Abstract
Prolactin and the prolactin receptors are members of a family of hormone/receptor pairs which include GH, erythropoietin, and other ligand/receptor pairs. The mechanisms of these ligand/receptor pairs have broad similarities, including general structures, ligand/receptor stoichiometries, and activation of several common signaling pathways. But significant variations in the structural and mechanistic details are present among these hormones and their type 1 receptors. The prolactin receptor is particularly interesting because it can be activated by three sequence-diverse human hormones: prolactin, GH, and placental lactogen. This system offers a unique opportunity to compare the detailed molecular mechanisms of these related hormone/receptor pairs. This review critically evaluates selected literature that informs these mechanisms, compares the mechanisms of the three lactogenic hormones, compares the mechanism with those of other class 1 ligand/receptor pairs, and identifies information that will be required to resolve mechanistic ambiguities. The literature describes distinct mechanistic differences between the three lactogenic hormones and their interaction with the prolactin receptor and describes more significant differences between the mechanisms by which other related ligands interact with and activate their receptors.
Introduction
-
Prolactin
Prolactin: sequence and structure
Prolactin: functional mechanisms
Prolactin: rate constants, affinities, and thermodynamics
-
Prolactin Receptor
Prolactin receptor: sequence, isoforms, and structure
What are the mechanisms that activate the prolactin receptor?
Conclusions
I. Introduction
Prolactin (PRL) is a class 1 helical protein hormone (1) produced in both autocrine/paracrine and endocrine systems and is responsible for a wide variety of physiological actions in vertebrates including fish, amphibians, reptiles, and mammals (2, 3). PRL activates target cells by interaction with a pair of single-transmembrane (TM) domain type 1 cytokine receptors located on the plasma membrane of target cells. Isoforms of the PRL receptor are derived by differential splicing of the RNA transcribed from a single gene (4) or by posttranslational modifications. Substantial progress has been made in identifying and characterizing the molecules involved in the human PRL (hPRL) signaling pathway (5); these pathways include the Janus kinase (JAK)/signal transducers and activators of transcription (STAT) system and others (6).
PRL has a wide variety of target tissues, including mammary gland, prostate, ovary, cells of the immune system, adipocytes, liver, and other tissues. PRL, growth factors, and steroids complement each other's actions to stimulate the growth and development of these tissues. The biological actions of PRL have been described by earlier review articles (7–9).
Lactogenic hormones and hPRL receptors are excellent models for studying the protein/protein interactions that initiate the molecular mechanisms that regulate the activities of the binding complex. Too few molecular interactions have been characterized to describe a classification system by which molecular interactions modulate activity. The hPRL receptor is an especially important model for cytokine/receptor interactions in that three ligands with significantly different chemical features bind and activate this protein. We must identify the appropriate physical models for this class of ligand/receptor interactions and gain an understanding of the binding order, kinetics, and energetics to derive how such interactions regulate the activities of these molecules. This information, obtained from a wide variety of binding pairs, is required to understand the principles of molecular function and provide the principles for rational protein engineering.
Because PRL is implicated in the initiation and progression of cancers of the breast and prostate (10, 11), the design of antagonists is a focus of current research (12, 13). But the rational design of either agonists or antagonists requires and awaits an accurate and detailed understanding of the molecular mechanisms by which PRL activates its receptor.
Considerable data have described the chemistry and biology of PRL and have led to identification of the molecules that carry out intracellular (IC) function (5). However, a less complete picture emerges when one examines the molecular mechanisms by which these molecules perform their functions. Similarities in the structures and utilization of common signaling pathways among PRL, GH, and erythropoietin (EPO) have been used to advance the belief that PRL will bind and activate its receptor by mechanisms similar to those described for GH and EPO (14). But data supporting this belief are scarce and have not been critically evaluated. The intention of this review is to identify and critically review the literature addressing the mechanisms by which PRL, GH, and placental lactogen (PL) bind and activate the PRL receptor. In addition, GH/receptor and erythropoietin/receptor pairs are compared with lactogen/receptor pairs. This review will largely focus on these systems in humans. This evaluation will provide insights into both the common and unique aspects of the mechanisms of receptor activation. I conclude that despite similar three-dimensional structures and the utilization of common signaling mechanisms, PRL binds and activates the PRL receptor by a mechanism unique from those used by other lactogens. I suggest that these mechanistic differences largely are a consequence of differences in the surface chemical topology provided by divergent amino acid sequences. Although the mechanism by which PRL binds and activates the PRL receptor is unsettled, it appears to be significantly different from those described for either the GH/receptor or EPO/receptor pair.
II. Prolactin
A. Prolactin: sequence and structure
PRL is a single peptide of approximately 200 amino acids. PRL orthologs have been identified in all classes of vertebrates (2, 3), suggesting that the coding sequence for PRL is an ancient gene evolved to modulate diverse activities. Based on sequence homology, hPRL can be considered a typical mammalian PRL. Compared with the seven mammalian species examined in Supplemental Fig. 1A (published on The Endocrine Society's Journals Online web site at http://edrv.endojournals.org), the hPRL sequence shares approximately a 44% identity and an additional 35% similarity. hPRL, human GH (hGH), and human PL (hPL) bind and activate several forms of the hPRL receptor and provide a rich system to compare the molecular mechanisms by which these hormones bind the hPRL receptor. These three human lactogenic hormones have varying sequence homologies (Supplemental Fig. 1B). hGH and hPL share an 85% sequence homology. In contrast, these two hormones share 21 and 22% homology with hPRL, respectively. The low sequence homologies between hPRL and either hGH or hPL produce unique chemical topologies on the surfaces of these lactogens that define their binding interfaces. Comparing the sequences of hPRL with hGH or hPL, the absence of homology beyond several contiguous residues does not argue for the presence of identical receptor binding surfaces. These observations suggest that the sequence differences displayed in their three-dimensional structures will provide unique molecular mechanisms by which hPRL, hGH, or hPL binds and activates hPRL receptor. This argument is supported by structural studies that identify a modest conservation of receptor-binding epitopes among these lactogens.
Structures of hPRL (15–20), hGH (21), and hPL (22), when either free or bound to the hPRL receptor, are rapidly providing a platform for the interpretation of structure/function studies. A nuclear magnetic resonance (NMR) structure of hPRL in the absence of receptor provided the first hPRL structure (15). The protein's four long helices fold into a parallel bundle with an up-up-down-down pattern (23) (Fig. 1A and Supplemental Fig. 2). This fold qualifies hPRL for membership in the long-helix-bundle group of approximately 20 cytokines (24) that includes its closest neighbors: hGH and hPL (Supplemental Fig. 1B). X-ray structures are available for mutant hPRL (G129R or Δ1–9/G129R) bound to a single extracellular (EC) domain of hPRL receptor (16, 17, 19). When bound by a single EC domain, hPRL retains its general fold, but several modest changes are observed including loop 1 (between helix 1 and 2). A 3.8-Å structure of hPRL bound to two EC domains of the rat PRL receptor (18, 20) provides the structure of the heterotrimeric hormone/EC domain complex and suggests that binding a second receptor does not induce further changes in the structure of hPRL. These structural studies describe two receptor-binding surfaces each on unique surfaces of hPRL, termed sites 1 and 2. This structure also shows a binding interface between the S2/S2 subdomains of the EC domain of the PRL receptor. The site 2 and S2/S2 interfaces constitute the binding surfaces for the second hPRL receptor as it binds the hPRL/receptor heterodimer. Similar studies of hGH (21) or hPL (25) bound to the EC domain of the PRL receptor reveal similar hormone/receptor binding surfaces. Structural, biochemical (26), and biophysical studies (27, 28) demonstrate that PRL binds two PRL receptors at these unique surfaces in an allostery-mediated obligate-ordered fashion.
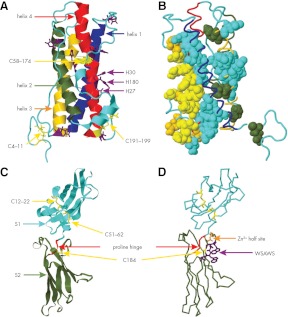
Figure 1.
Structures of hPRL and the EC domain of the hPRL receptor. A, hPRL, PDB no. 1RW5 (ribbon). Helix 1, residues 14–43 (blue); helix 2, residues 78–103 (green); helix 3, residues 111–137 (gold); and helix 4, residues 161–193 (red). Disulfides (yellow stick), residues 4–11, 58–174, and 191–195. Histidines (purple stick). B, hPRL (cyan backbone); residues displaying significant ΔΔG when mutated to alanine (solid). Solid gold, site 1 residues; solid yellow, site 1 residues noncovalently linked to the hPRL receptor. Site 1, Loop 1 (I55, N56, P66, E67, and Q73) and helix 4 (R177, H180, K181, D183, N184, K187, R192, N197, and N198). Solid green, site 2; helix 1 (Q12), and helix 3 (Q122, L126, and L132). Solid cyan, Coupling motif; helix 1 (F19, V23, S33, S34, and F37), loop 1 (S57 and L63), helix2/helix 3 junction (E110), helix 3 (E118, I119, E120, E121, T123, K124, and L127), helix 4 (Y169, N170, D178, S179, L189, and I193), and C terminus (I194 and N196). C, EC domain of the hPRL receptor (PDB no. 3NO6, ribbon residues 2–204). Cyan, S1 subdomain; green, S2 subdomain; yellow stick, cysteines (184) and disulfides (12–22, 51–62); red, proline hinge (PDPP, residues 103–106). D, EC domain of the hPRL receptor; yellow stick, cysteine 184; gold stick, Zn2+ half-site (gold stick residues D187 and H188); and blue stick, WSAWS motif (residues W191, S192, A193, W194, and S195).
X-ray studies have revealed 18 residues as structural epitopes for site 1 of hPRL (within 3.5 Å of the hPRL receptor) (17). Only nine of these residues are functional epitopes (29). A slightly larger set of site 1 functional epitopes are observed when either hGH or hPL is bound to hPRL receptor (21, 22). The structural epitopes for site 2 were identified in x-ray studies where hPRL bound the EC domain of rat receptor to form a heterotrimeric complex (18, 20). Finally, a series of heterodimeric structures of G129R-hPRL bound to a single EC domain of the hPRL receptor and containing various histidine mutations in either hPRL or the receptor domain have been published at a significantly higher resolution (2.0 to 2.5 Å) (19). The overall helical structure of these hPRL is retained, but modest structural changes are observed in the various receptor-bound hPRL.
Caution must be used when interpreting structural differences between various x-ray or standard NMR-based hPRL studies (30). Only comparisons of high resolution structures obtained under identical conditions are likely to provide valid interpretations. Even under optimal conditions, the influences of crystal packing and solute/solvent conditions may produce modest structural differences from the molecule in vivo. More consequential, structural information does not inform the dynamics of these proteins.
Mammalian lactogens contain both conserved and unique features (Supplemental Fig. 1B). The most obvious difference is that PRL contain an N-terminal sequence not present in either GH or hPL. Removal of between nine and 14 N-terminal residues of hPRL introduces functional changes (31). Deletion of nine N-terminal resides of hPRL in the presence of the G129R mutation produces a highly potent hPRL antagonist (32). hPRL has three disulfide bonds. hPRL contains a small eight-residue N-terminal disulfide loop (cysteines 4 and 11) located in the nonhelical portion of the protein. The N-terminal disulfide loop is retained in mammals, although it is slightly smaller in murine species. The N-terminal disulfide loop shows substantial mobility, as judged by the structural variations observed among the 20 calculated solutions for the NMR structure of hPRL (15). This N-terminal disulfide loop is not present in either GH or hPL. A large disulfide loop (cysteines 58 and 174) covalently links the N- and C-terminal portions of the protein and constrains their relative motions. Elimination of this large disulfide loop eliminates activity (33). Finally, a nine-residue disulfide loop between cysteines 191 and 199 in hPRL restricts motion of the C-terminal, as judged from the similarities of the various calculated NMR C-terminal structures (15). C191 is within helix 4, and disulfide formation with C199 reduces the movement of this C-terminus disulfide loop by fixing it to the C-terminus of helix 4. The central and C-terminal disulfide loops are retained in mammalian PRL (Supplemental Fig. 1A). All three human lactogens contain both the central and C-terminal disulfide loops but with modest size differences. Disruption of either the N- or C-terminal disulfide loops by selective chemical reduction or mutagenesis decreases, but does not eliminate, the biological activities of PRL, PL, or GH (33–35).
hPRL, hGH, and hPL each contain four long helices. Helices 1 (residues 14 through 43; 30 residues), 2 (residues 78 through 103; 26 residues), 3 (residues 111 through 137; residues 27), and 4 (residues 161 through 193; residues 33) of hPRL comprise the four-helix bundle (Fig. 2A). The arrangement of these helices (Supplemental Fig. 2) is typical for this class of four long helix bundle cytokines. In molecular dynamics simulations, we observed that the four large helices in hGH are quite rigid (J. M. Canfield and C. L. Brooks, unpublished observations); hPRL may share this property. Thermal denaturation studies of hPRL following helical structure by circular dichroism showed a melting temperature (Tm50%) of approximately 75 C (J. Patmastan and C. L. Brooks, unpublished observations). Based on Protein Data Bank (PDB) no. 3MZG, stabilizing salt bridges (carboxyl oxygen to amine nitrogen >4Å in side chains) are present in helix 3 (E120/K124: 3.93Å) and helix 4 (E161/R164: 3.77Å, and D183/K187: 2.85Å). Within the 116 residues in these long helical sequences, only two residues might disrupt or destabilize the helical structures: P94 and G129 in helices 2 and 3, respectively. Neither residue breaks the helices in which they reside as determined from x-ray data (15–20). P94 is conserved in most species (Fig. 1A). In bovine PRL, P94 is located near the major site of phosphorylation at S90 (36). G129 is critical to site 2 receptor binding (16). This glycine residue is located in the binding site for the second receptor and sits in the bottom of a pocket into which a critical receptor tryptophan is inserted. Replacement of this glycine with arginine fills this pocket with a large charged side chain and inhibits insertion of the receptor tryptophan, creating a steric antagonist that retains between 1 and 10% activity of the wild-type hormone. Based on the more favorable helix-forming properties of arginine, the G129R mutation also may increase the stability of helix 3. The helix 2 proline and the helix 3 glycine are conserved in both hGH and hPL (Supplemental Fig. 1B). In either hPL or hGH, replacement of G120 (corresponding to G129 of hPRL) with arginine produces antagonists for either the hPRL or hGH receptors (37, 38). These data indicate the functional centrality of this motif for these three members of this hormone/receptor family.
