Abstract
Insulin signaling plays a physiological role in traditional insulin target tissues controlling glucose homeostasis as well as in pancreatic β-cells and in the endothelium. Insulin signaling abnormalities may, therefore, be pathogenic for insulin resistance, impaired insulin secretion, endothelial dysfunction, and eventually, type 2 diabetes mellitus (T2DM) and cardiovascular disease. Tribbles homolog 3 (TRIB3) is a 45-kDa pseudokinase binding to and inhibiting Akt, a key mediator of insulin signaling. Akt-mediated effects of TRIB3 in the liver, pancreatic β-cells, and skeletal muscle result in impaired glucose homeostasis. TRIB3 effects are also modulated by its direct interaction with other signaling molecules. In humans, TRIB3 overactivity, due to TRIB3 overexpression or to Q84R genetic polymorphism, with R84 being a gain-of-function variant, may be involved in shaping the risk of insulin resistance, T2DM, and cardiovascular disease. TRIB3 overexpression has been observed in the liver, adipose tissue, skeletal muscle, and pancreatic β-cells of individuals with insulin resistance and/or T2DM. The R84 variant has also proved to be associated with insulin resistance, T2DM, and cardiovascular disease. TRIB3 direct effects on the endothelium might also play a role in increasing the risk of atherosclerosis, as indicated by studies on human endothelial cells carrying the R84 variant that are dysfunctional in terms of Akt activation, NO production, and other proatherogenic changes. In conclusion, studies on TRIB3 have unraveled new molecular mechanisms underlying metabolic and cardiovascular abnormalities. Additional investigations are needed to verify whether such acquired knowledge will be relevant for improving care delivery to patients with metabolic and cardiovascular alterations.
Introduction
-
TRIB3 Structure, Functions, and Expression
TRIB3 structure
TRIB3 functions
TRIB3 expression
-
TRIB3 and Human Metabolic and Cardiovascular Disturbances
TRIB3 expression, insulin resistance, and T2DM
Nonsynonymous TRIB3 Q84R polymorphism
Conclusions
I. Introduction
Insulin sensitivity (measured as insulin-mediated glucose uptake) has a quite large range of distribution in the general population (1). Subjects at the lower end of the spectrum (i.e. insulin-resistant individuals) need sustained insulin hypersecretion to maintain normoglycemia. When β-cells fail to secrete sufficient insulin to adequately counteract insulin resistance, type 2 diabetes mellitus (T2DM) ensues (2, 3).
Although compensatory hyperinsulinemia does help to prevent the occurrence of overt T2DM, it might contribute, especially in the presence of obesity, to a further deterioration of insulin sensitivity and development of dyslipidemia (4), hypertension (4, 5), and endothelial dysfunction (6). In this scenario, insulin-resistant individuals are undoubtedly at high risk of developing future cardiovascular events (7–9). Furthermore, other human diseases with high prevalence, such as polycystic ovary syndrome (10), nonalcoholic fatty liver disease (11), chronic neurodegenerative processes (e.g. Alzheimer's and Parkinson's disease) (12), and some forms of cancer (e.g. liver, colon-rectal, and breast cancer) (13) have lately been added to the list of conditions recognizing insulin resistance as a common pathogenic soil. Insulin resistance, then, represents a tremendous burden for healthcare systems as well as for a large number of patients and their relatives. Because the intrinsic mechanisms leading to insulin resistance are far from being fully elucidated, it is still very difficult to tackle this burden. Unraveling molecular abnormalities underlying defective insulin action is, therefore, urgently needed.
After binding to the α-subunit of its receptor, insulin stimulates a cascade of signaling events that ultimately mediate insulin action in several target tissues (14–16) (Fig. 1). The first such event is activation of the receptor's β-subunit via tyrosine phosphorylation. This switches on the receptor's intrinsic tyrosine kinase activity, which catalyzes phosphorylation of several insulin receptor substrates (IRS, all acting as docking molecules for Src homology 2 domain-containing proteins) and eventually initiates downstream signal transmission (14–16). Several, although not all, insulin effects are mediated by the phosphorylated forms of IRS-1 and IRS-2, which activate phosphatidyl-inositol-3-kinase (PI3K), thus producing phosphatidylinositol 3,4,5-triphosphate (PIP3) (14–16). PIP3, in turn, activates serine/threonine protein kinase Akt, a central mediator of several traditional insulin effects on the intermediate metabolism (14–16), including stimulation of glucose uptake and of both glycogen and fatty acid synthesis. Akt plays a pivotal role on insulin signaling in new, nontraditional target cells as well, being crucial for insulin-mediated nitric oxide synthase activation in endothelial cells (17) and insulin secretion and survival in pancreatic β-cells (18).
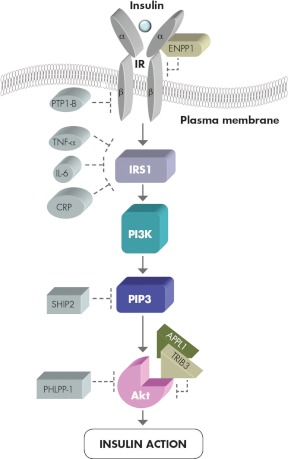
Figure 1.
Schematic representation of insulin signaling pathway. Insulin binding to its receptor, a transmembrane glycoprotein consisting of two α- and two β-subunits, which possesses tyrosine kinase activity, represents the first step in the insulin signaling cascade. After insulin binding to the α-subunit, β-subunit tyrosine residues become autophosphorylated. This increases intrinsic receptor kinase activity and accelerates tyrosine phosphorylation of the receptor substrates, the most important of which are IRS-1 and IRS-2. IRS-1 and IRS-2 bind the regulatory subunit of PI3K, which catalyzes the production of PIP3. PIP3 acts as a secondary messenger activating the serine/threonine protein kinase Akt, which finally mediates most insulin actions. Several negative modulators of insulin signaling have been described. A few of them act by protein-protein interaction: as shown on the right side of the insulin signaling cascade, the class II transmembrane glycoprotein ENPP1 binds and inactivates insulin receptor tyrosine kinase activity, whereas the pseudokinase TRIB3 binds Akt and inhibits downstream signaling. APPL1 competes with this latter binding, thus antagonizing TRIB3 inhibition of Akt-mediated insulin signaling. Other inhibitors act by modifying the phosphorylation state of signaling molecules: as shown on the left side of the insulin signaling cascade, these include phosphotyrosine phosphatase-1B (PTP-1B), a tyrosine phosphatase on the insulin receptor; TNF-α, IL-6, and C-reactive protein (CRP), chronic inflammation-related molecules that induce IRS-1 serine phosphorylation and subsequent inactivation; the inositol-5 phosphatase SHIP2, which dephosphorylates PIP3; and finally, phosphatase PH domain leucine-rich repeat protein phosphatase-1 (PHLPP-1), a specific Akt Ser473 phosphatase. Based on our present knowledge, it is entirely possible that, in some individuals, alterations to this redundant and complex network may be pathogenic for insulin resistance and related metabolic derangements.
