Abstract
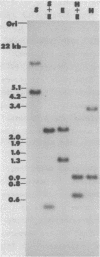
Analysis of total Drosophila melanogaster DNA by genomic blot hybridization indicates that two cytochrome c-like sequences exist in the Drosophila genome. These two sequences, DC3 and DC4, have been isolated from a Charon 4A-D. melanogaster genomic library. DC3 and DC4 are located within a 4 kb region of DNA, at position 36A 10-11, on the left arm of chromosome 2. The nucleotide sequence of these two clones has been determined. Both DC3 and DC4 can encode functional cytochrome c proteins. The polypeptide sequences predicted by these two genes, however, differ at 32 amino acid residues. DC4 is expressed at varying, but relatively high levels throughout Drosophila development. In contrast, DC3 is expressed at constant, but relatively low levels throughout development.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blattner F. R., Williams B. G., Blechl A. E., Denniston-Thompson K., Faber H. E., Furlong L., Grunwald D. J., Kiefer D. O., Moore D. D., Schumm J. W. Charon phages: safer derivatives of bacteriophage lambda for DNA cloning. Science. 1977 Apr 8;196(4286):161–169. doi: 10.1126/science.847462. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976 Sep;9(1):91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E. The structures of cytochrome c and the rates of molecular evolution. J Mol Evol. 1971;1(1):26–45. doi: 10.1007/BF01659392. [DOI] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Limbach K. J., Wu R. Isolation and characterization of two alleles of the chicken cytochrome c gene. Nucleic Acids Res. 1983 Dec 20;11(24):8931–8950. doi: 10.1093/nar/11.24.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARGOLIASH E. PRIMARY STRUCTURE AND EVOLUTION OF CYTOCHROME C. Proc Natl Acad Sci U S A. 1963 Oct;50:672–679. doi: 10.1073/pnas.50.4.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Hardison R. C., Lacy E., Lauer J., O'Connell C., Quon D., Sim G. K., Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978 Oct;15(2):687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Margoliash E., Schejter A. Cytochrome c. Adv Protein Chem. 1966;21:113–286. doi: 10.1016/s0065-3233(08)60128-x. [DOI] [PubMed] [Google Scholar]
- Matner R. R., Sherman F. Differential accumulation of two apo-iso-cytochromes c in processing mutants of yeast. J Biol Chem. 1982 Aug 25;257(16):9811–9821. [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Montgomery D. L., Leung D. W., Smith M., Shalit P., Faye G., Hall B. D. Isolation and sequence of the gene for iso-2-cytochrome c in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jan;77(1):541–545. doi: 10.1073/pnas.77.1.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan C., Margoliash E. Comparative aspects of primary structures of proteins. Annu Rev Biochem. 1968;37:727–790. doi: 10.1146/annurev.bi.37.070168.003455. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Russell P. R., Hall B. D. Structure of the Schizosaccharomyces pombe cytochrome c gene. Mol Cell Biol. 1982 Feb;2(2):106–116. doi: 10.1128/mcb.2.2.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Scarpulla R. C., Agne K. M., Wu R. Cytochrome c gene-related sequences in mammalian genomes. Proc Natl Acad Sci U S A. 1982 Feb;79(3):739–743. doi: 10.1073/pnas.79.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla R. C., Agne K. M., Wu R. Isolation and structure of a rat cytochrome c gene. J Biol Chem. 1981 Jun 25;256(12):6480–6486. [PubMed] [Google Scholar]
- Scarpulla R. C., Wu R. Nonallelic members of the cytochrome c multigene family of the rat may arise through different messenger RNAs. Cell. 1983 Feb;32(2):473–482. doi: 10.1016/0092-8674(83)90467-1. [DOI] [PubMed] [Google Scholar]
- Sherman F., Stewart J. W. Genetics and biosynthesis of cytochrome c. Annu Rev Genet. 1971;5:257–296. doi: 10.1146/annurev.ge.05.120171.001353. [DOI] [PubMed] [Google Scholar]
- Sherman F., Taber H., Campbell W. Genetic determination of iso-cytochromes c in yeast. J Mol Biol. 1965 Aug;13(1):21–39. doi: 10.1016/s0022-2836(65)80077-8. [DOI] [PubMed] [Google Scholar]
- Smith M., Leung D. W., Gillam S., Astell C. R., Montgomery D. L., Hall B. D. Sequence of the gene for iso-1-cytochrome c in Saccharomyces cerevisiae. Cell. 1979 Apr;16(4):753–761. doi: 10.1016/0092-8674(79)90091-6. [DOI] [PubMed] [Google Scholar]