Figure 2.
Schematic comparison of natural and man-made hPRL receptor isoforms. EC domain, blue; TM domain, yellow; and IC domain, green. Long, long form of the hPRL receptor; intermed, intermediate form of the hPRL receptor; short 1a, short 1a form of the hPRL receptor; short 1b, short 1b form of the hPRL receptor; ΔS1, long form of the hPRL receptor with the S1 subdomain deleted; ΔS2, long form of the hPRL receptor with the S2 subdomain deleted; ECD, extracellular domain; ΔICD, long form of the hPRL receptor with the intracellular domain deleted; ΔECD, long form of the hPRL receptor with the extracellular domain removed; Δ10-186, long form of the hPRL receptor with residues 10-186 removed; I76V, long form of the hPRL receptor with isoleucine 76 replaced with valine; l146L, long form of the hPRL receptor with isoleucine 146 replaced with leucine.
Two long sequences connect helices 1 and 2 (residues 44 through 77; 34 residues) and helices 3 and 4 (residues 138–160; 23 residues). A third short connection between helices 2 and 3 contains only seven residues. The sequence connecting helices 1 and 2 contains a short seven-residue α-helix, a four residue 3/10 helix, and cysteine 58, a member of the large disulfide loop coupling loop 1 and helix 4. Loop 1 contains site 1 structural epitopes. The second short helix in loop 1 of hPRL lays in a groove between the S1 and S2 subdomains of the hPRL receptor bound to site 1.
B. Prolactin: functional mechanisms
The similarities and differences in the molecular mechanisms by which hPRL and other ligands for type 1 receptors initiate their activities are only now coming to light. Two hPRL receptors bind sites 1 and 2 of the hormone, producing an active heterotrimeric complex (20, 26–28). We have proposed that the mechanism of receptor binding to hPRL is an obligate-ordered process where hPRL receptor cannot bind site 2 unless site 1 has previously bound a receptor (27, 28). These data imply that sites 1 and 2 are functionally coupled by an allosteric mechanism. Förster resonance energy transfer studies show that receptor binding at site 1 of hPRL induces a conformation change. Motifs coupling sites 1 and 2 have been identified in hGH and hPL (28,34) but in hPRL we failed to identify a similar coupling motif (T.J. Gordon, U. Sivaprasad, and C.L. Brooks, unpublished observations).
Recently, a set of functional epitopes for hPRL was identified by Rao and Brooks (29). Forty-one residues with significant changes in ΔG were identified by alanine mutagenesis (Fig. 2B) of 102 tested residues. These 41 residues were sorted into three groups. The first group contained structural residues within 3.5 Å of the hPRL receptor, with some forming noncovalent bonds with hPRL receptor. These nine functional residues (N56, E67, R177, H180, D183, N184, K187, N197, and N198) were found among the structural epitopes identified by Svensson et al. (17) and were located in loop 1, helix 4, and the C terminus of hPRL. Despite containing structural epitopes, no functional epitopes were found in helix 1 of hPRL. Six of these nine residues had contacts with the hPRL receptor that were consistent with noncovalent bonds. The second group also contained structural epitopes within 3.5 Å of the receptor. They provided noncovalent bonds within hPRL but did not provide noncovalent bonds to the receptor; these interactions appeared to couple the functional epitopes with the body of the hormone. The third and largest group of residues was not found among the structural epitopes, but their mutation to alanine changed the ΔG for site 1 binding. The residues of this third group formed a contiguous motif between site 1 and site 2 (Fig. 1B). Members of this group may form the motif that allosterically couples site 1 with site 2. In additional mutagenic studies, we identified similar but nonidentical sets of residues that functionally couple sites 1 and 2 of hGH (39). Comparison of the structural epitopes of hPRL (17), hGH (21), and hPL (22) identifies sets of modestly conserved residues with overlapping but not identical sets of functional epitopes. These data strongly indicate that the receptor binding residues are somewhat preserved, but the details of binding are unique.
When considering functional coupling of sites 1 and 2 in lactogenic hormones, one needs to consider the nature of allostery in a modern context. Decades ago, a general mechanism for allostery was proposed by Pauling (40) and Wyman (41, 42) and further developed into the Monod, Wyman, Changeux (43) and Koshland, Nemethy, Filmer (44) models. These models assumed allostery involved ligand-induced switching between two conformers of a protein. A linkage that communicated the structural change was contained within the residues of a motif coupling the two sites.
The current understanding of the dynamic nature of proteins has required evolution of allosteric models (45). Current views of allostery have several tenets. 1) Proteins occupy multiple conformational states, and this ensemble is influenced by ligand binding. 2) By classic structural studies, one may or may not be able to document a ligand-induced change in conformation; rather one may need to examine ligand-induced changes in the ensemble of conformers, a study that remains to be performed for this class of hormones. 3) A ligand-induced conformation change utilizes multiple structural pathways that functionally link sites 1 and 2. 4) These processes can be described by thermodynamics (combinations of entropy, enthalpy, and heat capacity). Allosteric coupling of sites 1 and 2 may be documented by the presence of several measurable physical characteristics, including changes in structure as measured by physical techniques including NMR (46) or x-ray crystallography (47), biophysical techniques such as Förster resonance energy transfer (48), and possibly statistical mechanics (49). Completion of double-mutation studies may more clearly identify residues that functionally couple sites 1 and 2 (50). Kossiakoff and colleagues have demonstrated that hGH undergoes a conformation change when binding the EC domain of the hGH receptor (51) or when binding the EC domain of the hPRL receptor (21).
hPRL binding to the EC domain of the hPRL receptor is pH dependent within the physiological range (52). pH-Dependent receptor binding is mediated by a cluster of three histidine residues located in site 1 of hPRL (53). hPRL contains six histidines (Fig. 1A and Supplemental Fig. 1B). Mutagenesis of H27, H30, or H180 reduces receptor binding by hPRL, but H180 appears to be the critical residue. Rao and Brooks (29) confirmed the importance of H180 in hPRL, weakening the affinity of site 1 by 339-fold, whereas alanine mutants of H27 and H30 produced small insignificant changes in affinity (in the absence of Zn2+). In addition, H188 of the hPRL receptor also contributes to the pH dependence of binding (19). A pH dependence is not observed in hGH (52), and no information is available for hPL. hGH contains only three histidines, two of which (H18 and H21) were homologous to H27 and H30 of hPRL, but hGH did not contain a histidine at the position equivalent to hPRL's H180 (D171; Supplemental Fig. 1B). In nonprimate, GH H170 (closely corresponding to H180 in hPRL) is present and functions as an important determinant for species specificity (54, 55). hPL contained six histidines, two of which (H18 and H21) are common to all lactogens. The remaining four histidines of hPL are unique when compared with hPRL, but one of these four histidines (H151) was retained in hGH. Based on the unique positions of histidines within the structures of lactogens, it is reasonable that each hormone has a unique pH dependence for binding and activation of the hPRL receptor. The histidine cluster of hPRL involved in binding and activation is close to the putative binding site for Zn2+. Based on these observations of histidine biochemistry, hPRL uses a different molecular mechanism to bind hPRL receptors than either hGH or hPL.
Zn2+ binds hPRL utilizing residue H27 (56). The structure of the dimeric hGH/hPRL receptor complex demonstrates that the Zn2+ binding site is shared by half-sites in the hormone and receptor (21). The receptor half-site is within the S2 subdomain of the hPRL receptor and uses adjacent residues: D187 and H188. The conserved half-site in bovine PRL bound Zn2+ with a micromolar affinity (57). Zn2+ modestly weakened the site 1 affinity of hPRL for the EC domain of hPRL receptor but strengthens the observed global affinity (58). In contrast, Zn2+ strengthened both the global and site 1 affinities of hGH or hPL.
Although total plasma concentrations of Zn2+ are in the micromolar range (59), free plasma Zn2+ concentrations are in the nanomolar range (60); thus, in the circulation PRL is unlikely to be bound by Zn2+. The affinity of the complete Zn2+-binding site, composed of both hormone and receptor half-sites, remains to be determined and may describe a role for Zn2+ in the ligand/receptor complex. The molecular mechanism by which Zn2+ binding influences the activity of the hormone/receptor complex is currently unknown. Both hGH and hPL contain a Zn2+ half-site (H18 and E174). Zn2+ binding increases the strength of hGH (37) binding to hPRL receptor and is required for the binding and activity of hPL (61). The mechanism by which Zn2+ produces different behaviors of hPRL and the other two lactogens remains to be fully understood. Zn2+ does not influence binding of hGH to the hGH receptor.
The role of Zn2+ may not be at the target cell but may instead be in the pituitary acidophil. Millimolar Zn2+ concentrations are present in the acidophilic lactogen-secreting cells of the anterior pituitary (62) and likely provide Zn2+-bound hormones at the time of secretion. Unfortunately, the stoichiometry of Zn2+ and lactogen at the time of secretion has not been reported. The H27A mutant of hPRL reduces both Zn2+ binding (56) and the ability of GH4C1 cells to secrete this mutant hormone (63), and this is consistent with data indicating that Zn2+ played a role in PRL secretion (62). Once secreted, the persistence of hormones containing Zn2+ is unclear because the rate of Zn2+ dissociation is unknown. The loss of Zn2+ from hormones may modulate hormone activity after secretion, but this hypothesis remains to be explored.
C. Prolactin: rate constants, affinities, and thermodynamics
The kinetics, affinities, and free energies for hPRL receptor binding to either site 1 or site 2 of hPRL are different. It must be remembered that what is frequently described as site 2 binding is actually the sum of receptor binding to site 2 of the ligand plus binding of the S2/S2 receptor interfaces. Before the last several years, a nanomolar global affinity for hPRL/receptor binding was described. When global affinities were analyzed by Scatchard analysis, the graphical plots often were nonlinear and suggested that both high and low affinity binding events were present. Early studies supported PRL/receptor binding as a two-step process (64). The individual affinities of both sites 1 and 2 for EC domains of hPRL receptors can be measured by carefully constructed surface plasmon resonance (SPR) experiments or perhaps by isothermal titration calorimetry. Bimolecular studies best discriminate binding at site 1, but site 2 binding is best discerned by trimolecular experiments (65). Several models of hPRL binding to its two receptors are available, but the simplest bidirectional mechanism is used most commonly.
Using this simple model and the trimolecular method, Goffin and colleagues (16) reported a site 1 Kd of 6.5 nm and a site 2 Kd of 32.9 μm. An earlier SPR study from the Gertler and Djiane groups (66) used a different approach for SPR studies where hormones were fixed to the SPR surface and receptor binding rates and affinities were deconvoluted from the combined receptor-binding signals. This study reported similar conclusions, but higher affinities were observed for site 1 than site 2. Binding of hGH or hPL to site 1 also provided nanomolar affinities (28). Data for hGH or hPL binding the hPRL receptor at site 2 are not available. Finally, hGH binding to the EC domain of the hGH receptor shows a strong affinity (∼1 nm) (67) at site 1 and a marginally weaker affinity at site 2 (3.8 nm) (68) with a functional coupling between sites 1 and 2 (51). Serum concentrations of hGH typically vary between 0 and 1 nm (59). Thus, modulation of hGH within this range would be well within the binding isotherms of both sites 1 and 2 and would appear to provide long-lived heterotrimers that function in the same time frame as observed for JAK/STAT phosphorylation and activation of other downstream signaling mechanisms. This mechanism would provide roles for hGH dissociation, suppressors of cytokine signaling proteins, or other modulators to regulate receptor activity. Site 1 affinity for the hGH/receptor pair is similar to that for the hPRL/receptor pair. In contrast, the site 2 affinity for the hGH/receptor pair is approximately 8000 times stronger that that for hPRL/receptor pair, and the site 2 kinetics are dramatically different. Clearly, the hGH/receptor pair has a significantly different mechanism of ligand/receptor heterotrimer formation and activation than does the hPRL/receptor pair.
Interpretation of the rate constants and affinities for sites 1 and 2 in the context of physiological hPRL concentrations is necessary to evaluate the mechanism by which hPRL activates the hPRL receptor. Reference blood concentrations of hPRL in nonpregnant adult women vary between 3.8 and 23.2 ng/ml (0.15 to 0.99 nm) (59). Changes of hPRL concentrations within this range are observed to influence human biology and therefore must modulate hPRL receptor activities. In vitro biological studies of the activity of hPRL generally provide ED50 values in the nanomolar range. Clearly, the nanomolar affinity of site 1 is compatible with changes in blood hormone concentrations modulating the amount of hormone/receptor heterodimers. The modest on rate constant is compatible with the modulation of serum hPRL concentrations in the order of several minutes. The half-lives of these heterodimeric complexes, as defined by a slow off rate constant, is compatible with the time frame of receptor activation. Thus, site 1 binding and the observed biochemistry and biology appear to be compatible.