Several molecules have been reported to inhibit the insulin signaling pathway (Fig. 1). Such inhibition takes place via at least two mechanisms: 1) protein-protein interaction and 2) alteration of the phosphorylation state of some of the signaling molecules described above. Examples of the first mechanism are binding between class II transmembrane glycoprotein ectonucleotide pyrophosphatase phosphodiesterase 1 (ENPP1) and insulin receptor (19) and binding between the catalytically inactive pseudokinase termed tribbles homolog 3 (TRIB3) and Akt (20). Examples of the second mechanism include insulin receptor dephosphorylation (as described for several tyrosine phosphatases, such as phosphotyrosine phosphatase-1B) (21), IRS-1 serine phosphorylation (induced by cytokines such as TNF-α and IL-6 as well as by C-reactive protein) (22–24), PIP3 5′-dephosphorylation (hydrolyzed by Src homology 2 domain-containing inositol 5 phosphatase) (25), and finally, dephosphorylation of Akt by the specific phosphatase pleckstrin homology (PH) domain leucine-rich repeat protein phosphatase-1 (26, 27). In the end, therefore, insulin modulation of cell metabolism largely results from the opposite effects of molecules acting as enhancers and molecules acting as inhibitors of the insulin signaling pathway. Dysfunctions in such a complex network are likely to influence insulin sensitivity and foster insulin resistance and related metabolic derangements as well as adverse clinical outcomes (28, 29).
Among the many molecules acting on this network, in this review we will specifically focus on TRIB3, a molecule capable of impairing insulin signaling by affecting insulin-induced Akt activation in several insulin target tissues. To this end, a literature search was performed using MEDLINE PubMed with the following search terms: NIPK or SKIP or tribble or TRB or TRIB or TRIB3 or TRIB3 R84 variant or Q84R polymorphism. A total of 175 different papers were found. An additional search on MEDLINE PubMed was performed using TRIB1 or tribbles homolog 1 (85 different papers found) or TRIB2 (37 papers found) as search terms. We will review and comment on studies (both in animal models and in humans) indicating that changes in TRIB3 expression levels induce cellular and systemic insulin resistance. In this context, we will also review studies on nonsynonymous TRIB3 Q84R polymorphism (i.e. rs2295490), a gain-of-function amino acid substitution representing a natural model of human insulin signaling dysfunction. Studying this model has provided both novel insights into the physiological significance of insulin signaling in nontraditional insulin target tissues as well as a better understanding of how abnormalities in this pathway underlie metabolic defects of potentially high clinical relevance.
II. TRIB3 Structure, Functions, and Expression
A. TRIB3 structure
Human TRIB3 gene (gene ID 57761; also known as NIPK/SKIP3/TRB3), spanning 16.9 kb of genomic region on chromosome 20p13-p12.2 (Fig. 2), belongs to the tribbles family (first identified in Drosophila melanogaster) (30), which, in mammals, also includes TRIB1 and TRIB2. The amino acid sequences of tribbles are highly conserved between human and mouse (amino acid identities: TRIB1, 97.5%; TRIB2, 99.2%; TRIB3, 81.2%), and a high degree of similarity is also noted within the human family of sequences (TRIB1/TRIB2, 71.3%; TRIB1/TRIB3, 53.3%; TRIB2/TRIB3, 53.7%) (31).
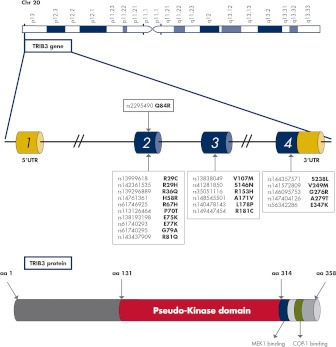
Figure 2.
TRIB3 genomic location, gene organization, and protein structure. The human TRIB3 gene spans 16.9 kb on chromosome 20p13-p12.2 and is organized in four exons. Several nonsynonymous SNP have been identified in the coding sequence of the TRIB3 gene (exons 2–4) as indicated by black arrows from the white boxes. Their biological relevance, if any, has never been investigated. The rs2295490 in exon 2 (highlighted in red), which results in a glutamine (Q) to arginine (R) substitution at codon 84 (i.e. Q84R), is the only relatively frequent and well-characterized SNP. TRIB3 gene encodes a 45-kDa, 358-amino-acid protein, comprising different domains acting as adaptor molecules in several signaling pathways. Among these are a pseudokinase domain (highlighted in red) by which TRIB3 interacts with C/EBP transcription factors and retinoic acid receptors, a mitogen-activated protein kinase kinase 1 (MEK1) binding site (highlighted in violet) involved in the interaction with MAPK kinases (MAPKK) and an E3 ubiquitin ligase COP1 binding site (highlighted in green) important for a core function of tribbles as proteasome-mediated degradation of the C/EBP family of proteins. UTR, Untranslated region.
Human TRIB3 is organized as four exons, encoding a 358-amino-acid protein. Due to alternative transcription initiation and alternative splicing of the first exon, several human TRIB3 mRNA isoforms have been reported (32). Such variation at the 5′-untranslated region of the gene may have a strong impact on translational efficiency and serves the important function of regulating gene expression levels (33).
Beside the prevalent Q84R single-nucleotide polymorphism (SNP), which has been extensively characterized (as described in later sections of this article), several additional infrequent nonsynonymous SNP have been identified in the genomic region encompassing the human TRIB3 gene (dbSNP www.ncbi.nlm.nih.gov/snp) (Fig. 2) whose function and potential biological relevance have never, so far, been addressed.
The protein encoded by TRIB3 (as well as those encoded by TRIB1 and TRIB2) is characterized by a well-conserved central domain (34) consisting of 12 subdomains belonging to the protein kinase superfamily (31) (Fig. 2). However, whereas tribbles proteins' predicted three-dimensional structure does resemble that of a protein kinase (35), they lack consensus sequences for protein phosphorylation and therefore do not possess intrinsic kinase activity (34), thus being named pseudokinases. According to the evidence available, tribbles proteins act as adaptors in signaling pathways for several important cellular processes (34).
B. TRIB3 functions
By interacting with a host of molecules listed in Table 1, TRIB3 affects a number of cellular functions, such as insulin signaling and action, pancreatic β-cell survival and insulin secretory capacity, adipose and muscle cell differentiation, and endoplasmic reticulum (ER) stress. Thus, TRIB3 plays an instrumental role in modulating several metabolic pathways related to insulin sensitivity, glucose homeostasis, and vascular function, as will be discussed in Section II.B.
Table 1.
Direct modulators and related pathways involved in TRIB3 action
| Modulators | Pathway | Main effect | Refs. |
|---|---|---|---|
| Akt | Akt-mediated insulin signaling | Decreased insulin action | 20 |
| APPL1 | Akt-mediated insulin signaling | Decreased insulin action | 39 |
| MLK3 | Akt-mediated insulin signaling | Increased β-cell apoptosis | 50 |
| MAPKK | ERK1/2 and JNK-mediated signaling | Not addressed | 40 |
| C/EBPβ | PPARγ-mediated gene expression | Reduced adipogenesis | 51 |
| COP1 | Ubiquitination-dependent ACC degradation | Reduced fatty acid oxidation | 52 |
| ATF4/CHOP | ER-dependent gene expression | Modulation of ATF4/CHOP-mediated apoptosis | 65 |
| ATF4 | Gene expression | Reduced insulin exocytosis | 71 |
The mechanisms through which the interaction of TRIB3 with any of the listed modulators affect specific pathways and, produce, eventually specific effects are described into details in the main text. Akt is the murine thymoma viral oncogene homolog. MAPKK, MAPK kinase; MLK3, mixed lineage kinase 3.
