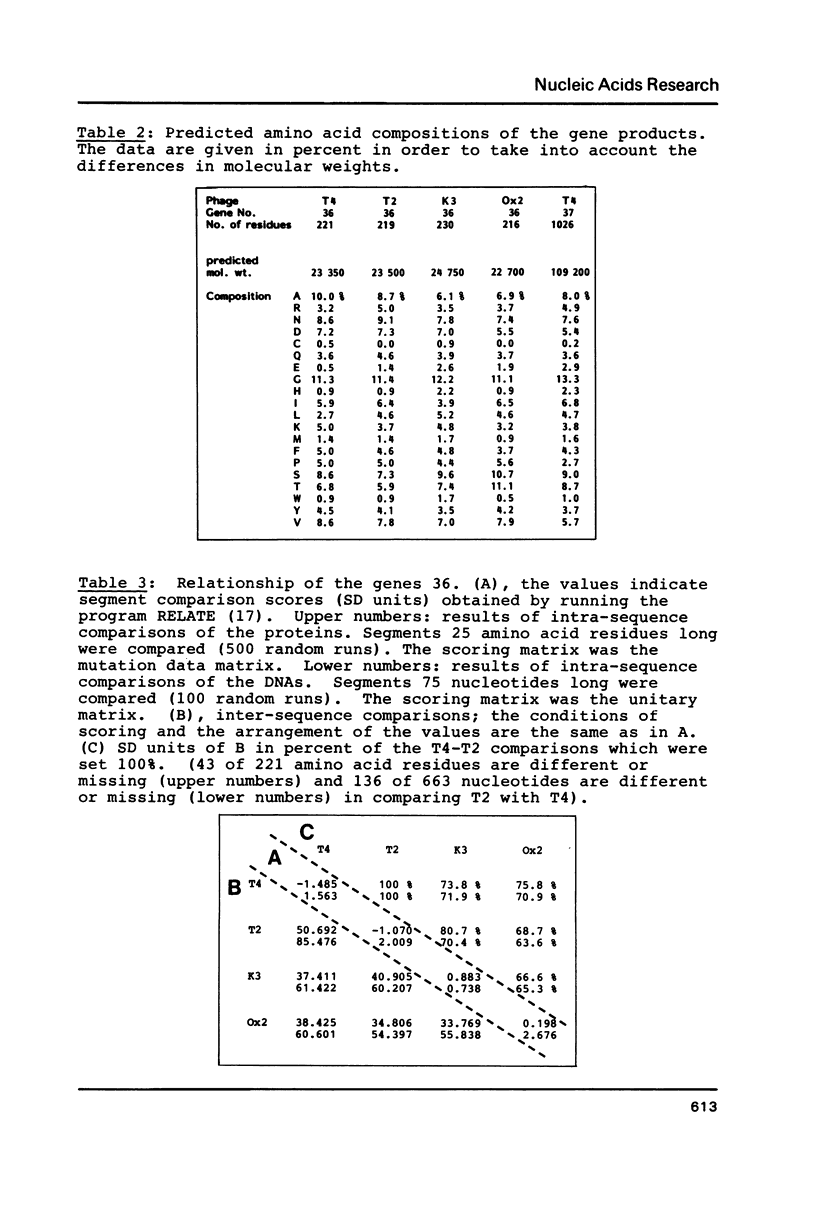
Genes 36 have been cloned from phage T2 and the T-even type phages K3 and Ox2. The products of these genes are part of the long tail fibers of the phages, they form the proximal moiety of the distal half fiber. The genes have been sequenced, the nucleotide sequence of gene 36 of phage T4 is known (Oliver, D.B. & Crowther, R.A. (1981) J.Mol.Biol. 153, 545-568). Comparison of the deduced amino acid sequences of the four proteins revealed a surprising pattern. These sequences can be divided into two highly conserved and one very variable region. The former consist of about 60 NH2-terminal and 70 CO2H-terminal residues flanking the variable middle region of about 100 residues. Thus, an identical and unique morphology can be formed by a number of different primary structures. It is proposed that the conserved areas are involved in binding of the proteins to the neighboring products of genes 35 and 37 and that this function has put constraints on the variability of the primary protein structure. The overall amino acid composition of the proteins is rather similar; the codon usage is that known for phage T4. The intercistronic region between genes 35 and 36 consisting of 62 base pairs and containing a presumed terminator for g35 transcription and the 'late type' promoter for transcription of genes 36, 37, and 38, is almost completely identical in the four phages.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beckendorf S. K., Kim J. S., Lielausis I. Structure of bacteriophage T4 genes 37 and 38. J Mol Biol. 1973 Jan;73(1):17–35. doi: 10.1016/0022-2836(73)90156-3. [DOI] [PubMed] [Google Scholar]
- Beckendorf S. K. Structure of the distal half of the bacteriophage T4 tail fiber. J Mol Biol. 1973 Jan;73(1):37–53. doi: 10.1016/0022-2836(73)90157-5. [DOI] [PubMed] [Google Scholar]
- Dayhoff M. O., Barker W. C., Hunt L. T. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- Elliott T., Geiduschek E. P. Defining a bacteriophage T4 late promoter: absence of a "-35" region. Cell. 1984 Jan;36(1):211–219. doi: 10.1016/0092-8674(84)90091-6. [DOI] [PubMed] [Google Scholar]
- King J., Laemmli U. K. Polypeptides of the tail fibres of bacteriophage T4. J Mol Biol. 1971 Dec 28;62(3):465–477. doi: 10.1016/0022-2836(71)90148-3. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Oliver D. B., Crowther R. A. DNA sequence of the tail fibre genes 36 and 37 of bacteriophage T4. J Mol Biol. 1981 Dec 15;153(3):545–568. doi: 10.1016/0022-2836(81)90407-1. [DOI] [PubMed] [Google Scholar]
- Revel H. R. Molecular cloning of the T4 genome: organization and expression of the tail fiber gene cluster 34--38. Mol Gen Genet. 1981;182(3):445–455. doi: 10.1007/BF00293934. [DOI] [PubMed] [Google Scholar]
- Riede I., Eschbach M. L., Henning U. DNA sequence heterogeneity in the genes of T-even type Escherichia coli phages encoding the receptor recognizing protein of the long tail fibers. Mol Gen Genet. 1984;195(1-2):144–152. doi: 10.1007/BF00332737. [DOI] [PubMed] [Google Scholar]
- Rüther U., Koenen M., Otto K., Müller-Hill B. pUR222, a vector for cloning and rapid chemical sequencing of DNA. Nucleic Acids Res. 1981 Aug 25;9(16):4087–4098. doi: 10.1093/nar/9.16.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz H., Riede I., Sonntag I., Henning U. Degrees of relatedness of T-even type E. coli phages using different or the same receptors and topology of serologically cross-reacting sites. EMBO J. 1983;2(3):375–380. doi: 10.1002/j.1460-2075.1983.tb01433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurray R. A., Hancock R. E., Reeves P. Con--mutants: class of mutants in Escherichia coli K-12 lacking a major cell wall protein and defective in conjugation and adsorption of a bacteriophage. J Bacteriol. 1974 Sep;119(3):726–735. doi: 10.1128/jb.119.3.726-735.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]