The weak affinity of the hPRL/receptor heterodimeric complex for a second hPRL receptor presents a problem. The greater than micromolar affinity of site 2 in hPRL and the rapid apparent on and off rate constants measured in solution by SPR provide a vanishingly small population of hPRL/receptor heterotrimers in the context of nanomolar changes in hPRL concentrations. Given these observations, it is difficult to explain how hPRL can create a sufficient population of active heterotrimeric ligand/receptor complexes to modulate the activities of downstream signaling systems within the observed time frame. Two mechanisms may be available to reconcile these data. Observed changes in hormone concentrations will modulate the densities of site 1-bound hPRL/receptor heterodimers providing a population of lactogens with activated site 2. This first interaction may serve as the concentration-dependent gatekeeper. As for binding the second receptor, one possible mechanism assumes that the rapid site 2 kinetics are both real and relevant and that activation of the trimeric complex can be accomplished very rapidly during transient hPRL interactions with a second receptor. This is compatible with JAK2 being constitutively associated with the hPRL receptor (69). Perhaps formation of a heterotrimeric hPRL/receptor complex rapidly places the two receptors in a stable active conformer that once formed does not require site 2 binding by hPRL. This would require hPRL receptors to provide the interaction that maintains their activity. Perhaps this is accomplished by the S2/S2 receptor binding interface, but constitutive activities of the ΔS2 and Δ10–186 hPRL receptors (70–72) where this interface is removed indicates that the S2/S2 does not play this role. Rather other yet to be identified interfaces are required. Currently, there are no data to indicate that once activated, receptor dimers without bound hormone either remain homodimers or display activity. Thus, this mechanism might be hard pressed to generate biologically effective concentrations of activated IC second messengers. A second mechanism requires preformed ligand-free hPRL receptor dimers. Such dimers are present in target cells (73–75). With a nanomolar site 1 affinity, increases in hPRL concentrations in the physiological range would provide significant numbers of hPRL/receptor trimeric complexes where hPRL was only bound at site 1. But these heterotrimeric complexes would provide high local concentrations of specifically oriented receptors that subsequently might bind the activated site 2 of hPRL. In this later case, site 2 rates and affinities previously measured by SPR for site 2 are not relevant. Potentially, this second mechanism could provide significant densities of activated heterotrimeric complexes. Although hPRL receptor dimers have been documented in several cellular models (73–75), the ability of these receptor dimers to bind hPRL and stimulate activity remain to be directly demonstrated. This second mechanism also requires ligands to convert inactive dimeric receptor pairs to active conformers. Studies showing hormone-induced conversion of inactive (hPRL unbound) to active (hPRL bound) homodimeric forms of the hPRL receptor have yet to be reported.
The heterotrimeric complex of hPRL and two EC receptor domains joins these domains through the S2/S2 subdomain interface (20). No data have been reported to show that these interfaces can join receptors in the absence of ligand. Clearly some mechanism must be present that prevents S2/S2 receptor interactions when ligands are absent. Furthermore, receptor dimers were observed in the absence of hPRL even when the EC domains have been removed (73), suggesting that the mechanisms available for receptor dimerization may differ in the presence or absence of ligand. Clearly, more studies are required to identify the structural features required for such a ligand-mediated mechanism.
Several additional studies utilizing the G129R hPRL antagonists are relevant when considering the mechanism by which hPRL activates the hPRL receptor. The G129R mutation in hPRL eliminates a critical cavity for receptor binding at site 2 (16). SPR experiments show that the Δ1–9/G129R mutant hPRL does not bind the receptor at site 2 even at high receptor concentrations, and this ligand does not form heterotrimeric species (16). In in vitro bioluminescence resonance energy transfer (BRET) binding experiments, the signal was increased when cells were treated with wild-type hPRL but not by the G129R mutant (76), indicating that binding by both site 1 and site 2 is required for orientating receptors appropriately for activation. Thus, a site 2 binding event clearly is required for receptor activation. Taken together, the mechanism by which binding of site 2 of hPRL activates the hPRL receptor is unclear.
Finally, the observation of biphasic dose-response curves for cellular activation by hPRL (77–79) requires reinterpretation in light of the affinities for sites 1 and 2 and the existence of dimeric ligand-free hPRL receptors. Biphasic dose-response curves, containing both an agonist and antagonist phase, were first interpreted to support a mechanism where hPRL sequentially bound two hPRL receptors creating an active complex. Very high concentrations of hPRL were sufficient to bind single receptors and deplete the population of free receptors to such an extent that free receptors were not available to create active heterotrimeric complexes. If activation of hPRL receptors occurs by hPRL binding to preformed receptor dimers, then this explanation for biphasic dose-response curves is not appropriate. The initial binding event would create a heterotrimeric complex where site 2 of the receptor-bound hPRL is active, juxtaposed with a second receptor, and likely to be activated.
Substantial differences in the sequences of hPRL, hGH, and hPL (Fig. 1B) provide significant functional differences. Although these three lactogens all form similar long four-helix bundles, the residues' side chains displayed on the surfaces and within the interior of these hormones create unique chemical mosaics that define their ability to bind and activate receptors. These unique chemistries also define the conformers populated by the protein and make it unlikely that hPRL shares a similar conformer ensemble when compared with those of either hGH or hPL. Although the general features of receptor binding by these lactogens are similar, these differences express themselves as specific and unique binding mechanisms and indicate that even when binding the same receptor (or the hGH receptor), hPRL, hGH, or hPL cannot be used as reliable models for each other and that an understanding of their mechanisms must be discerned by studies of individual hormones. Neither hGH/hGH receptor nor hEPO/receptor can provide instructive models for the mechanisms by which hPRL binds the hPRL receptor.
III. Prolactin Receptor
The hPRL receptor undergoes a life cycle. Various isoforms of the hPRL receptors are produced in specific cell types during their differentiation, and the receptors are matured and transported to the plasma membrane. Receptors bind lactogenic hormones creating a heterotrimeric complex that changes the structure of the EC domains and promotes transmission of a structural change to the IC domain. These structural changes modulate receptor activities. Activated IC domains control various signaling pathways, amplifying IC signaling. This amplification process is subsequently regulated by various processes. Finally, the receptor is removed from the plasma membrane. In this section, we will discuss the structure of the hPRL receptor and how ligand binding promotes activation.
Description of structures, identification of functionally important motifs, and identification of the molecules of signal transduction have made important contributions to our understanding of the hPRL receptor's function in the last several decades. The identity of molecules involved in these processes has shown continual progress (5) and is likely to continue to evolve. But the molecular mechanisms by which these molecules interact and modulate function have not been well described. Such information is required to place the hPRL receptor into the broader mechanistic framework of this protein family, to design agonists and antagonists that influence signaling pathways, and to understand the mechanisms by which mutations influence function.
A. Prolactin receptor: sequence, isoforms, and structure
The hPRL receptor is a class 1 receptor composed of three major domains; EC, TM, and IC. The hPRL receptor is found in several isoforms produced by differential RNA splicing or posttranslational cleavage (Fig. 2) (70, 80–85). These structural variations are observed in either the EC or IC domains, with the TM domain being identical in all reported isoforms. Differential splicing of the IC domain has created the long (80), intermediate (81), and two short forms of the hPRL receptor (82, 84). The mature long form is the largest of the hPRL receptors, containing 598 residues including a 210-residue EC domain, a 24-residue TM domain, and a 364-residue IC domain. Variants of IC domains of the hPRL receptor appear to modulate the IC signaling systems in distinct fashion. These properties of the receptors are currently being identified and have been recently summarized (86). Significant functional differences are observed for EC domain variants that lack either the S1 subdomain (ΔS1) (83) or the S2 subdomain (ΔS2) (70). Single amino acid mutations are reported for the hPRL receptor and are associated with functional changes (87, 88). In addition, EC domains are found in the blood where they may function to buffer rapid changes in the concentration of free hPRL (85). Fewer splice variants have been reported for hGH (89–93) or hEPO (94) receptors, and these isoforms are largely dissimilar to those described for the hPRL receptor. These structural differences among the receptor isoforms imply that variants of each class 1 receptor may perform unique functions.
The EC domain of the hPRL receptor is well conserved among mammals (Supplemental Fig. 3A). It is composed of two subdomains (S1 and S2, also referred to as D1 and D2, respectively). Each subdomain contains a seven-strand antiparallel β-sheet fibronectin III motif (95) (Fig. 2C and Supplemental Fig. 4). A short proline-rich sequence (residues 101–106, VQPDPP) is believed to be a nonstructured hinge linking a terminal one-turn helix in the S1 subdomain and the β-strand A′ initiating the S2 subdomain. The flexibility of the hinge, restrictions introduced by the prolines, and the utility of this motif remain to be explored. The S1 subdomain contains two highly conserved disulfide bonds (cysteines 12 and 22 and cysteines 51 and 62). These disulfides are retained among class 1 receptors, but their locations are not identical among hPRL, hGH, and hEPO receptors (Supplemental Fig. 3B). hGH receptors contain a third disulfide loop. Removal of any of the disulfide bonds in hPRL receptors eliminates both ligand-mediated activation (96) and biological actions (97). In addition, an unpaired cysteine is present in the EC domain of the hPRL receptor at position 184 in the S2 subdomain, 26 residues from the TM domain but close to both the receptor's Zn2+ half-site (D187 and H188) and the WSAWS motif (residues 191–195). Unpaired cysteines in the hPRL receptor at positions 184, 225, and 242 are involved in the formation of redox-sensitive hPRL receptor dimers (74). hPRL treatment does not appear to increase either the population of redox-sensitive dimeric hPRL receptors or increase their tyrosine phosphorylation (W. Liu and C. L. Brooks, submitted for publication). The S2 subdomain of hGH and hEPO receptors contain a single unpaired cysteine, but at different positions than the hPRL receptor. Cysteine 240 of hGH is only four residues from the TM domain (residues 245–267) and participates in the formation of hGH-induced disulfide-linked receptor dimers (98–100). The single unpaired cysteine in hEPO is 45 residues from the TM domain (residues 228–249). Disulfide-linked hEPO receptor dimers have been reported (101). Neither hGH nor hEPO contains cysteines either in the TM domain (analogous to C225) or between the TM domain and Box 1 (analogous to C242) as observed in the hPRL receptor. Thus, it is not surprising that hPRL or hGH disulfide-linked receptor dimers may serve different functions. The hPRL receptor's WSXWS motif (residues 191–195) is found in both hGH (residues 221–225) (although a less than canonical sequence) and hEPO receptors (residues 212–216) (Supplemental Fig. 3B).
Several functional epitopes for the receptor's interface for site 1 of hPRL have been identified by mutagenesis (102). Structural epitopes of the hPRL receptor that form an interface with site 1 are present in strands E and G as well as loops 1, 3, and 5 of S1 subdomain; several additional epitopes are found in the S2 subdomain in loops 2′ and 6′ (20). Loop 6′ includes the Zn2+ half-site and the WSAWS motif. The structural epitopes for site 2 and the receptor/receptor interface in the S2 subdomains are described for hPRL bound to the EC domain of the rat receptor (18). Overlapping but nonidentical sets of residues form the rat receptor's structural interfaces for sites 1 and 2 for hPRL. Similar but nonidentical sets of receptor residues form the S2/S2 subdomain interface when the rat receptor is bound by hPRL (20), ovine PL (25), or a hybrid ligand (18). The sequence (residues 155–173) containing many of the residues of the S2/S2 subdomain interface in the rat receptor are modestly conserved in the human hPRL receptor (63%). The role of this interface in signal transduction is unclear, but several mutations are shown to reduce the S2/S2 subdomain affinities of EC domains of the rat receptor (20). In the absence of ligand, there are no reports of receptor/receptor binding through S2 subdomains of the hPRL receptor, implying that these surfaces are either inaccessible or disorganized in the absence of bound ligand. The S2/S2 interface residues of the hPRL receptor are not conserved in the hGH receptor (103). Identification of the hPRL receptor's structural and functional epitopes for both site 2 and the S2/S2 subdomain interfaces as well as their free energy contributions remain to be determined for the human proteins. In these interfaces, several tryptophans and H188 are important for hPRL receptor function (19, 20).
The EC domain of the PRL receptor is glycosylated. Attachment of sugars to the rat PRL receptor was first indicated by its binding to concanavalin A-Sepharose (104) and was confirmed during sequencing of the protein (105). The EC domain of the hPRL receptor contains six asparagines: residues 16, 35, 60, 80, 83, and 209. Asparagines 16 and 209 are poorly conserved in other mammalian PRL (Supplemental Fig. 3A), but the other four asparagines are generally conserved. When asparagines 35, 80, and 108 in the rat PRL receptor are mutated to aspartic acid, the activation of the receptor is not eliminated, but the transport of the protein to the plasma membrane is restricted (106). Potential N-glycosylation sites are not conserved in receptors for hPRL, hGH, and hEPO (Supplemental Fig. 3B). Neither the structure nor variation of the polysaccharides associated with the hPRL receptor has been reported.