1. In vitro insulin signaling
TRIB3 interacts with and inhibits activation of Akt, a Thr/Ser kinase playing a crucial role in the insulin signaling pathway (Fig. 1). This key feature has been demonstrated in several cell types where TRIB3 knockdown enhances and TRIB3 overexpression impairs Akt activation and subsequent insulin signaling and action. Thus, knocking down TRIB3 by RNA interference in human HepG2-cultured hepatoma cells increases insulin-induced phosphorylation of Akt and of its substrates glycogen synthase kinase 3 and forkhead box class O factor 1 (FoxO1) (20), resulting (as shown by a different study) in enhanced insulin-stimulated ribosomal protein S6 kinase activity and phosphorylation of both S6 ribosomal protein and eukaryotic translation initiation factor 4E binding protein 1 (36). On the other hand, TRIB3 overexpression inhibits insulin-stimulated Akt phosphorylation and downstream insulin signaling (36) in primary mouse hepatocytes and almost completely abolishes the insulin-inhibitory effect on glucose output in rat cultured hepatoma cells.
Consistent with results obtained in liver cells, a TRIB3 inhibitory effect on insulin-mediated Akt activation has also been reported in skeletal muscle cells. Thus, in L6 rat skeletal muscle cells, TRIB3 overexpression reduces insulin-stimulated Akt and ERK1/2 phosphorylation and blunts insulin-induced glucose transporter 4 (GLUT4) translocation and glucose transport (37). Similarly, insulin-stimulated Akt phosphorylation is reduced in mouse myoblast C2C12 cells overexpressing TRIB3 (38).
Very preliminary data have suggested that TRIB2 likewise inhibits Akt activation in human embryonic kidney (HEK293) cells (20).
In summary, data obtained from both liver and skeletal muscle cells strongly suggest that TRIB3 is a negative modulator of insulin-mediated Akt activation and downstream insulin signaling and action. However, as discussed in the following sections, also other mechanisms, besides modulation of Akt activation, are operating as well in mediating some TRIB3 effects.
It is worth noting that the TRIB3 inhibitory effect on Akt activation appears to be modulated by adaptor protein, phosphotyrosine interaction, PH domain, and leucine zipper-containing 1 (APPL1), an adaptor protein containing an NH2-terminal Bin/Amphiphysin/Rvs domain (39). Immunoprecipitation experiments in primary rat hepatocytes have shown that Akt interacts with both APPL1 and TRIB3 in a competitive manner (Fig. 1). The insulin action impairment observed by overexpressing TRIB3 was abolished by APPL1 overexpression, suggesting that APPL1 can directly antagonize the insulin-desensitizing effects of TRIB3 on Akt signaling. In liver cells from several models of obese, insulin-resistant mice, coimmunoprecipitation analysis detected an increased association between Akt and TRIB3, which was accompanied by a decreased association between Akt and APPL1 (39). Adenoviral overexpression of APPL1 in the liver of db/db diabetic insulin-resistant mice suppressed the interaction between Akt and TRIB3, which was paralleled by a marked reduction in blood glucose levels and by amelioration of insulin resistance. Analysis of subcellular compartmentalization of Akt showed that insulin-mediated Akt activation was finely tuned by the relative APPL1 and TRIB3 expression levels (39). Thus, TRIB3 inhibited Akt activation by arresting it at the cytosol and blocking its membrane translocation. By contrast, APPL1 competed with TRIB3 for binding to Akt and subsequently promoted Akt translocation to the cell membrane for further activation.
Finally, in addition to modulating Akt, TRIB3 appears also to bind to MAPK kinase and to modulate the cascade of downstream molecules among which are ERK1/2 and Jun N-terminal kinases (JNK). Thus, high TRIB3 expression levels inhibit both ERK1/2 and JNK, whereas low TRIB3 expression levels have the opposite effect leading to ERK1/2 and JNK activation (40). Whether these TRIB3 effects on MAPK cascades have a functional impact on cell cycle and/or insulin signaling and action requires to be ascertained by additional studies.
2. In vivo insulin action
TRIB3 emerged as a possibly important player in the regulation of in vivo insulin action when it was found that, compared with control mice, TRIB3 expression was increased in db/db diabetic insulin-resistant mice (20). At the same time, it was shown that, in control mice, TRIB3 overexpression ensuring protein content levels comparable to those observed in db/db mice resulted in hyperglycemia in the fed state, glucose intolerance, and insulin resistance (20, 36).
Data gathered from other animal models of insulin resistance and/or diabetes also point to enhanced TRIB3 expression as a pathogenic factor of metabolic disturbances. Thus, enhanced TRIB3 expression has been observed in the skeletal muscle of Zucker fatty rats and streptozotocin-treated diabetic rats (37) as well as in the adipose tissue of rats with metabolic syndrome induced by a high-fructose diet (41) and in the liver of rats with nonalcoholic fatty liver disease (42). Taken altogether, these data indicate that in rodents, TRIB3 overexpression plays a very important role in modulating whole-body insulin sensitivity and is possibly involved in the pathogenesis of insulin resistance-related metabolic abnormalities (Table 2).
Table 2.
Relationship between TRIB3 expression levels, insulin resistance, glucose homeostasis, and related traits in animal models
| Animal model | Phenotype | Tissue | TRIB3 expression | Refs. |
|---|---|---|---|---|
| db/db mice | Obesity | Liver | Increased | 20 |
| Hyperglycemia | ||||
| Insulin resistance | ||||
| TRIB3-overexpressing mice | Fasting hyperglycemia | Liver | Increased | 20 |
| Impaired glucose tolerance | ||||
| TRIB3-overexpressing mice | Impaired glucose tolerance | Liver | Increased | 36 |
| Insulin resistance | ||||
| ob/ob mice | Obesity | Liver | Increased | 46 |
| Hyperglycemia | ||||
| Insulin resistance | ||||
| DIO mice | Obesity | Liver | Increased | 46 |
| Hyperglycemia | ||||
| Insulin resistance | ||||
| NAFLD rats | Liver steatosis | Liver | Increased | 42 |
| db/db mice | Obesity | Skeletal muscle | Increased | 37 |
| Hyperglycemia | ||||
| Zucker fatty rats | Obesity | Skeletal muscle | Increased | 37 |
| Insulin resistance | ||||
| Streptozotocin-treated rats | Hyperglycemia | Skeletal muscle | Increased | 37 |
| ob/ob mice | Obesity | Skeletal muscle | Increased | 45 |
| Hyperglycemia | ||||
| Insulin resistance | ||||
| Goto-Kakizaki rats | Hyperglycemia | Pancreatic islets | Increased | 70 |
| Pancreatic β-cell dysfunction | ||||
| Pancreatic β-cell-specific TRIB3-overexpressing mice | Impaired glucose tolerance | Pancreatic β-cell | Increased | 71 |
| Impaired insulin secretion | ||||
| Reduced β-cell mass | ||||
| Adipose tissue-specific TRIB3-overexpressing mice | Protection from DIO | Adipose tissue | Increased | 52 |
| Increased fatty acid oxidation | ||||
| HFD rats | Metabolic syndrome | Adipose tissue | Increased | 41 |
| Cardiac specific TRIB3-overexpressing mice | Decreased cardiac glucose oxidation | Heart | Increased | 67 |
| Liver specific | Increased liver insulin sensitivity | Liver | Decreased | 47 |
| Liver specific TRIB3-deficient mice | Improved glucose tolerance | Liver | Decreased | 47 |
| Acutely exercised | Decreased fasting glucose | Liver | Decreased | 46 |
| DIO mice | Increased insulin sensitivity | |||
| Acutely exercised | Decreased fasting glucose | Liver | Decreased | 46 |
| ob/ob mice | Increased insulin sensitivity | |||
| Skeletal muscle specific | Decreased fasting glucose | Skeletal muscle | Decreased | 38 |
| LKB1-deficient mice | Decreased fasting insulin | |||
| Increased insulin-stimulated muscle glucose uptake | ||||
| Acutely exercised | Decreased fasting glucose | Skeletal muscle | Decreased | 45 |
| ob/ob mice | Increased insulin sensitivity | |||
| TRIB3-deficient mice | Unmodified glucose tolerance | Liver, skeletal muscle, adipose tissue | Decreased | 43 |
| Unmodified insulin sensitivity | ||||
| TRIB3 ASO-treated rats | Increased insulin sensitivity | Liver, white adipose tissue | Decreased | 44 |
DIO, Diet-induced obesity; HFD, high-fat diet; NAFLD, nonalcoholic fatty liver disease.
If, as discussed, TRIB3 overexpression impairs insulin action, one might expect that reducing TRIB3 expression would, on the contrary, increase insulin sensitivity. However, Okamoto et al. (43) reported that TRIB3 genetic deletion in the mouse did not alter insulin-stimulated activation of Akt or of specific downstream substrates, nor did it change the expression of genes involved in hepatic glucose production (PEPCK, G6P, PGC1α, and glucokinase). Consistent with this, serum glucose, insulin, and lipid levels as well as glucose tolerance, insulin sensitivity, and energy metabolism were not altered in these TRIB3 knockout animals (43). By contrast, knocking down TRIB3 in rats by antisense oligonucleotides (ASO), although not affecting insulin-stimulated Akt phosphorylation levels in the liver, adipose tissue, or skeletal muscle (44), did enhance insulin-stimulated glucose disposal during euglycemic-hyperinsulinemic clamp, largely by improving skeletal muscle glucose uptake (44). It should be noted that in this study, TRIB3 knockdown increased white adipose tissue mass by 70% and enhanced the expression of PPAR-γ and its key target genes CCAAT/enhancer-binding protein α (CEBPα), CD36, and acyl-coenzyme A oxidase 1 (ACOX1). This raises the possibility that the observed improvement in insulin sensitivity in TRIB3 ASO-treated animals was primarily dependent upon peroxisome proliferator-activated receptor-γ (PPAR-γ) activation. This hypothesis is supported by experiments showing that when these animals were cotreated with the PPAR-γ antagonist bisphenol A diglycidyl ether (BADGE), not only were the expansion of white adipose tissue and the increased expression of PPAR-γ and of its downstream targets, CEBBPα, CD36, and ACOX1, prevented but the increased insulin-stimulated peripheral glucose disposal was also annulled (44). Thus, these data on the one hand indicate that reducing TRIB3 expression does improve insulin action and on the other hand suggest that this effect is largely due to PPAR-γ activation.
In support of the notion that TRIB3 down-regulation exerts a positive effect on insulin resistance-related metabolic disturbances, we find the results of animal studies showing that intervention strategies aimed at improving insulin sensitivity are associated with decreased TRIB3 expression levels. Thus, in ob/ob obese diabetic mice, acute exercise reduced skeletal muscle TRIB3 expression, which was accompanied by increased Akt phosphorylation, GLUT4 translocation to plasma membrane, and glycogen content and, most importantly, enhanced whole-body insulin sensitivity (45). Likewise, in mice with diet-induced obesity and in ob/ob obese diabetic mice as well, acute exercise reduced hepatic TRIB3 expression, which was accompanied by increased phosphorylation of Akt and downstream target nuclear FoxO1; this, in turn, reduced the expression of the key gluconeogenic enzyme phosphoenolpyruvate carboxykinase (PEPCK), increased glycogen content, and decreased fasting glucose levels (46).
Finally, in animals genetically modified so as to increase insulin sensitivity, TRIB3 appears to be down-regulated. Koo et al. (47), for instance, demonstrated that in mice with PPAR-γ coactivator-1 (PGC-1) hepatic deficiency, the observed increase in liver insulin sensitivity was accompanied by reduced TRIB3 expression and, conversely, reversed by TRIB3 overexpression. These results suggest that PGC-1 might inhibit hepatic insulin sensitivity during fasting by inducing TRIB3 expression.
Again, mice lacking liver kinase B1 (LKB1) (a pleiotropic serine/threonine kinase involved in several cellular functions) in skeletal muscle, another model of genetic modification leading to enhanced insulin sensitivity, exhibited lower fasting blood glucose and serum insulin levels and enhanced insulin-stimulated muscle glucose uptake; these changes were paralleled by TRIB3 (and PGC1α and PPARα) down-regulation and by increased Akt phosphorylation (38).
In summary, as shown in Table 1, the data available in animal models do clearly indicate that in vivo TRIB3 overexpression inhibits insulin-stimulated Akt activation and, therefore, induces insulin resistance. Conversely, TRIB3 down-regulation seems to be associated with increased insulin sensitivity. This last phenomenon might equally be due to modulation of Akt activation, although additional pathways, such as those mediated by PPAR-γ activation, may also be operating.
3. Pancreatic β-cell survival
The progressive loss of functional pancreatic insulin-secreting β-cell mass appears to be a key pathogenic factor for T2DM (48). Increased cytokine release and the related low-grade chronic inflammation characterizing metabolic syndrome and prediabetes contribute to β-cell-accelerated apoptosis, decreased survival, and mass loss (49). TRIB3, by binding Akt, might also play a role in this process. In several cell types, Akt exerts prosurvival effects, such as inhibition of conformational change and mitochondrial translocation of BCL2-associated X protein, a proapoptotic member of the BCL-2 family of proteins (50). In pancreatic β-cells, TRIB3 knockdown has been shown to prevent cytokine-induced BCL2-associated X protein conformational change and to inhibit apoptosis (50). Similar apoptosis inhibition is obtained by knocking down MLK3 (the serine-threonine of MAP3K mixed-lineage kinase-3), which directly interacts with and stabilizes TRIB3, resulting in Akt inhibition and increased cytokine-induced apoptosis (50). These data suggest that, in the β-cell, TRIB3 facilitates cytokine-induced apoptosis by inhibiting Akt protective effects. Furthermore, as will be discussed later, it is very likely that TRIB3 also affects β-cell apoptosis through mechanisms controlling ER stress.