Evaluation of natural and man-made variants of the EC domain of the hPRL receptor is instructive regarding the structural elements necessary for hPRL receptor function. Removal of the S1 subdomain [ΔS1, residues 1–100 (83) or residues 4–102 (70)] (Fig. 2) provides a receptor with either reduced or no apparent affinity for hPRL, respectively. This is consistent with structural data showing that the majority of ligand contacts are provided by the S1 subdomain (19). But despite deletion of the S1 subdomain, the receptor still contains all features necessary for activation, albeit at significantly higher ligand concentrations. These data indicate that the S1 domain is largely involved in ligand binding and may not contain structures required for subsequent receptor activation. When residues 106 through 206 (ΔS2, Fig. 2) are removed, the hPRL receptor gains constitutive activity when measured by BRET signaling or by cellular proliferation assays (70), suggesting that the S2 subdomain (which includes the S2/S2 interface, several residues binding sites 1 or 2 of hPRL, C184, the Zn2+ half-site, and the WSAWS motif) is not required for receptor activity. When both the S1 and S2 subdomains are removed (ΔEC, residues 1–210 removed) (70, 73), the receptor cannot be activated but is capable of forming ligand-independent dimers (73). Surprisingly, when residues 10–186 are removed (Δ10–186), the hPRL receptor is constitutively active (71). Thus, removal of residues constituting the interfaces for sites 1 and 2, the S2/S2 subdomain interface, and C184 (but not the removal of either the Zn2+ binding site or the WSAWS motif) indicates that these features are not required for receptor activity. Removal of residues 10–186 negates the requirement for ligand, suggesting that a ligand-specified orientation of two receptors is not required for activation. Furthermore, if one assumes that dimerization of receptors is required for activity, these data demonstrate that formation of active hPRL receptor dimers can occur without ligand binding and does not require most of the EC domain to form active dimers. Similar results were observed with deletion of residues 103 through 203 of the rabbit PRL receptor producing a constitutively active receptor (72). This deletion in the rabbit PRL receptor leaves the S1 subdomain as well as the Zn2+ binding site and the WSVWS motif but eliminates the conserved single cysteine of the S2 subdomain. Finally, both the I76V and I146L mutations of the hPRL receptor provide a species with constitutive activity (87, 88); these residues fall within sequences whose elimination displays constitutive activity. The mechanisms remain to be identified by which these mutations produce constitutive activity. These various deletion and mutation experiments suggest that the many elements of the EC domain are not required for activation of the receptor. They also allow for the possibility that activity of the PRL receptor is inhibited by elements of the EC domain in the absence of ligand; either removal or changes within these elements or ligand binding provide activity. Finally, if receptor dimerization is required for activity and ligand-directed dimerization is not required, then other structural features must provide an interface that creates functional receptor dimers.
These data suggest that the precise orientation of receptor dimers needed to activate receptor pairs is not provided by the binding of the ligand. The nonspecificity of the EC domain in receptor activation is also supported by the observation that chimeric receptors, where the native EC domain has been replaced with that from the human granulocyte-macrophage colony stimulating factor (GM-CSF), can be activated by exposure to GM-CSF (107). An imprecise connection between EC and IC domains is consistent with our observation that alanine additions at the interface of the EC and TM domains of hPRL do not influence the ability of hPRL to activate the receptor (75). Although the EC domain clearly influences activity of the IC domain, none of these data provide the mechanism by which the EC domains physically couple receptors or transmit a signal to the IC domain.
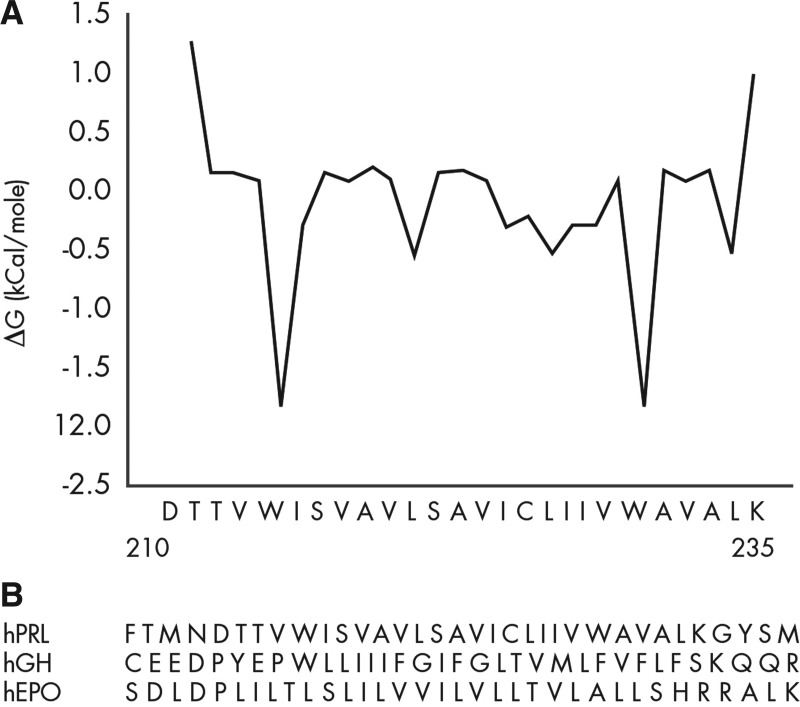
The TM domain (residues 211 through 234) of the hPRL receptor is predicted to be a helix, as are all TM domains, but no data are available for the hPRL receptor to confirm this prediction, provide structural detail, determine stability, or describe changes in the TM domain associated with hPRL receptor activation. The TM sequence of hPRL receptor is unique when compared with the TM of either hGH or hEPO receptors (Fig. 3). The hPRL receptor TM domain provides a hydrophobicity plot with distinctly symmetrical features including conserved charged residues at the N (D210) and C (K235) termini, tryptophans at positions 214 and 230, and a central cysteine at position 225. These features are not retained in the TM of either the hGH or hEPO receptors (Fig. 3 and Supplemental Fig. 3B) but are retained in PRL receptors from various mammalian species (Supplemental Fig. 3A). Binding between TM domains provides one mechanism to create ligand-independent receptor dimers (73). Removal of either the EC or IC domain does not eliminate ligand-independent hPRL receptor dimers; thus, it is believed that the TM is most important when forming ligand-free hPRL receptor dimers. The TM sequence contains elements of a leucine zipper (108, 109), a motif capable of dimer formation (110). The hPRL receptor residues responsible for dimerization remain to be identified, and their role in transmission of the lactogen signal from the EC domain to the IC domain remains to be identified. Elements of a leucine zipper motif are also observed in the TM domain of other cytokine receptors (111, 112). In the hEPO receptor, abolishing receptor dimerization with mutations in the TM domain reduces the efficiency of signal transduction (113). Several studies have resulted in different views regarding the importance of TM domain dimerization for the hGH receptor (111, 114).
Figure 3.
A, Hydrophobicity plot of hPRL receptor TM domain. B, Sequence comparison of hPRL, hGH, and hEPO receptor TM domains.
A discussion of the structures and activities of the various IC domain isoforms of the hPRL receptor is beyond the scope of this review. Thus, I will limit discussion to the long form (598 residues) of the hPRL receptor. There are no x-ray or NMR structures for the IC domain for any of the isoforms of the hPRL receptor (or of the IC domains for any class-1 cytokine receptors). The sequence of the long-form IC domain analyzed by computational methods suggests that several areas of secondary structure may exist (my unpublished observations) primarily in the N-terminal sequence of the IC domain, but no x-ray, NMR, or biophysical data are available to support this prediction. The IC domain is predicted to contain large areas of unstructured sequence when analyzed by the method of Uversky et al. (Ref. 115; and my unpublished observations). Furthermore, Ren et al. (116) have observed that motifs such as those binding SH2 domains are preferentially found in unstructured sections of proteins. These observations may explain the lack of structural information for the IC domain and raise the question whether activation or binding of various signaling proteins promotes structuring of the IC domain.
Approximately the first 60 residues of the IC domain are predicted to contain secondary structures (117, 118) (my unpublished observations) and are known to contain Box 1, V Box, and Box 2. The first 100 residues of the IC domain are largely conserved in mammalian hPRL receptors (Supplemental Fig. 3A), and several motifs have been described that are conserved in other class 1 receptors (119, 120), including Box 1 and Box 2. The proline-rich Box 1 motif (IFPPVPGP, residues 243–250) is required for JAK2 binding (121). Multiple cis-trans proline conformers have been observed in model Box 1 peptides (122), but the role of these proline conformers in modulating JAK2 binding or receptor function has not been determined. A role for proline isomerization has been suggested to regulate hPRL receptor activity (14, 123).
Box 2 (VEYLEVDDSEDQH, residues 288–300) is located C-terminal to Box 1 and may influence the binding of JAK2 to the hPRL receptor, but this remains to be investigated in hPRL receptors. The 37 residues between Box 1 and Box 2 are called the V Box, and the residues immediately C-terminal to Box 2 are termed the X Box.
Activation of the long form of the hPRL receptor by ligand binding is mediated by tyrosine kinases including JAK2 (124), Fyn (81, 125), and Raf-1 (126), as well as other signaling pathways (5), most notably the STAT, MAPK, and inositol trisphosphate pathways, and VAV (127). The IC domain of the long-form hPRL receptor contains 10 tyrosine residues. Phosphorylation of IC tyrosines creates binding sites for various members of the 110-member Src homology protein 2 (SH2) protein family (128, 129). The specificity of SH2 protein binding to phosphotyrosines is influenced by the surrounding amino acid sequences. Unfortunately, our understanding of the amino acid sequences influencing binding of specific SH2 proteins is incomplete; thus, we cannot predict which members of the SH2 protein family are likely to bind the phosphorylated hPRL receptor. Based on what is known of binding sites for SH2 proteins, it is highly probable that variations in the binding sites' amino acid sequences observed in various receptor types and among various species will define the kinetics and half-lives for specific SH2 protein binding to various phosphotyrosines in class 1 receptors and provide unique functional characteristics to each type of receptor. The number of tyrosines, their positions, and sequences surrounding the various IC domain tyrosines in hPRL, hGH, and hEPO receptors (Fig. 4) are not conserved; thus, identification of the receptor binding sites for SH2 proteins will be impossible to translate between the various class 1 receptors. hPRL receptor binding by several SH2 proteins has been identified largely by coimmunoprecipitation experiments. The inability of this technique to identify weakly bound or rapidly dissociating proteins leaves open the possibility that additional SH2 proteins may be members of the hPRL receptor complex. Work in the rat hPRL receptor has identified several sites for tyrosine phosphorylation (130, 131). To date, only two tyrosine residues have been identified as phosphorylation sites (Y522 and Y587) in rat PRL receptor (132). Both of these sites are conserved in the hPRL receptor. Based on the number of SH2 proteins known to associate with the hPRL receptor and their binding sequences, these tyrosines are unlikely to represent the complete in vivo set of phosphorylations of the hPRL receptor. No studies have addressed this problem in the hPRL receptor using state-of-the-art peptide mapping and mass spectrometry (133, 134). Thus, mapping the proteins that compose the hPRL receptor complex and identification of their receptor-binding sites by either a better-informed predictive approach or an experimental approach needs further investigation.
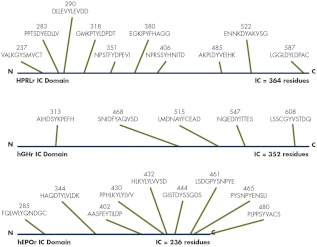
Figure 4.
Comparison of IC tyrosines in hPRL, hGH, and hEPO. The relative lengths of the IC domains are shown in blue. The location of each tyrosine in the complete sequence of the mature protein is shown and underlined in the associated sequence of the surrounding residues. The relative locations of each tyrosine are indicated by the green lines.
Of the 10 tyrosines in the IC domain of the hPRL receptor (Fig. 4 and Supplemental Fig. 3), approximately half are retained between the aligned sequences of mammalian hPRL receptors (Y237, Y406, Y485, Y522, and Y587). The amino acid sequences surrounding these IC tyrosines are highly varied. Of the 10 residues surrounding each conserved tyrosine, between two and eight are retained (average, 5.6) in mammals, whereas between zero and eight of the surrounding residues were retained for the nonconserved tyrosines (average, 4.6). These data indicate significant variation between species in the possible phosphorylation sites and serve as a caution when projecting findings between the PRL receptors from various species.
There are no recognizable similarities between the pattern of IC tyrosines or their surrounding sequences in the hPRL receptor when compared with sequences in either the hGH or hEPO receptors (Fig. 4). Considering that these receptors use several common signaling systems, it is surprising to find so few similarities. Such sequence differences suggest that the molecular recognition for the various IC signaling systems are likely to display highly nuanced kinetics that are unique for each class 1 receptor. Finally, threonine residues in the hPRL receptor are phosphorylated, including T349 (135) and T515 (136); phosphorylation of these residues creates binding sites for 14-7-7 proteins (137) and cAMP-responsive element-binding protein (CREB protein) (138), respectively. Serine phosphorylation increases with hPRL stimulation (T. J. Gordon and C. L. Brooks, unpublished observations). The identity and functional role of these nontyrosine phosphorylations remains to be completely understood. Serine/threonine phosphorylations adjacent to tyrosines may function as regulatory processes that influence phosphorylation of nearby tyrosines or binding of SH2 proteins.
The pattern of cysteine residues in PRL receptors from various mammalian species is maintained in both the EC and TM domains but are poorly conserved in the IC domain (only three of the 10 cysteines are conserved) (Supplemental Fig. 3A). The cysteines found in the IC domain of the hPRL receptor are not conserved in either the hGH or hEPO receptors (Supplemental Fig. 3B).
The picture that emerges from these similarities and differences is that the hPRL receptor has substantially evolved from the other human class 1 receptors. Although the hPRL receptor retains similarities to those of other mammals and to other class 1 human receptors, the details of the functionally important IC tyrosines are quite different. It remains to be determined whether these differences represent a fine-tuning of the various signaling systems or whether these differences are irrelevant to function.
B. What are the mechanisms that activate the prolactin receptor?
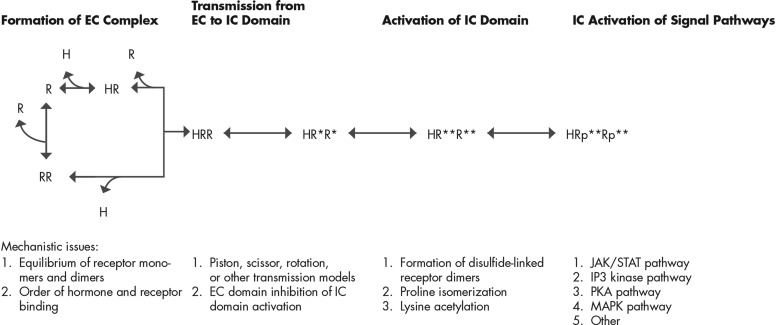
Activities of the hPRL receptor can be divided into four related sets of processes. First, increases in hPRL concentrations promote binding to monomeric or dimeric forms of the hPRL receptor and induce structural changes in the EC domain of the hPRL receptor. The sum of these reactions forms a ligand/receptor complex. Second, the ligand-induced structural changes in the EC domain transmit structural changes to the IC domain. These changes in the structure of the IC domain promote several processes that allow activation of the receptor. Third, the activated IC domain engages the molecules of various signaling systems that modulate cellular processes. These reactions produce stoichiometric amplification of the various IC manifestations of the hPRL signal. Fourth, the sum of these reactions are modulated and eventually terminated. The complete set of these processes is not understood for any class 1 ligand/receptor system and to a greater or lesser extent may be unique to each member of this receptor class. I intend to address the first two sets of processes: complex formation and signal transmission/activation.