4. Adipose and muscle cell differentiation
Because insulin enhances both adipogenesis in preadipocytes and lipid accumulation in mature adipocytes, it is not surprising that TRIB3, an inhibitor of insulin action, acts as a negative regulator of both processes (51, 52). Adipogenesis is a complex process: it involves the interplay of several negative and positive effectors, modulated by a number of hormonal and growth factor signals including insulin, glucocorticoid, and cAMP signaling pathways. Downstream of this network, a programmed series of transcriptional events occur, resulting in the induction of two members of the CCAAT/enhancer-binding protein family, C/EBPβ and C/EBPδ. These, in turn activate the expression of PPARγ and C/EBPα, the two main transcription factors implicated in adipogenesis. TRIB3 is expressed in preadipocytes and is transiently down-regulated during the clonal expansion phase of adipogenesis induced by both glucocorticoids and cAMP-dependent mechanisms (51). TRIB3 overexpression in 3T3-L1 preadipocytes or in embryonic fibroblasts blocks terminal adipogenesis by preventing C/EBPβ-mediated C/EBPα and PPARγ induction. TRIB3 inhibits C/EBPβ activity through two distinct mechanisms: 1) directly, through a physical interaction, thus preventing C/EBPβ from binding DNA and transactivating adipogenic genes; and 2) indirectly, by inhibiting ERK1/2 phosphorylation and activation, which, in turn, activates C/EBPβ by phosphorylating its regulatory phospho-acceptor sites (51). Interestingly, TRIB2 overexpression in 3T3-L1 preadipocytes likewise blocks adipogenesis via several mechanisms, including C/EBPβ inhibition (53).
Consistent with a potential TRIB3 role in slowing down adipogenesis, it has been reported that TRIB3 expression is blunted at an early stage of adipocyte differentiation, similarly to what happens with other antiadipogenic factors (54). By contrast, at a later stage in adipocyte differentiation, TRIB3 expression is increased, acting as a brake on the adipogenic program induced by PPAR-γ and thus preventing excessive progression of the differentiation process (54).
TRIB3 not only is a negative regulator of differentiation but also acts as a negative regulator of lipid accumulation in mature adipocytes (52). Transgenic mice overexpressing TRIB3 in adipose tissue are protected from diet-induced obesity due to an enhanced rate of fatty acid oxidation and increased energy expenditure. Furthermore, TRIB3 induces inactivation of acetyl coenzyme A carboxylase (ACC), a key regulatory enzyme in the fatty acid synthesis pathway during fasting, by recruiting ubiquitin E3 ligase's constitutive photomorphogenic protein 1 (COP1) and thus promoting ubiquitinated ACC proteasomal degradation (52). Taken together, these data show that TRIB3 definitely contributes in modulating several molecular pathways related to adipose tissue differentiation and lipid storage.
Interestingly, studies on animal models have suggested that TRIB1 too is a regulator of several aspects of lipid metabolism, including lipogenesis and lipoprotein production (55).
TRIB3, moreover, plays an important role in skeletal muscle differentiation as well (56). During differentiation of C2C12 mouse myoblast cells, activated Akt increases the ability of myogenic differentiation factor D, the master regulator of muscle differentiation, to recruit transcriptional coactivators, including cAMP response element-binding binding protein, protein 300 and cAMP response element-binding binding protein-associated factor. In C2C12 cells undergoing differentiation, TRIB3 overexpression impairs Akt activation and myogenic differentiation factor D-mediated transcriptional activity, thus leading to a reduction in myotube formation. In contrast, TRIB3 knockdown enhances muscle differentiation. Thus, TRIB3 not only affects insulin action in skeletal muscle (37) but also acts as a modulator of skeletal muscle differentiation. It could, therefore, be considered an important player in the overall regulation of muscle physiology.
5. ER stress
The ER is a critical organelle where secreted and transmembrane proteins are synthesized, folded into their correct three-dimensional structures, modified, and transported to their final cellular destinations. When an excess of (or mutant) proteins reach the ER, the unfolded protein accumulation (i.e. ER stress) activates an adaptive mechanism called unfolded protein response (UPR) aimed at reestablishing homeostasis. When prolonged or excessive ER stress overwhelms the UPR, cell death may eventually occur. To alleviate ER stress, UPR activation initiates downstream signaling events aimed at reducing protein synthesis and increasing protein degradation and folding. UPR thus promotes global translation blockade as well as transcription and translation of selected genes encoding transcription factors, chaperones, and folding enzymes. Reviewing the complex network involved in ER stress and UPR is beyond the scope of this article. How this can impact metabolic diseases has recently been reviewed (57). Here we will briefly illustrate only those mechanisms underlying ER stress and UPR that are specifically related to TRIB3.
TRIB3 expression is induced by several ER stressors including neurotrophic factor deprivation (58), Ca2+ release from the ER in response to the endoplasmic reticular Ca2+-ATPase inhibitor thapsigargin, glutathione depletion (59), cytokines (60), alcohol (61), the ER load of mutant proteins (62), palmitate (63), and both low and high glucose concentrations (37, 64). Activating transcription factor-4 (ATF4), a molecule able to bind to and activate a second transcription factor named C/EBP homologous protein (CHOP), is among the transcription factors whose expression is increased under ER stress. Several studies have indicated that ER stress-dependent TRIB3 induction occurs downstream of the ATF4/CHOP axis. CHOP overexpression in HEK293 cells activated TRIB3 promoter activity via heterodimerization with endogenous proteins, whereas dominant-negative forms of CHOP suppressed the tunicamycin-induced TRIB3 promoter activation (65). Again, ATF4 overexpression in HEK293 cells or human hepatoma HepG2 cells induced TRIB3 promoter transcriptional activation (32). Compared with the expression of either ATF4 or CHOP alone, ATF4 and CHOP coexpression resulted in a significantly greater activation of TRIB3 promoter (65). Moreover, ATF4 knockdown in HEK293 cells repressed the expression of both CHOP and TRIB3, whereas CHOP knockdown repressed only tunicamycin-induced TRIB3 expression (65). In addition, in HEK293 cells, TRIB3 up-regulation increased whereas TRIB3 down-regulation decreased tunicamycin-dependent cell death (65). Thus, it appears that ATF4 and CHOP cooperate in inducing the expression of TRIB3, which, in turn, mediates CHOP-dependent cell death.
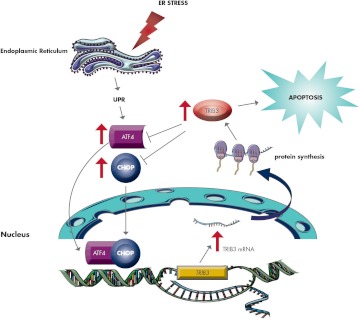
On the other hand, TRIB3 binds to and inhibits transcriptional activity of ATF4 (66) and CHOP (65) while it efficiently suppresses both ATF4- and CHOP-induced TRIB3 promoter activation (32, 66). Based on these results, it is tempting to speculate that TRIB3 is at one and the same time a target of the ATF4/CHOP axis and also a modulator of its own induction by ATF4/CHOP in a sort of negative feedback mechanism (32, 65, 66). In this scenario, TRIB3 may be envisioned as a sensor for ER stress-induced cell death. If the ER stress is mild and transient, TRIB3 is induced, binds to and inhibits ATF4/CHOP, and plays a protective role for cell survival. However, when prolonged and intense ER stress occurs, a marked and prolonged TRIB3 overexpression leads to apoptosis (Fig. 3).
Figure 3.
ER stress and TRIB3. Under ER stress, ATF4 and CHOP cooperate in inducing TRIB3 promoter activity and eventually increasing TRIB3 gene expression and protein content. Increased TRIB3 protein not only activates cell apoptosis but also binds to and inhibits transcriptional activity of ATF4 and CHOP, thus efficiently suppressing their TRIB3 promoter activation, in a sort of negative feedback mechanism. TRIB3 might, therefore, be envisioned as an ER stress-induced cell death sensor. If the ER stress is mild and transient, TRIB3 is induced, binds to and inhibits ATF4/CHOP, and plays a protective role for cell survival. On the contrary, when prolonged and intense ER stress occurs, a marked and prolonged TRIB3 overexpression leads to apoptosis.
TRIB3 might be an important regulator of ER stress-dependent cellular apoptosis in the cardiac tissue as well, which might be relevant for the potential associations between genetic variability at the TRIB3 locus and cardiovascular risk factors, which will be discussed later. In cardiac tissue, ER stress occurs in response to various stress stimuli, including ischemia. Exposing HL-1 murine cardiomyocytes to chemical stressors such as tunicamycin and thapsigargin or culturing cardiomyocytes in medium at low glucose concentrations (0.5 mmol/liter) resulted in increased expression of both TRIB3 and CHOP (67). These changes were associated with impaired insulin-stimulated Akt activation, which was reversed by TRIB3 knockdown. Cardiac samples from mice subjected to experimental myocardial infarction showed increased expression of TRIB3. Histological examination of left ventricular sections obtained after myocardial infarction surgery showed that the infarct area was significantly greater in transgenic mice with cardiac-specific overexpression of TRIB3 than in wild-type littermates. These alterations were accompanied by increased cardiomyocyte apoptosis in the infarct border zone, indicating that, in these cells, TRIB3 induction is involved in the ER stress response to stimuli such as ischemia (67). In addition, transgenic mice with cardiac-specific TRIB3 overexpression exhibited impaired insulin-induced activation of Akt and of its downstream target glycogen synthase kinase-3β, and this was associated with a reduced glucose oxidation rate (67). All in all, it appears from these data that increasing TRIB3 levels has a profound negative effect on glucose metabolism in the heart muscle and adversely affects cardiomyocyte survival under stress conditions.
ER stress is also an important factor in the β-cell apoptosis and mass loss characterizing T2DM (68). TRIB3 is probably involved in modulating the ER stress effects in pancreatic β-cells because ATF4 and CHOP overexpression, as well as increased ATF4/CHOP binding to TRIB3 promoter, TRIB3 overexpression and impaired Akt activation, have all been observed in pancreatic islets from postpregnant rats undergoing a transient increase in pancreatic islet apoptosis during early lactation (69). In the same study, the chemical chaperone 4-phenyl butyric acid, an inhibitor of ER stress, was shown to reverse the increased CHOP and TRIB3 expression, thus leading to increased Akt phosphorylation and decreased pancreatic islet apoptosis (69). At least in this model, therefore, UPR, through CHOP/ATF4-induced TRIB3 expression and subsequent impairment of prosurvival Akt activity, triggers a physiological and transitory burst of apoptosis in pancreatic islets (69). Further support for the notion that TRIB3 is involved in death/survival signaling is provided by the increased TRIB3 expression observed in islets from hyperglycemic Goto-Kakizaki rats and in INS-1 pancreatic β-cells exposed to either high glucose concentrations or the ER stressor thapsigargin (70). Furthermore, in INS-1 pancreatic β-cells, TRIB3 overexpression increased high-glucose-induced apoptosis, whereas TRIB3 knockdown resulted in a marked reduction in apoptosis induced by high glucose or thapsigargin (70).
Finally, in a very elegant study on rat MIN6 pancreatic β-cells, Liew and co-workers (71) were able to demonstrate that TRIB3 down-regulates Akt activation and, by binding to ATF4, the expression of synaptosomal-associated protein 25 (SNAP25) and the Ras-like GTP-binding protein 3D (RAB3D) with both proteins participating in insulin exocytosis. Of note, increased TRIB3 expression, paralleled by reduced expression of SNAP25 and RAB3D, was observed in human islets from patients with T2DM as compared to islets from nondiabetic controls (71). It is conceivable that when ER stress occurs in the course of diabetes, UPR-mediated ATF4 and TRIB3 up-regulation switches on the above-described cascade of events, eventually leading to decreased insulin secretion (71).
Taken altogether, these findings support the notion that ER stress-induced TRIB3 up-regulation could be a common mechanism underlying 1) insulin resistance in peripheral target tissues (37), 2) increased apoptosis of pancreatic β-cells (69–71), and 3) β-cell dysfunction with impaired insulin secretion in response to hyperglycemia (70). Importantly, these abnormalities constitute the main pathophysiological events leading to T2DM in humans.
C. TRIB3 expression
TRIB3 is very likely ubiquitous, having been described in a range of tissues and cells (40). TRIB3 expression is modulated by insulin in a cell-type-specific manner. Although insulin has no effect on TRIB3 expression in TC3 pancreatic β-cells, L6 myotubes, or 3T3-L1 fibroblasts, the hormone does promote TRIB3 expression in FAO hepatoma cells and 3T3-L1 adipocytes (72). In insulin-stimulated expression of TRIB3 is mediated by PI3K and protein kinase Cζ (72). Additional studies have shown that insulin promotes TRIB3 expression through C/EBPβ induction in both hepatoma cells and 3T3-L1 adipocytes (73). Under both basal and insulin-stimulated conditions, C/EBPβ ectopic expression increased TRIB3 expression by enhancing transcriptional activity of the TRIB3 gene, whereas silencing C/EBPβ strikingly reduced insulin-induced TRIB3 expression (73).
Although insulin appears to enhance TRIB3 expression, Akt, a central mediator of insulin signaling, seems to exert a negative effect on it. Thus, chemical inhibition or genetic inactivation of Akt enhanced TRIB3 expression in liver cells, suggesting that Akt may negatively regulate insulin-induced TRIB3 expression (72). This negative Akt effect on TRIB3 expression is further supported by the finding that induction of TRIB3 expression by insulin is markedly impaired by transfection of a constitutively active Akt (72). These findings may explain why TRIB3 expression is elevated both in the fasting state and in conditions of insulin resistance, two situations characterized by low Akt activation (20, 36). The available data on the impact of insulin signaling upon TRIB3 expression are thus difficult to reconcile and call for additional investigations to unravel the underlying biology.
As mentioned above, other molecules such as PGC-1 and LKB1 can also positively modulate TRIB3 expression, whereas adenoviral delivery of constitutively active FoxO1 to mouse liver reduced liver TRIB3 expression (74), thus pointing to FoxO1 as a negative modulator of TRIB3 expression.