1. Ligand/receptor binding and formation of an active EC complex
Twenty years ago, the mechanism for class 1 receptor activation was believed to be a ligand-driven dimerization of two receptors (139–141). Receptor proximity was the key feature required for JAK2-mediated transphosphorylation of the IC domains. Several lines of evidence, including structural studies (18, 20), stoichiometric studies (26), biophysical studies (27, 28), mutagenic studies (16), and functional studies (77–79), are interpreted to support this mechanism of complex formation.
Several lactogenic hormones (142), dimeric wild-type or G129R hPRL, dimeric wild-type or G120R hGH (143), and several dimeric antibodies (144, 145) are able to activate the hPRL receptor. In addition, replacement of the EC domains of hPRL receptors with those from other class 1 receptors produced hybrid receptors able to be activated by these alternative ligand/EC domain pairs (107, 146). Despite the diverse structures of these ligands, each can form an EC complex capable of initiating biological actions. These data suggest that a degree of structural freedom is available in creating EC complexes capable of signal transmission to the IC domain. Taken together, these studies are consistent with a mechanism where the key is ligand-mediated formation of receptor dimers. The observation of a biphasic dose-response curve in in vitro cellular studies (77–79) is consistent with the ligand-mediated receptor dimerization model. In the cases of hGH and PL and their receptors, structural (25, 103), stoichiometric (141), and mutagenic (147) studies also support the requirement for ligand-mediated receptor dimerization. Similar phenomena are observed with dimers of wild-type and G120R that activate the hGH receptor (100).
More recently, preformed ligand-independent hPRL receptor homodimers and heterodimers have been suggested as a mechanism by which hPRL activates receptors (14, 148, 149). These dimeric hPRL receptors may be in equilibrium with receptor monomers in the plasma membrane. The speed by which ligands activate receptors is said to support this mechanism; it is argued that without preformed receptor dimers, the time required for hPRL/receptor heterodimers to laterally diffuse within the plasma membrane, subsequently bind a second receptor, and activate the complex is too long to account for the observed time of receptor activation. This is a plausible argument, but no studies have provided supporting evidence. Would ligands activate hPRL receptors in a similar time frame if mutations created receptors that do not form dimers in the absence of ligands? Alternatively, hPRL-free receptor dimers may be formed by ligand binding and subsequent ligand dissociation, presumably creating inactive receptor dimers. Such complexes may await removal from the plasma membrane by ubiquitination and internalization (150). Additional studies are needed to resolve the role of ligand-free dimers of hPRL receptor. Perhaps receptor activation occurs through both mechanisms—ligand-mediated receptor dimerization and the ligand activation of preformed receptor dimers.
Based on the evidence, the TM domain provides the primary interface for ligand-independent hPRL receptor dimer formation (73). Qazi et al. (74) have shown formation of both homo- and heterodimer receptors of long and short forms of the hPRL receptors. hGH receptors (111), hEPO receptors (112, 113), and other receptors also form ligand-independent dimers through leucine zipper motifs (110) located in the TM domain. The residues responsible for formation of hPRL receptor dimers remain to be identified.
The mechanism for the ligand-mediated activation of preformed hPRL receptor dimers must be considered. Preformed hPRL receptor dimers must be held in “off” (inactive) conformers presumably by the TM/TM binding interface. Ligand binding to the EC domain would be required to provide the energy to break this inhibitory TM/TM domain interface and allow formation of active conformers of the dimeric hPRL receptor. The “on” conformers of hPRL receptors may be defined by the site 1, site 2, and S2/S2 subdomain interfaces. This model relies on the formation of ligand-free hPRL receptor dimers (73, 74) and offers a mechanism where rapid kinetics and differential affinities for receptors at sites 1 and 2 of hPRL are reasonable. The creation of constitutively active hPRL receptors when residues 10 through 186 are removed does not support this model because the residues that form the receptor interfaces for sites 1 and 2 as well as between the two S2 subdomains are not present. These data argue for a more complex mechanism or that receptors can be activated by two independent mechanisms. Finally, hPRL binding to preformed hPRL receptor dimers has not been demonstrated.
A variation of the preformed dimer mechanism may explain a greater portion of the current data. This mechanism would require the EC domains to prevent formation of active hPRL receptor dimers in the absence of ligand. This inhibition of receptor activity would be removed either by hPRL binding or by removal of elements of the EC domain necessary for inhibition. The structure of the EC domain may support such a hypothesis where the hinged S1 subdomain could function as an inhibitory domain that is removed from an inhibitory position by ligand binding. This theory is supported by the observation that removal of most of the EC domain (Δ10–186), including the residues involved in binding sites 1 and 2 interfaces as well as residues of the S2/S2 interface, provides a constitutively active hPRL receptor (71). But this notion is not supported by the observation that removal of the S1 subdomain does not create constitutive receptor activity (70, 83). Furthermore, removal of the entire EC domain (Δ1–210) eliminates both constitutive and hPRL-induced activity. These data suggest that some elements of the EC domain are necessary for hPRL receptor activity. The activity of the Δ10–186 hPRL receptor also demonstrates that ligand binding is not required to create active hPRL receptors when the majority of the S1 subdomain is removed. Currently, it is not known whether Δ10–186 hPRL receptors form dimers in the absence of ligand. Based on the constitutive activity of Δ10–186 receptors, one would assume ligand-independent dimer formation as previously shown for the Δ1–210 hPRL receptor (73), but this ligand-independent dimerization remains to be demonstrated. Because the structural differences between the Δ10–186 and Δ1–210 hPRL receptors must account for their functional differences, then elements within these short sequences (residues 1–9 and 187–210) appear necessary for hPRL receptor activity; they include a proline-rich N-terminal, C185, the Zn2+ half-site, and the WSAWS motif.
The structural differences between hormone-bound and hormone-free hPRL receptor dimers need to be determined. Description of the hPRL-induced structural changes may reveal the mechanism by which the receptor transmits its signal from the EC to the IC domains. Additional biophysical and mutagenic studies are required to support or dismiss these various mechanisms. Perhaps both hPRL receptor monomers and preformed dimers can be activated by ligand binding.
2. What mechanisms transmit the hPRL signal from the EC to the IC domains?
Activation of hPRL receptors requires a signal be transmitted from the EC domain to the IC domain. Normally, ligand binding must induce structural changes in the EC domain that either induce or allow structural changes in the IC domain that promote the well-described IC biochemistry. Generally, we have thought about potential mechanisms as nanoscale machines. Several mechanisms have been proposed for hGH and hEPO receptors: a piston model, a scissor model, and a rotation model. The piston model is based on structural studies of ligand/EC domain heterotrimers (103). Two receptors are offset along the z-axis (perpendicular to the presumed plane of the plasma membrane), suggesting that ligand binding causes a piston movement that may be associated with receptor activation. A similar piston-like offset can be observed in the hPRL-bound rat dimeric EC domain structure (18). But it remains to be shown that ligand binding induces a piston movement relative to the plasma membrane or whether receptor dimers display a similar offset in the absence of ligands. Thus, piston movements of the hPRL receptor have yet to be related to ligand-induced receptor activation.
The scissor model is based on two structures of dimeric EC domain of the hEPO receptor with or without bound ligand (151, 152). The structures show large changes in the orientation of hEPO's EC domains that are consistent with hEPO-mediated repositioning of the IC domains. There are no structural studies of the hPRL receptor that support this mechanism, but BRET data (76) showing an increased proximity of the receptors might be consistent with the scissor mechanism.
Finally, a rotation model has been proposed as the mechanism by which ligand binding activates several class 1 receptors. This model argues that a ligand-induced z-axis rotation of the receptor dimers' IC domains relative to each other is responsible for receptor activation. Data supporting this rotational model are based on mutagenic studies performed in the hGH and hEPO systems. Addition of alanine, a residue with a high propensity for helix formation, extends the TM domain helices rotating the IC domains relative to the TM domain (111, 153–155). This mechanism assumes that noncovalent interactions in TM/TM interfaces hold the receptors in either “off” or, in the presence of ligand, “on” conformers in the absence of ligand. Thus, alanine additions rotate the IC domains relative to the dimeric TM domains and rotate the IC domains relative to each other to provide the “on” or “off” conformers. With a specific number of inserted alanines, the activities of hGH-free hGH receptors are increased (111), and the ligand-induced activity of hEPO receptors are reduced by specific alanine additions (156).
Some alanine additions to either the hGH or hEPO receptors provided changes in receptor activity, but many of the alanine additions did not change either the constitutive or ligand-induced activity of these receptors. These data suggest that only specific orientations of the IC domains provide receptor activity (113, 157). The structural specificity required to activate hGH receptors appears to be inconsistent with the promiscuity of the hPRL receptor suggested by the variety of ligands and mechanisms known to induce activity.
To investigate the rotation model in the hPRL receptor, we have performed expanded alanine addition experiments (75). Between one and four alanines were introduced at the junctions of either the EC/TM or TM/IC domains, or immediately N-terminal to Box 1 to extend the TM helices and rotate the domains relative to each other. In a separate experiment, we added glycines at the junction of the TM/IC domains, providing increased flexibility between these domains to test whether a fixed orientation of the IC domains is required for activity. When these various hPRL receptors were expressed in 293T cells, constitutive activity was not observed for any of these mutants, indicating that the data did not support a rotation mechanism at the orientations investigated. When each of these mutant hPRL receptors was stimulated by hPRL, phosphorylation of the hPRL receptor tyrosines was observed, demonstrating that none of the receptor orientations provided by the various alanine insertions produced receptors unable to be activated by ligand. This is particularly important for the glycine insertions where fixed structural relationships between the TM and IC domains as well as the structural relationships between the two IC domains of dimeric hPRL receptors would be disrupted.
But the amount of spatial freedom between IC domains of hPRL receptors does have limits; although increasing glycine additions in hPRL receptors all can be activated by hPRL, each additional glycine reduces the extent of receptor tyrosine phosphorylation. Hormone-induced proximity appears to be less stringent for hPRL receptors than for either hGH or hEPO receptors.
3. What mechanisms activate the IC domain?
Once IC domains are positioned to create active hPRL receptor conformers several additional processes for IC activation have been identified.
a. Prolyl isomerase-mediated activation.
Peptidyl prolyl isomerase-A (124, 158) has been shown by coimmunoprecipitation and Western analysis to bind the hPRL receptor (159). Furthermore, inhibition of prolyl isomerases with cyclosporine or suppression of prolyl isomerase-D expression with inhibitory RNA reduces hPRL-mediated signaling (148, 160). These data imply that prolyl isomerase may be involved in regulating hPRL receptor conformers, but it is not clear whether prolyl isomerases are required for the proper folding and delivery of hPRL receptors to the plasma membrane (a chaperone function) or whether prolyl isomerases are a mechanism for modulation of the activities of the hPRL receptor (a molecular switch function) (124); prolyl isomerases may be required for both functions. Proline 334 (P310 in the mature protein) appears to be a site of peptidyl prolyl isomerase-A action in the activation of the hPRL receptor. P334 is located in the X Box of the IC domain. A P344A mutation reduces the intensity of STAT5A phosphorylation when expressed in 293 cells (160). If prolyl isomerases participate in hPRL-induced receptor activation, then several issues need to be resolved to support this hypothesis. First, the mechanism by which lactogen binding modulates the activity of hPRL receptor-associated prolyl isomerases and induces changes in receptor activities needs to be determined. Second, the hPRL-mediated ability of prolyl isomerase to change the proline conformers must be demonstrated in the hPRL receptor.
b. Acetylation-mediated activation.
Recent work by Ma et al. (136) identified a new hPRL-induced dimerization mechanism mediated by lysine acetylation of the IC domain. The authors argue that acetylation-mediated neutralization of positively charged lysines removes a repulsion of the IC domains and allows their juxtaposition and activation. Acetylation of the IC domain lysines requires phosphorylation of T539 (T515 in the mature protein) to bind CREB-binding protein, which contains the necessary acetylase activity (138). The identity of the serine/threonine kinase that phosphorylates T539 and the mechanism by which hPRL regulates T539 phosphorylation remains to be described. Earlier work by the same group identified a similar CREB-binding protein-mediated acetylation of interferon receptors (161). No similar mechanism has been described for either the hGH or hEPO systems.
c. Disulfide activation.
The hGH receptor requires formation of a ligand-induced intermolecular disulfide bond immediate to the EC/TM junction (98–100). The hPRL receptor does not have a similar mechanism. We have replaced each of the 12 C-terminal cysteines with serines, leaving the required four N-terminal disulfide cysteines unmodified, to determine the requirement for these residues (W. Liu and C. L. Brooks, submitted for publication). Replacement of these cysteines eliminated the formation of redox-sensitive hPRL receptor dimers, but these mutations did not reduce the ability of hPRL to increase tyrosine phosphorylation of these receptors. These data suggest that the hPRL receptor does not require ligand-induced disulfide-mediated formation of receptor dimers as does the hGH/receptor pair.