III. TRIB3 and Human Metabolic and Cardiovascular Disturbances
A. TRIB3 expression, insulin resistance, and T2DM
As previously discussed, a wealth of cellular and animal studies have clearly established a significant role for TRIB3 in the regulation of insulin action and insulin secretion and in general cell metabolism. It is thus entirely conceivable that TRIB3 expression and or action might be altered in individuals suffering from insulin resistance and/or T2DM. A number of studies have addressed the possible role of TRIB3 expression levels on insulin resistance-related abnormalities and T2DM in humans. In a study comprising 10 insulin-sensitive, 10 insulin-resistant, and 10 untreated patients with T2DM, TRIB3 abundance was determined in skeletal muscle biopsies obtained from the vastus lateralis (37). Muscle TRIB3 protein levels were significantly increased by approximately 2-fold in individuals with T2DM compared with insulin-sensitive subjects; in nondiabetic insulin-resistant subjects, TRIB3 expression levels were in between those observed in the diabetics and in the insulin-sensitive controls. In the whole study sample, TRIB3 protein levels were inversely related to whole-body insulin sensitivity, as assessed by euglycemic-hyperinsulinemic glucose clamp, and positively correlated with fasting plasma glucose, thus reinforcing the hypothesis that TRIB3 plays a role in both insulin resistance and T2DM.
In a study on 93 obese individuals undergoing bariatric surgery, TRIB3 mRNA levels were measured in biopsies obtained from liver as well as visceral and sc adipose tissue (75). Obese subjects were then stratified into two groups, on the basis of the homeostasis model assessment of insulin resistance index (HOMA-IR). As compared with insulin-sensitive subjects, insulin-resistant individuals had increased hepatic TRIB3 mRNA levels, whereas Akt phosphorylation was reduced. Furthermore, in the whole sample, hepatic TRIB3 mRNA levels strongly correlated with plasma insulin and triglyceride levels as well as with the degree of hepatic steatosis. Finally, hepatic TRIB3 expression exhibited a significant correlation with mRNA levels of PEPCK, a key gluconeogenic enzyme. Notably, increased TRIB3 mRNA levels were observed in the visceral adipose tissue of insulin-resistant individuals as well. A similar trend, albeit not significant, was also detected in sc adipose tissue specimens.
Finally, TRIB3 is also expressed in human pancreatic islets, where it is specifically localized in insulin-secreting β-cells (71). Islets from patients with T2DM showed an approximately 4-fold increase in TRIB3 mRNA expression and an approximately 3-fold increase in TRIB3 protein levels compared with pancreatic islets from nondiabetic controls. This was paralleled by reduced expression of SNAP25 and RAB3D, two mediators of insulin exocytosis (71), suggesting that increased TRIB3 expression might be implicated in impaired insulin secretory capacity.
Thus, the data presently available, although limited, all support the hypothesis that TRIB3 overexpression might have a detrimental role in individuals suffering from insulin resistance and T2DM, contributing to the pathophysiology of these abnormal metabolic states (Fig. 4). Interestingly, TRIB3 overexpression in such conditions seems to occur in all the main tissues playing a central role in the modulation of insulin sensitivity and glucose homeostasis, such as skeletal muscle, liver, adipose tissue, and pancreatic β-cells. It remains to be established whether changes in TRIB3 expression should be interpreted as an early and causal defect or, on the contrary, as a late epiphenomenon secondary to metabolic abnormalities. In either case, what needs to be addressed is whether TRIB3 expression and action might be considered a target for both pharmacological and nonpharmacological treatment strategies aimed at improving insulin resistance and glucose homeostasis.
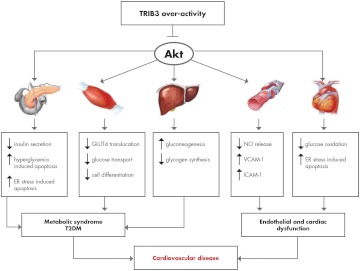
Figure 4.
Consequences of TRIB3 overactivity. TRIB3 exerts a host of metabolic effects in several tissues. Most of these effects are mediated by Akt signaling inhibition and are detectable in all the main tissues involved in the regulation of whole-body glucose homeostasis (liver, pancreatic β-cells, and skeletal muscle) as well as in endothelial cells. TRIB3 overactivity may be the result of overexpression or may be secondary to the presence of the gain-of-function R84 variant. The overall outcome of TRIB3 overactivity is the deterioration of glucose homeostasis as a consequence of increased gluconeogenesis and decreased glycogen synthesis in the liver, defective pancreatic insulin-secreting β-cell function and survival under stress conditions, and decreased glucose transport in skeletal muscle cells. Akt inhibition also mediates deleterious TRIB3 effects on glucose oxidation and stress-induced apoptosis in the cardiomyocyte as well as reduced nitric oxide (NO) release and increased vascular cell adhesion molecule-1 (VCAM-1) and intercellular adhesion molecule-1 (ICAM-1) production from endothelial cells. Thus, TRIB3 overactivity and subsequent Akt inhibition might be linked to increased risk of cardiovascular disease by two mechanisms. One is indirect and related to the deleterious systemic milieu caused by the insulin resistance-related metabolic syndrome and/or T2DM. The other one is direct and related to endothelial and/or cardiac dysfunction. The two mechanisms are not mutually exclusive; actually, it is entirely possible that in some individuals, they are simultaneously at work.
B. Nonsynonymous TRIB3 Q84R polymorphism
As reviewed above, changes in TRIB3 abundance are associated with modifications in insulin signaling and cellular and whole-body metabolism in rodents as well as with metabolic human diseases such as insulin resistance and T2DM. We will here review data from several human studies showing that changes in TRIB3 function might also be associated with abnormal insulin signaling, insulin resistance, and related abnormalities.
A TRIB3 missense SNP (i.e. Q84R, where arginine replaces glutamine at position 84; rs2295490) has recently been described (76). This SNP has a global minor allele frequency of 14.4% (as estimated by the 1000 Genome phase 1 genotype data from 1094 worldwide individuals, released in the May 2011 dataset), varying from 13% in both European and African individuals (from HapMap CEU and YRI, respectively) to 25–27% in Japanese and Chinese individuals (from HapMap JPT and HCB, respectively).
A certain amount of evidence points to the fact that this amino acid change acts as a gain-of-function substitution in several insulin target cells (71, 76, 77). Thus, according to in silico bioinformatic analysis, the glutamine (Q) to arginine (R) amino acid change at position 84 alters intramolecular salt bridge formation which eventually makes TRIB3 R84 a more avid Akt binder and inhibitor than the Q84 variant (77). In the following sections, we will review the data available relating to 1) the effects of this polymorphism on insulin action in various different cell types; 2) the association of this polymorphism with insulin resistance, impaired insulin secretion, and abnormal glucose homeostasis; and c) the association of this polymorphism with increased cardiovascular risk.
1. In vitro data on insulin signaling in different cell types
The effects of TRIB3 R84 variant on insulin signaling and insulin action have been explored in several cell types. Transfection of TRIB3 R84 variant into HepG2 human liver cells resulted in a greater reduction of insulin-induced Ser473-Akt phosphorylation than was observed after transfecting the Q84 variant (i.e. 45 vs. 22% reduction, respectively). These data provided the first biological evidence that the TRIB3 R84 variant is a stronger inhibitor of insulin signaling than the more common Q84 variant and that it might thus play a role in human insulin resistance (76).