IV. Conclusion
The proteins that constitute the system by which hPRL induces its actions in various cells are largely described, but the mechanisms by which their interactions change their structures and associated functions are poorly documented. I have critically reviewed selected literature regarding the interaction and the associated structural and functional changes of hPRL and its receptor. I have compared the properties of the hPRL/receptor system with those of hGH and hEPO. The current literature indicates that the general features of lactogen binding to the hPRL receptor are conserved. These similarities include general domain structures of the ligand and receptor, the binding stoichiometry of the ligand and receptor, and the signaling pathways modulated by ligand binding. But the detailed molecular mechanisms by which hPRL activates its receptor are unique from those of either hGH or hPL. These include differences in the key interface residues and their bonding patterns both between and within ligand and receptor, differences in the receptor isoforms, differences in the nondisulfide cysteines, differences in the histidines within the sequence of the ligand or receptor, differences in the potential phosphotyrosine binding sites for SH2 proteins, and differences in the mechanisms of signal transduction. These differences suggest that an elegant evolutionary development has produced unique mechanisms that provide a nuanced regulation of IC signaling by the three human lactogenic hormones and the hPRL receptor. In contrast, many of the structural features of the hPRL/receptor pair are retained among mammalian species, suggesting that careful application of data from animals may facilitate our understanding of the human molecules. The current literature suggests that several molecular mechanisms for hPRL/receptor activation can be supported, but descriptions of the various mechanisms for the critical molecular interactions are incomplete (Fig. 5). Thus, a molecular mechanism describing the interaction of hPRL and its receptor is not available to guide the rational design of hPRL agonists and antagonists. Information from other class 1 hormone/receptor systems may not serve as accurate mechanistic models for hPRL/receptor activation.
Figure 5.
Model for formation of the hormone-bound EC domains, transmission of the hormone-induced signal from the EC to the IC domains, activation of the IC domain, and IC activation of the signaling pathways for four-helix cytokines and their receptors. Proposed mechanisms for each of these four steps are listed beneath the schematic. H, Hormone; R, receptor; HRR, heterotrimeric complex; HR*R*, heterotrimeric complex where EC complex has orientated the IC domains where they may be activated; HR**R**, heterotrimeric complex where the IC domains have been activated; HRP**RP**, heterotrimeric complex that has activated the IC signaling pathways; IP3, inositol trisphosphate; PKA, protein kinase A.
Supplementary Material
Acknowledgments
Portions of my work discussed in this manuscript were supported by the National Institutes of Health (Grant DK080450) and the American Cancer Society, Ohio Division.
Disclosure Summary: C.L.B. has previously consulted for and received grants from the National Institutes of Health of the U.S. Department of Health and Human Services. He holds U.S. patent no. 6995244.
Footnotes
- BRET
- Bioluminescence resonance energy transfer
- CREB
- cAMP-responsive element-binding
- EC
- extracellular
- EPO
- erythropoietin
- GM-CSF
- granulocyte-macrophage colony stimulating factor
- hGH
- human GH
- hPL
- human PL
- hPRL
- human PRL
- IC
- intracellular
- JAK
- Janus kinase
- NMR
- nuclear magnetic resonance
- PL
- placental lactogen
- PRL
- prolactin
- SH2
- Src homology protein 2
- SPR
- surface plasmon resonance
- STAT
- signal transducers and activators of transcription
- TM
- transmembrane.
References
- 1. Huising MO, Kruiswijk CP, Flik G. 2006. Phylogeny and evolution of class-1 helical cytokines. J Endocrinol 189:1–25 [DOI] [PubMed] [Google Scholar]
- 2. Forsyth IA, Wallis M. 2002. Growth hormone and prolactin—molecular and functional evolution. J Mammary Gland Biol Neoplasia 7:291–312 [DOI] [PubMed] [Google Scholar]
- 3. Wallis OC, Mac-Kwashie AO, Makri G, Wallis M. 2005. Molecular evolution of prolactin in primates. J Mol Evol 60:606–614 [DOI] [PubMed] [Google Scholar]
- 4. Kelly PA, Djiane J, Postal-Vinay MC, Edery M. 1991. The prolactin/growth hormone receptor family. Endocr Rev 12:235–251 [DOI] [PubMed] [Google Scholar]
- 5. Clevenger CV, Kline JB. 2001. Prolactin receptor signal transduction. Lupus 10:706–718 [DOI] [PubMed] [Google Scholar]
- 6. Watson CJ, Burdon TG. 1996. Prolactin signal transduction mechanisms in the mammary gland: The role of the Jak/Stat pathway. Rev Reprod 1:1–5 [DOI] [PubMed] [Google Scholar]
- 7. Nicoll CS. 1974. Physiological actions of prolactin. In: Knobil E, Sawyer WH, eds. Handbook of physiology. Section 7. Endocrinology. Washington, DC: American Physiological Society; 253–292 [Google Scholar]
- 8. Bernichtein S, Touraine P, Goffin V. 2010. New concepts in prolactin biology. J Endocrinol 206:1–11 [DOI] [PubMed] [Google Scholar]
- 9. Grattan DR, Kokay IC. 2008. A pleiotropic neuroendocrine hormone. J Neuroendocrinol 20:752–763 [DOI] [PubMed] [Google Scholar]
- 10. Clevenger CV, Furth PA, Hankinson SE, Schuler LA. 2003. The role of prolactin in mammary carcinoma. Endocr Rev 24:1–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ben-Jonathan N, Liby K, McFarland M, Zinger M. 2002. Prolactin as an autocrine/paracrine growth factor in human cancer. Trends Endocrinol Metab 13:245–250 [DOI] [PubMed] [Google Scholar]
- 12. Goffin V, Bernichtein S, Touraine P, Kelly PA. 2005. Development and potential clinical uses of human prolactin receptor antagonists. Endocr Rev 26:400–422 [DOI] [PubMed] [Google Scholar]
- 13. Jacobson EM, Hugo ER, Tuttle TR, Papoian R, Ben-Jonathan N. 2010. Unexploited therapies in breast and prostate cancer: blockade of the prolactin receptor. Trends Endocrinol Metab 21:691–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kossiakoff AA, Clevenger CV. 2010. Growth hormone and prolactin family of hormone receptors: the structural basis of receptor activation and regulation. In: Bradshaw RA, Dennis EA. eds. Handbook of cellular signaling. 2nd ed Vol 3 Chap 36 Amsterdam: Elsevier; 237–243 [Google Scholar]
- 15. Teilum K, Hoch JC, Goffin V, Kinet S, Martial JA, Kragelund BB. 2005. Solution structure of human prolactin. J Mol Biol 351:810–823 [DOI] [PubMed] [Google Scholar]
- 16. Jomain JB, Tallet E, Broutin I, Hoos S, van Agthoven J, Ducruix A, Kelly PA, Kragelund BB, England P, Goffin V. 2007. Structural and thermodynamic bases for the design of pure prolactin receptor antagonists: x-ray structure of Del1–9-G129R-hPRL. J Biol Chem 282:33118–33131 [DOI] [PubMed] [Google Scholar]
- 17. Svensson LA, Bondensgaard K, Nørskov-Lauritsen L, Christensen L, Becker P, Andersen MD, Maltesen MJ, Rand KD, Breinholt J. 2008. Crystal structure of a prolactin receptor antagonist bound to the extracellular domain of the prolactin receptor. J Biol Chem 283:19085–19094 [DOI] [PubMed] [Google Scholar]
- 18. Broutin I, Jomain JB, Tallet E, van Agthoven J, Raynal B, Hoos S, Kragelund BB, Kelly PA, Ducruix A, England P, Goffin V. 2010. Crystal structure of an affinity-matured prolactin complexed to its dimerized receptor reveals the topology of hormone binding site 2. J Biol Chem 285:8422–8433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kulkarni MV, Tettamanzi MC, Murphy JW, Keeler C, Myszka DG, Chayen NE, Lolis EJ, Hodsdon ME. 2010. Two independent histidines, one in human prolactin and one in its receptor, are critical for pH-dependent receptor recognition and activation. J Biol Chem 285:38524–38533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van Agthoven J, Zhang C, Tallet E, Raynal B, Hoos S, Baron B, England P, Goffin V, Broutin I. 2010. Structural characterization of the stem-stem dimerization interface between prolactin receptor chains complexed with the natural hormone. J Mol Biol 404:112–126 [DOI] [PubMed] [Google Scholar]
- 21. Somers W, Ultsch M, De Vos AM, Kossiakoff AA. 1994. The x-ray structure of a growth hormone-prolactin receptor complex. Nature 372:478–481 [DOI] [PubMed] [Google Scholar]
- 22. Walsh ST, Kossiakoff AA. 2006. Crystal structure and site 1 binding energetics of human placental lactogen. J Mol Biol 358:773–784 [DOI] [PubMed] [Google Scholar]
- 23. Kamtekar S, Hecht MH. 1995. The four helix bundle: what determines a fold? FASEB J 9:1013–1022 [DOI] [PubMed] [Google Scholar]
- 24. Chaiken IM, Williams WV. 1996. Identifying structure-function relationships in four-helix bundle cytokines: towards de novo mimetics design. Trends Biotechnol 14:369–375 [DOI] [PubMed] [Google Scholar]
- 25. Elkins PA, Christinger HW, Sandowski Y, Sakal E, Gertler A, de Vos AM, Kossiakoff AA. 2000. Ternary complex between placental lactogen and the extracellular domain of the prolactin receptor. Nat Struct Biol 7:808–815 [DOI] [PubMed] [Google Scholar]
- 26. Hooper KP, Padmanabhan R, Ebner KE. 1993. Expression of the extracellular domain of the rat prolactin receptor and its interaction with ovine prolactin. J Biol Chem 268:22347–22352 [PubMed] [Google Scholar]
- 27. Sivaprasad U, Canfield JM, Brooks CL. 2004. Mechanism for ordered receptor binding by human prolactin. Biochemistry 43:13755–13765 [DOI] [PubMed] [Google Scholar]
- 28. Voorhees JL, Brooks CL. 2010. Obligate ordered binding of human lactogenic cytokines. J Biol Chem 285:20022–20030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rao GV, Brooks CL. 2011. Functional epitopes for site 1 of human prolactin. Biochemistry 50:1347–1358 [DOI] [PubMed] [Google Scholar]
- 30. DePristo MA, de Bakker PI, Blundell TL. 2004. Heterogeneity and inaccuracy in protein structures solved by x-ray crystallography. Structure 12:831–838 [DOI] [PubMed] [Google Scholar]
- 31. Bernichtein S, Jomain JB, Kelly PA, Goffin V. 2003. The N-terminus of human prolactin modulates its biological properties. Mol Cell Endocrinol 208:11–21 [DOI] [PubMed] [Google Scholar]
- 32. Bernichtein S, Kayser C, Dillner K, Moulin S, Kopchick JJ, Martial JA, Norstedt G, Isaksson O, Kelly PA, Goffin V. 2003. Development of pure prolactin receptor antagonists. J Biol Chem 278:35988–35999 [DOI] [PubMed] [Google Scholar]
- 33. Doneen BA, Bewley TA, Li CH. 1979. Studies on prolactin. Selective reduction of the disulfide bonds of ovine hormone. Biochemistry 18:4851–4860 [DOI] [PubMed] [Google Scholar]
- 34. Schleyer M, Etzrodt H, Trah TJ, Voigt KH. 1982. Dependence of human somatotropin activity on interchain disulfide bridges. Hoppe Seylers Z Physiol Chem 363:1111–1116 [DOI] [PubMed] [Google Scholar]
- 35. Handwerger S, Pang EC, Aloj SM, Sherwood LM. 1972. Correlations in the structure and function of human placental lactogen and human growth hormone. I. Modification of the disulfide bonds. Endocrinology 91:721–727 [DOI] [PubMed] [Google Scholar]
- 36. Kim BG, Brooks CL. 1993. Isolation and characterization of phosphorylated bovine prolactin. Biochem J 296:41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dattani MT, Hindmarsh PC, Brook CG, Robinson IC, Kopchick JJ, Marshall NJ. 1995. G120R, a human growth hormone antagonist, shows zinc-dependent agonist and antagonist activity on Nb2 cells. J Biol Chem 270:9222–9226 [DOI] [PubMed] [Google Scholar]
- 38. Chen WY, Chen NY, Yun J, Wagner TE, Kopchick JJ. 1994. In vitro and in vivo studies of antagonistic effects of human growth hormone analogs. J Biol Chem 269:15892–15897 [PubMed] [Google Scholar]
- 39. Duda KM, Brooks CL. 2003. Identification of residues outside of the two binding sites that are critical for activation of the lactogenic activity of human growth hormone. J Biol Chem 278:22734–22739 [DOI] [PubMed] [Google Scholar]
- 40. Pauling L. 1935. The oxygen equilibrium of hemoglobin and its structural interpretation. Proc Natl Acad Sci USA 21:186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wyman J., Jr 1948. Heme proteins. Adv Protein Chem 4:407–531 [DOI] [PubMed] [Google Scholar]
- 42. Wyman J, Gill SJ. 1990. Binding and linkage. Mill Valley, CA: University Science Books [Google Scholar]
- 43. Mondo J, Wyman J, Changeux JP. 1965. On the nature of allosteric transitions: a plausible model. J Mol Biol 12:88–118 [DOI] [PubMed] [Google Scholar]
- 44. Koshland DE, Jr, Némethy G, Filmer D. 1966. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry 5:365–385 [DOI] [PubMed] [Google Scholar]
- 45. Tsai CJ, del Sol A, Nussinov R. 2008. Allostery: absence of a change in shape does not imply that allostery is not at play. J Mol Biol 378:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Clore GM. 2011. Exploring sparsely populated states of macromolecules by dimagnetic and paramagnetic NMR relaxation. Prot Sci 20:229–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Laskowski RA, Gerick F, Thornton JM. 2009. The structural basis of allosteric regulation in proteins. FEBS Lett 583:1692–1698 [DOI] [PubMed] [Google Scholar]
- 48. Lakowicz JR. 2006. Principles of fluorescence spectroscopy. 3rd ed New York: Springer [Google Scholar]
- 49. Wrabl JO, Gu J, Liu T, Schrank TP, Whitten ST, Hilser VJ. 2011. The Role of protein conformational fluctuations in allostery, function, and evolution. Biophys Chem 159:129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. di Cera E. 1998. Site-specific analysis of mutational effects in proteins. Adv Protein Chem 51:59–119 [DOI] [PubMed] [Google Scholar]
- 51. Walsh ST, Sylvester JE, Kossiakoff AA. 2004. The high- and low-affinity receptor binding sites of growth hormone are allosterically coupled. Proc Natl Acad Sci USA 101:17078–17083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Keeler C, Jablonski EM, Albert YB, Taylor BD, Myszka DG, Clevenger CV, Hodsdon ME. 2007. The kinetics of binding human prolactin, but not growth hormone, to the prolactin receptor vary over a physiological pH range. Biochemistry 46:2398–2410 [DOI] [PubMed] [Google Scholar]
- 53. Tettamanzi MC, Keeler C, Meshack S, Hodsdon ME. 2008. Analysis of site-specific histidine protonation in human prolactin. Biochemistry 47:8638–8647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Behncken SN, Rowlinson SW, Rowland JE, Conway-Campbell BL, Monks TA, Waters MJ. 1997. Engineering porcine growth hormone to activate the human receptor. J Biol Chem 272:27077–27083 [DOI] [PubMed] [Google Scholar]
- 55. Peterson FC, Brooks CL. 2000. The species specificity of growth hormone requires the cooperative interaction of two motifs. FEBS Lett 472:276–282 [DOI] [PubMed] [Google Scholar]
- 56. Sun Z, Li PS, Dannies PS, Lee JC. 1996. Properties of human prolactin and H27A-PRL, a mutant that does not bind Zn++. Mol Endocrinol 10:265–271 [DOI] [PubMed] [Google Scholar]
- 57. Permyakov EA, Veprintsev DB, Deikus GY, Permyakov SE, Kalinichenko LP, Grishchenko VM, Brooks CL. 1997. pH-induced transition and Zn2+-binding properties of bovine prolactin. FEBS Lett 405:273–276 [DOI] [PubMed] [Google Scholar]
- 58. Voorhees JL, Rao GV, Gordon TJ, Brooks CL. 2011. Zinc binding to human lactogenic hormones and the human prolactin receptor. FEBS Lett 585:1783–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Burtis CA, Ashwood ER. 2001. Teitz fundamentals of clinical chemistry. 5th ed Philadelphia: Saunders [Google Scholar]
- 60. Magneson GR, Puvathingal JM, Ray WJ., Jr 1987. The concentrations of free Mg2+ and free Zn2+ in equine plasma blood. J Biol Chem 262:11140–11148 [PubMed] [Google Scholar]
- 61. Lowman HB, Cunningham BC, Wells JA. 1991. Mutational analysis and protein engineering of receptor binding determinates of human placental lactogen. J Biol Chem 266:10982–10988 [PubMed] [Google Scholar]
- 62. Greenan J, Lorenson MY, Conconi MV, Walker AM. 1990. Alterations in in situ prolactin secretory granule morphology and immunoactivity by thiols and divalent cations. Endocrinology 126:512–518 [DOI] [PubMed] [Google Scholar]
- 63. Sun Z, Lee MS, Rhee HK, Arrandale JM, Dannies PS. 1997. Inefficient secretion of human H27A-prolactin, a mutant that does not bind Zn2+. Mol Endocrinol 11:1544–1551 [DOI] [PubMed] [Google Scholar]
- 64. Brooks CL, Welsch CW. 1978. Effects of the association time of in vivo bound prolactin on the [125I] prolactin receptor assays of female rat livers. Mol Cell Endocrinol 11:145–151 [DOI] [PubMed] [Google Scholar]
- 65. Joss L, Morton TA, Doyle ML, Myszka DG. 1998. Interpreting kinetic rate constants from optical biosensor data recorded on a decaying surface. Anal Biochem 261:203–210 [DOI] [PubMed] [Google Scholar]
- 66. Gertler A, Grosclaude J, Strasburger CJ, Nir S, Djiane J. 1996. Real-time kinetic measurements of the interaction between lactogenic hormones and prolactin-receptor extracellular domains from several species support the model of hormone-induced transient receptor dimerization. J Biol Chem 271:24482–24491 [DOI] [PubMed] [Google Scholar]
- 67. Cunningham BC, Wells JA. 1993. Comparison of a structural and a functional epitope. J Mol Biol 234:554–563 [DOI] [PubMed] [Google Scholar]
- 68. Walsh ST, Jevitts LM, Sylvester JE, Kossiakoff AA. 2003. Site 2 binding energetics of the regulatory step of growth hormone-induced receptor homodimerization. Protein Sci 12:1960–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rui H, Kirken RA, Farrar WL. 1994. Activation of receptor-associated tyrosine kinase JAK2 by prolactin. J Biol Chem 269:5364–5368 [PubMed] [Google Scholar]
- 70. Tan D, Huang KT, Ueda E, Walker AM. 2008. S2 Deletion variants of human PRL receptors demonstrate that extracellular domain can alter conformation of the intracellular signaling domain. Biochemistry 47:479–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lee RC, Walters JA, Reyland ME, Anderson SM. 1999. Constitutive activation of the prolactin receptor results in the induction of growth factor-independent proliferation and constitutive activation of signaling molecules. J Biol Chem 274:10024–10034 [DOI] [PubMed] [Google Scholar]
- 72. Gourdou I, Gabou L, Paly J, Kermabon AY, Belair L, Djiane J. 1996. Development of a constitutively active mutant form of the prolactin receptor, a member of the cytokine receptor family. Mol Endocrinol 10:45–56 [DOI] [PubMed] [Google Scholar]
- 73. Gadd SL, Clevenger CV. 2006. Ligand-independent dimerization of the human prolactin receptor isoforms: functional implications. Mol Endocrinol 20:2734–2746 [DOI] [PubMed] [Google Scholar]
- 74. Qazi AM, Tsai-Morris CH, Dufau ML. 2006. Ligand-independent homo- and heterodimerization of human prolactin receptor variants: inhibitory action of the short forms by heterodimerization. Mol Endocrinol 20:1912–1923 [DOI] [PubMed] [Google Scholar]
- 75. Liu W, Brooks CL. 2011. Functional impact of manipulation on the relative orientation of human prolactin receptor domains. Biochemistry 50:5333–5344 [DOI] [PubMed] [Google Scholar]
- 76. Tan D, Johnson DA, Wu W, Zeng L, Chen YH, Chen WY, Vonderhaar BK, Walker AM. 2005. Unmodified prolactin (PRL) and S179D PRL-initiated bioluminescence resonance energy transfer between homo- and hetero-pairs of long and short human PRL receptors in living human cells. Mol Endocrinol 19:1291–1303 [DOI] [PubMed] [Google Scholar]
- 77. Ilondo MM, Damholt AB, Cunningham BA, Wells JA, De Meyts P, Shymko RM. 1994. Receptor dimerization determines the effects of growth hormone in primary rat adipocytes and cultured human IM-9 lymphocytes. Endocrinology 134:2397–2403 [DOI] [PubMed] [Google Scholar]
- 78. Goffin V, Kinet S, Ferrag F, Binart N, Martial JA, Kelly PA. 1996. Antagonistic properties of human prolactin analogs that show paradoxical agonistic activity in the Nb2 bioassay. J Biol Chem 271:16573–16579 [DOI] [PubMed] [Google Scholar]
- 79. Duda KM, Brooks CL. 1999. Human growth hormone site 2 lactogenic activity requires a distant tyrosine 164. FEBS Lett 449:120–124 [DOI] [PubMed] [Google Scholar]
- 80. Boutin JM, Edery M, Shirota M, Jolicoeur C, Lesueur L, Ali S, Gould D, Djiane J, Kelly PA. 1989. Identification of a cDNA encoding a long form of prolactin receptors in human hepatoma and breast cancer cells. Mol Endocrinol 3:1455–1461 [DOI] [PubMed] [Google Scholar]
- 81. Kline JB, Roehrs H, Clevenger CV. 1999. Functional characterization of intermediate isoforms of the human prolactin receptor. J Biol Chem 274:35461–35468 [DOI] [PubMed] [Google Scholar]
- 82. Hu ZZ, Meng J, Dufau ML. 2001. Isolation and characterization of two novel forms of the human prolactin receptor generated by alternative splicing of a newly identified exon 11. J Biol Chem 276:41086–41094 [DOI] [PubMed] [Google Scholar]
- 83. Kline JB, Rycyzyn MA, Clevenger CV. 2002. Characterization of a novel and functional human prolactin receptor isoform (ΔS1PRLr) containing only one extracellular fibronectin-like domain. Mol Endocrinol 16:2310–2322 [DOI] [PubMed] [Google Scholar]
- 84. Trott JF, Hovey RC, Koduri S, Vonderhaar BK. 2003. Alternative splicing of exon 11 of human prolactin receptor gene results in multiple isoforms including a secreted prolactin-binding protein. J Mol Endocrinol 30:31–47 [DOI] [PubMed] [Google Scholar]
- 85. Kline JB, Clevenger CV. 2001. Identification and characterization of the prolactin-binding protein (PRLBP) in human serum and milk. J Biol Chem 276:24760–24766 [DOI] [PubMed] [Google Scholar]
- 86. Binart N, Bachelot A, Bouilly J. 2010. Impact of prolactin receptor isoforms on reproduction. Trends Endocrinol Metab 21:362–368 [DOI] [PubMed] [Google Scholar]
- 87. Bogorad RL, Courtillot C, Mestayer C, Bernichtein S, Harutyunyan L, Jomain JB, Bachelot A, Kuttenn F, Kelly PA, Goffin V, Touraine P. 2008. Identification of a gain-of-function mutation of the prolactin receptor in women with benign breast tumors. Proc Natl Acad Sci USA 105:14533–14538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Goffin V, Bogorad RL, Touraine P. 2010. Identification of gain-of-function variants of the human prolactin receptor. Methods Enzymol 484:329–355 [DOI] [PubMed] [Google Scholar]
- 89. Urbanek M, Russell JE, Cooke NE, Liebhaber SA. 1993. Functional characterization of the alternatively spliced placental human growth hormone receptor. J Biol Chem 268:19025–19032 [PubMed] [Google Scholar]
- 90. Dastot F, Sobrier ML, Duquesnoy P, Duriez B, Goossens M, Amselem S. 1996. Alternatively spliced forms in the cytoplasmic domain of the human growth (GH) receptor regulate its ability to generate a soluble GH-binding protein. Proc Natl Acad Sci USA 93:10723–10728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ross RJ, Esposito N, Shen XY, Von Laue S, Chew SL, Dobson PR, Postel-Vinay MC, Finidori J. 1997. A short isoform of the human growth hormone receptor functions as a dominate negative inhibitor of the full length receptor and generates large amounts of binding protein. Mol Endocrinol 11:265–273 [DOI] [PubMed] [Google Scholar]
- 92. Amit T, Bergman T, Dastot F, Youdim MB, Amselem S, Hochberg Z. 1997. A membrane-fixed, truncated form of the human growth hormone receptor. J Clin Endocrinol Metab 82:3813–3817 [DOI] [PubMed] [Google Scholar]
- 93. Ayling RM, Ross R, Towner P, Von Laue S, Finidori J, Moutoussamy S, Buchanan CR, Clayton PE, Norman MR. 1997. A dominate negative mutation of the growth hormone receptor causes familial short stature. Nat Genet 16:13–14 [DOI] [PubMed] [Google Scholar]
- 94. Arcasoy MO, Jiang X, Haroon ZA. 2003. Expression of erythropoietin receptor splice variants in human cancer. Biochem Biophys Res Comm 307:999–1007 [DOI] [PubMed] [Google Scholar]
- 95. Bazan JF. 1990. Structural design and molecular evolution of a cytokine receptor superfamily. Proc Natl Acad Sci USA 87:6934–6938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Rozakis-Adcock M, Kelly PA. 1991. Mutational analysis of the ligand-binding domain of the prolactin receptor. J Biol Chem 266:16472–16477 [PubMed] [Google Scholar]
- 97. Xie YL, Hassan SA, Qazi AM, Tsai-Morris CH, Dufau ML. 2009. Intramolecular disulfide bonds of the prolactin receptor short form are required for its inhibitory action on the function of the long form of the receptor. Mol Cell Biol 29:2546–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Frank SJ, Gilliland G, Van Epps C. 1994. Treatment of IM-9 cells with human growth hormone (GH) promotes rapid disulfide linkage of GH receptor. Endocrinology 135:148–156 [DOI] [PubMed] [Google Scholar]
- 99. Zhang Y, Jiang J, Kopchick JJ, Frank SJ. 1999. Disulfide linkage of growth hormone (GH) receptors (GHR) reflects GH-induced GHR dimerization. Association of JAK2 with GHR is enhanced by dimerization. J Biol Chem 274:33072–33084 [DOI] [PubMed] [Google Scholar]
- 100. Yang N, Langenheim JF, Wang X, Jiang J, Chen WY, Frank SJ. 2008. Activation of growth hormone receptors by growth hormone and growth hormone antagonist dimers: insights into receptor triggering. Mol Endocrinol 22:978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Miura O, Ihle JN. 1993. Dimer- and oligomerization of the erythropoietin receptor by disulfide bond formation and significance of the region near the WSXWS motif in intracellular transport. Arch Biochem Biophys 306:200–208 [DOI] [PubMed] [Google Scholar]
- 102. Rozakis-Adcock M, Kelly PA. 1992. Identification of ligand binding determinates of the prolactin receptor. J Biol Chem 267:7428–7433 [PubMed] [Google Scholar]
- 103. de Vos AM, Ultsch M, Kossiakoff AA. 1992. Human growth hormone and the extracellular domain of its receptor: crystal structure of the complex. Science 255:306–312 [DOI] [PubMed] [Google Scholar]
- 104. Mitani M, Dufau ML. 1986. Purification and characterization of prolactin receptors from the ovary. J Biol Chem 261:1309–1315 [PubMed] [Google Scholar]
- 105. Okamura H, Raguet S, Bell A, Gagnon J, Kelly PA. 1989. Purification and protein sequence analysis of rat liver prolactin receptor. J Biol Chem 264:5904–5911 [PubMed] [Google Scholar]
- 106. Buteau H, Pezet A, Ferrag F, Perrot-Applanat M, Kelly PA, Edery M. 1998. N-Glycosylation of the prolactin receptor is not required for activation of gene transcription but is critical for its cell surface targeting. Mol Endocrinol 12:544–555 [DOI] [PubMed] [Google Scholar]
- 107. Chang WP, Ye Y, Clevenger CV. 1998. Stoichiometric structure-function analysis of prolactin receptor signaling domain by receptor chimeras. Mol Cell Biol 18:896–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Landschulz WH, Johnson PF, McKnight SL. 1988. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240:1759–1764 [DOI] [PubMed] [Google Scholar]
- 109. Miller M. 2009. The importance of being flexible: the case of basic region leucine zipper transcription regulators. Curr Protein Pept Sci 10:244–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Ryadnov MG, Papapostolou D, Woolfson DN. 2008. The leucine zipper as a building block for self-assembled protein fibers. Methods Mol Biol 474:35–51 [DOI] [PubMed] [Google Scholar]
- 111. Brown RJ, Adams JJ, Pelekanos RA, Wan Y, McKinstry WJ, Palethorpe K, Seeber RM, Monks TA, Eidne KA, Parker MW, Waters MJ. 2005. Model for growth hormone receptor activation based on subunit rotation within a receptor dimer. Nat Struct Mol Biol 12:814–821 [DOI] [PubMed] [Google Scholar]
- 112. Ruan W, Becker V, Klingmüller U, Langosch D. 2004. The interface between self-assembling erythropoietin receptor transmembrane segments corresponds to a membrane-spanning leucine zipper. J Biol Chem 279:3273–3279 [DOI] [PubMed] [Google Scholar]
- 113. Kubatzky KF, Ruan W, Gurezka R, Cohen J, Ketteler R, Watowich SS, Neumann D, Langosch D, Klingmüller U. 2001. Self assembly of the transmembrane domain promotes signal transduction through the erythropoietin receptor. Curr Biol 11:110–115 [DOI] [PubMed] [Google Scholar]
- 114. Yang N, Wang X, Jiang J, Frank SJ. 2007. Role of the growth hormone (GH) receptor transmembrane domain in receptor predimerization and GH-induced activation. Mol Endocrinol 21:1642–1655 [DOI] [PubMed] [Google Scholar]
- 115. Uversky VN, Gillespie JR, Fink AL. 2000. Why are “natively unfolded” proteins unstructured under physiological conditions? Proteins 41:415–427 [DOI] [PubMed] [Google Scholar]
- 116. Ren S, Uversky VN, Chen Z, Dunker AK, Obradovic Z. 2008. Short linear motifs recognized by SH2, SH3 and Ser/Thr kinase domains are conserved in disordered protein regions. BMC Genomics 9(Suppl 2):S26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Rost B. 1996. PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol 266:525–539 [DOI] [PubMed] [Google Scholar]
- 118. Rost B, Yachdav G, Liu J. 2004. The ProteinPredict server. Nucleic Acids Res 32:W321–W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Goujon L, Allevato G, Simonin G, Paquereau L, Le Cam A, Clark J, Nielsen JH, Djiane J, Postel-Vinay MC, Edery M. 1994. Cytoplasmic sequences of the growth hormone receptor necessary for signal transduction. Proc Natl Acad Sci USA 91:957–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Venkitaraman AR, Cowling RJ. 1992. Interleukin 7 receptor functions by recruiting the tyrosine kinase p59fyn through a segment of its cytoplasmic tail. Proc Natl Acad Sci USA 89:12083–12087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Pezet A, Buteau H, Kelly PA, Edery M. 1997. The last proline of box 1 is essential for association with JAK2 and the functional activation of the prolactin receptor. Mol Cell Endocrinol 129:199–208 [DOI] [PubMed] [Google Scholar]
- 122. O'Neal KD, Chari MV, Mcdonald CH, Cook RG, Yu-Lee LY, Morrisett JD, Shearer WT. 1996. Multiple cis-trans conformers of the prolactin receptor proline-rich motif (PRM) peptide detected by reverse-phase HPLC, CD and NMR spectroscopy. Biochem J 315:833–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Lu KP, Finn G, Lee TH, Nicholson LK. 2007. Prolyl cis-trans isomerization as a molecular timer. Nat Chem Biol 3:619–629 [DOI] [PubMed] [Google Scholar]
- 124. Campbell GS, Argetsinger LS, Ihle JN, Kelly PA, Rillema JA, Carter-Su C. 1994. Activation of JAK2 tyrosine kinase by prolactin receptors in Nb2 cells and mouse mammary gland explants. Proc Natl Acad Sci USA 91:5232–5236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Clevenger CV, Medaglia MV. 1994. The protein tyrosine kinase p59fyn is associated with the prolactin (PRL) receptor and is activated by prolactin stimulation of T-lymphocytes. Mol Endocrinol 8:674–681 [DOI] [PubMed] [Google Scholar]
- 126. Clevenger CV, Torigoe T, Reed JC. 1994. Prolactin induces rapid phosphorylation and activation of prolactin receptor associated Raf-1 kinase in a T-cell line. J Biol Chem 269:5559–5565 [PubMed] [Google Scholar]
- 127. Clevenger CV, Ngo W, Sokol DL, Luger SM, Gewirtz AM. 1995. VAV is necessary for prolactin-stimulated proliferation and is translocated into the nucleus of a T-cell line. J Biol Chem 270:13246–13253 [DOI] [PubMed] [Google Scholar]
- 128. Schlessinger J, Lemmon MA. 2003. SH2 and PTB domains in tyrosine kinase signaling. Sci STKE 2003:RE12. [DOI] [PubMed] [Google Scholar]
- 129. Liu BA, Jablonowski K, Raina M, Arcé M, Pawson T, Nash PD. 2006. The human and mouse complement of SH2 domain proteins—establishing the boundaries of phosphotyrosine signaling. Mol Cell 22:851–868 [DOI] [PubMed] [Google Scholar]
- 130. Pezet A, Ferrag F, Kelly PA, Edery M. 1997. Tyrosine docking sites of the rat prolactin receptor required for association and activation of STAT5. J Biol Chem 272:25043–25050 [DOI] [PubMed] [Google Scholar]
- 131. Lebrun JJ, Ali S, Goffin V, Ullrich A, Kelly PA. 1995. A single phosphotyrosine residue of the prolactin receptor is responsible for activation of gene transcription. Proc Natl Acad Sci USA 92:4031–4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. 2012. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in men and mouse. Nucleic Acids Res 40:D261–D270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Choudhary C, Mann M. 2010. Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol 11:427–439 [DOI] [PubMed] [Google Scholar]
- 134. Kosako H, Nagano K. 2011. Quantitative phosphoproteomics strategies for understanding protein kinase-mediated signaling transduction pathways. Expert Rev Proteomics 8:81–94 [DOI] [PubMed] [Google Scholar]
- 135. Olayioye MA, Guthridge MA, Stomski FC, Lopez AF, Visvader JE, Lindeman GJ. 2003. Threonine 391 phosphorylation of human prolactin receptor mediates a novel interaction with 14–3-3 proteins. J Biol Chem 278:32929–32935 [DOI] [PubMed] [Google Scholar]
- 136. Ma L, Gao JS, Guan Y, Shi X, Zhang H, Ayrapetov MK, Zhang Z, Xu L, Hyun YM, Kim M, Zhuang S, Chin YE. 2010. Acetylation modulates prolactin receptor dimerization. Proc Natl Acad Sci USA 107:19314–19319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Aitken A. 2006. 14-3-3 proteins: a historic overview. Semin Cancer Biol 16:162–172 [DOI] [PubMed] [Google Scholar]
- 138. McManus KJ, Hendzel MJ. 2001. CBP, a transcriptional coactivator and acetyltransferase. Biochem Cell Biol 79:253–266 [PubMed] [Google Scholar]
- 139. Goffin V, Kelly PA. 1997. The prolactin/growth hormone receptor family: structure/function relationships. J Mammary Gland Biol Neoplasia 2:7–17 [DOI] [PubMed] [Google Scholar]
- 140. Kossiakoff AA, Somers W, Ultsch M, Andow K, Muller YA, De Vos AM. 1994. Comparison of the intermediate complexes of human growth hormone bound to the human growth and prolactin receptors. Protein Sci 3:1697–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Cunningham BC, Ultsch M, De Vos AM, Mulkerrin MG, Clauser KR, Wells JA. 1991. Dimerization of the extracellular domain of the human growth hormone receptor by a single hormone molecule. Science 254:821–825 [DOI] [PubMed] [Google Scholar]
- 142. Kossiakoff AA. 2004. The structural basis for biological signaling, regulation, and specificity in the growth hormone/prolactin system of hormones and receptors. Adv Protein Chem 68:147–169 [DOI] [PubMed] [Google Scholar]
- 143. Langenheim JF, Tan D, Walker AM, Chen WY. 2006. Two wrongs can make a right: dimers of prolactin and growth hormone receptor antagonists behave as agonists. Mol Endocrinol 20:661–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Rui H, Lebrun JJ, Kirken RA, Kelly PA, Farrar WL. 1994. JAK2 activation and cell proliferation induced by antibody-mediated prolactin receptor dimerization. Endocrinology 135:1299–1306 [DOI] [PubMed] [Google Scholar]
- 145. Djiane J, Houdebine LM, Kelly PA. 1981. Prolactin-like activity of anti-prolactin receptor antibodies on casein and DNA synthesis in the mammary gland. Proc Natl Acad Sci USA 78:7445–7448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Dusanter-Fourt I, Muller O, Ziemiecki A, Mayeux P, Drucker B, Djiane J, Wilks A, Harpur AG, Fischer S, Gisselbrecht S. 1994. Identification of JAK protein tyrosine kinases as signaling molecules for prolactin. Functional analysis of prolactin receptor and prolactin-erythropoietin receptor chimera expressed in lymphoid cells. EMBO J 13:2583–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Cunningham BC, Wells JA. 1989. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science 244:1081–1085 [DOI] [PubMed] [Google Scholar]
- 148. Clevenger CV, Gadd SL, Zheng J. 2009. New mechanism for PRL action in breast cancer. Trends Endocrinol Metab 20:223–229 [DOI] [PubMed] [Google Scholar]
- 149. Walker AM. 2007. S179D prolactin: antagonistic agony. Mol Cell Endocrinol 276:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Varghese B, Barriere H, Carbone CJ, Banerjee A, Swaminathan G, Plotnikov A, Xu P, Peng J, Goffin V, Lukacs GL, Fuchs SY. 2008. Polyubiquitination of prolactin receptor stimulates its internalization, postinternalization sorting, and degradation via the lysosomal pathway. Mol Cell Biol 28:5275–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Livnah O, Stura EA, Middleton SA, Johnson DL, Jolliffe LK, Wilson IA. 1999. Crystallographic evidence for performed dimers of erythropoietin receptor before ligand activation. Science 283:987–990 [DOI] [PubMed] [Google Scholar]
- 152. Remy I, Wilson IA, Michnick SW. 1999. Erythropoietin receptor activation by a ligand-induced conformation change. Science 283:990–993 [DOI] [PubMed] [Google Scholar]
- 153. Watowich SS. 1999. Activation of erythropoietin signaling by receptor dimerization. Int J Biochem Cell Biol 31:1075–1088 [DOI] [PubMed] [Google Scholar]
- 154. Watowich SS, Yoshimura A, Longmore GD, Hilton DJ, Yoshimura Y, Lodish HF. 1992. Homodimerization and constitutive activation of the erythropoietin receptor. Proc Natl Acad Sci USA 89:2140–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Waters MJ, Hoang HN, Fairlie DP, Pelekanos RA, Brown RJ. 2006. New insights into growth hormone action. J Mol Endocrinol 36:1–7 [DOI] [PubMed] [Google Scholar]
- 156. Constantinescu SN, Huang LJ, Nam H, Lodish HF. 2001. The erythropoietin receptor cytosolic juxtamembrane domain contains an essential, precisely oriented, hydrophobic motif. Mol Cell 7:377–385 [DOI] [PubMed] [Google Scholar]
- 157. Syed RS, Reid SW, Li C, Cheetham JC, Aoki KH, Liu B, Zhan H, Osslund TD, Chirino AJ, Zhang J, Finer-Moore J, Elliott S, Sitney K, Katz BA, Matthews DJ, Wendoloski JJ, Egrie J, Stroud RM. 1998. Efficiency of signaling through cytokine receptors depends critically on receptor orientation. Nature 395:511–516 [DOI] [PubMed] [Google Scholar]
- 158. Ellgaard L, Ruddock LW. 2005. The human protein disulfide isomerase family: Substrate interactions and functional properties. EMBO Rep 6:28–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Syed F, Rycyzyn MA, Westgate L, Clevenger CV. 2003. A novel and functional interaction between cyclophilin A and prolactin receptor. Endocrine 20:83–90 [DOI] [PubMed] [Google Scholar]
- 160. Zheng J, Koblinski JE, Dutson LV, Feency YB, Clevenger CV. 2008. Prolyl isomerase cyclophilin A regulation of Janus-activated kinase 2 and the progression of human breast cancer. Cancer Res 68:7769–7778 [DOI] [PubMed] [Google Scholar]
- 161. Tang X, Gao JS, Guan YJ, McLane KE, Yuan ZL, Ramratnam B, Chin YE. 2007. Acetylation-dependent signal transduction for type 1 interferon receptor. Cell 131:93–105 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.