Similar data have also been reported in human vein endothelial cells (HUVEC) naturally carrying the TRIB3 Q84 or R84 variant obtained from umbilical cords of healthy women (77). Thus, insulin-induced Akt phosphorylation on both Ser473 and Thr308 was sharply reduced in HUVEC naturally carrying the TRIB3 Q84R or R84R genotype compared with those carrying the Q84Q genotype. In addition, as a likely consequence of reduced Akt activation, cells carrying the R84 variant also exhibited impaired insulin-induced endothelial nitric oxide synthase (eNOS) Ser1177 phosphorylation and Thr495 dephosphorylation (77). In keeping with these findings, the R84 cells also showed a blunted response to insulin in terms of eNOS activation and nitric oxide (NO) release, whose deficiency is an established early step in the development of atherosclerosis (17). It is worth noting that other naturally occurring genetic polymorphisms that affect insulin signaling at different levels, namely IRS-1 G972R (78) and ENPP1 K121Q (79), have likewise been reported to affect eNOS activation and NO release, thus reinforcing the hypothesis that reduced insulin signaling has the potential to directly affect human endothelial cell function. Quite similar results have recently been obtained in animal models of genetically impaired endothelial insulin signaling (80).
Furthermore, compared with Q84Q cells, HUVEC carrying the R84 variant showed constitutive MAPK kinase-MAPK activation and increased cell-surface expression of both vascular cell adhesion molecule-1 and intercellular adhesion molecule-1 (81). These proatherogenic changes are a likely consequence of R84 variant-induced selective impairment of the PI3K/Akt axis, which tips the balance of insulin signaling toward the MAPK-dependent pathway, thus increasing the potential of this variant to be a promoter of atherosclerosis.
Finally, transfection of TRIB3 R84 full-length cDNA into dispersed human islet cells, as well as into rat MIN6 cultured cells, resulted in a stronger inhibitory effect on Akt activation than was found with the Q84 variant (71). Given the central role of Akt in modulating insulin secretion (18), it is not surprising that, under the same experimental conditions, cells transfected with the R84 variant also showed impaired insulin secretion (71). Along much the same line, defective glucose-induced insulin secretion has been observed in isolated human pancreatic islets from TRIB3 R84-carrying donors but not in islets from QQ donors (29). Taken altogether, these data strongly support the concept that TRIB3 R84 induces β-cell insulin resistance and, eventually, abnormal insulin secretion. It is worth noting that IRS-1 G972R (82–84) and ENPP1 K121Q (85) genetic polymorphisms, both affecting insulin signaling, have likewise been reported to play a detrimental role for insulin secretion in cultured β-cells and human isolated pancreatic islets, thus corroborating the hypothesis that reduced insulin signaling has the ability to directly affect insulin secretion. This fascinating new scenario was first proposed by some very elegant studies carried out in animal models of genetically reduced β-cell insulin signaling (86).
In three different human tissues, therefore, namely the liver, the endothelium, and pancreatic islets, in vitro studies have very consistently reported that TRIB3 R84 acts as a gain-of-function variant that affects insulin signaling and consequently several insulin actions that are specific for different cell types.
The exacerbated inhibitory activity exerted on insulin signaling by the R84 variant may well be the molecular basis for the observed epidemiological association between this variant and several human metabolic abnormalities, including insulin resistance, defective insulin secretion, T2DM, and cardiovascular disease, as will be discussed in the following sections.
2. In vivo data on insulin sensitivity, insulin secretion, and glucose homeostasis
a. Insulin sensitivity.
The TRIB3 R84 variant is a stronger inhibitor of insulin signaling in human cells than is the more prevalent Q84. Thus, one would expect it also to be associated with human insulin resistance. Several studies have addressed this issue, and from analyzing their results, it appears that indeed the presence of TRIB3 R84 is associated with impaired insulin sensitivity in humans. The main evidence for this stems from a recent study comprising 394 nondiabetic offspring of T2DM patients from Italy (87). In this cohort, insulin-mediated glucose disposal, as measured by the euglycemic-hyperinsulinemic glucose clamp technique, was progressively reduced from QQ to QR and RR individuals. In the same study, it was also reported that a similar trend of decreasing insulin sensitivity from QQ to RR individuals was obtained in 791 unrelated, mostly obese, individuals, in whom insulin sensitivity was determined by a validated oral glucose tolerance test (OGTT)-derived index (87). In addition, among Italian T2DM subjects, the R84 variant was associated with an approximately 3-fold increased risk of presenting several insulin resistance-related abnormalities, clustering within the metabolic syndrome (76). Furthermore, among 513 Chinese individuals, carriers of the TRIB3 R84 variant exhibited an approximately 2-fold higher risk of presenting insulin resistance as detected by HOMA-IR and/or metabolic syndrome (88). A similar association of TRIB3 R84 variant with HOMA-IR was also reported in an independent Chinese sample of 177 patients with T2DM but not in 245 nondiabetic individuals (89). However, in this Chinese cohort, both in the diabetic and in the nondiabetic subgroup,TRIB3 R84 carriers presented higher fasting insulin levels, a proxy of insulin resistance (89).
Taken altogether, these studies do support the hypothesis that the TRIB3 R84 variant might foster insulin resistance in humans. Because, as previously discussed, increased TRIB3 expression is also associated with insulin resistance, two different and complementary mechanisms by which TRIB3 affects insulin sensitivity, namely overexpression and expression of the gain-of-function R84 variant in insulin target tissues, can be envisioned (Fig. 4). It is entirely possible, although so far not demonstrated, that in some individuals both mechanisms can be operating concomitantly, thus further increasing the risk of insulin resistance and related metabolic disorders.
b. Insulin secretion.
Several data suggest that the TRIB3 R84 variant is also associated with impaired insulin secretion. In 645 nondiabetic individuals from Italy, early insulin secretion adjusted for the level of insulin resistance [i.e. the disposition index (DI)] was assessed from OGTT-derived data. The DI was significantly reduced in individuals carrying the R84 variant compared with individuals homozygous for the Q84 variant (29). Similar data were obtained by a second study carried out on an independent sample of 791 individuals from Italy (87). The meta-analysis of these two studies (comprising a total of 1436 individuals) confirmed that compared with that observed in individuals not carrying the R84 variant (i.e. QQ individuals), the DI was reduced by about 25% in QR and 50% in RR subjects (87).
Finally, Liew and co-workers (71), in a sample of 766 patients with T2DM from Poland, reported that individuals homozygous for the R84 variant exhibited a 30% lower plasma C peptide level than Q84Q subjects.
Therefore, the available in vivo data (29, 71, 87), although far from definitive, are consistent with the previously discussed in vitro data showing reduced insulin release in human islets isolated from carriers of the R84 variant (29). Similar data have been obtained with other genetic polymorphisms affecting insulin signaling, namely IRS1 G972R and ENPP1 K121Q with carriers of IRS1 R972 or ENPP1 Q121 showing reduced in vivo insulin secretion (90, 91). Altogether, these genetic association studies reinforce the idea that reduced insulin signaling impairs insulin secretion.
c. Glucose homeostasis.
Because, as discussed, the TRIB3 R84 variant appears to negatively affect both insulin sensitivity and insulin secretion, it is very tempting to conjecture that this variant might also play a role in determining individual susceptibility to abnormalities of glucose homeostasis and T2DM. Taking advantage of the GENIUS (Genetics of Type 2 Diabetes in Italy and United States) consortium, which combines the efforts of a number of institutions from the two countries, Prudente et al. (87) carried out a case-control study for impaired glucose regulation (IGR: i.e. either T2DM or impaired fasting glucose and/or impaired glucose tolerance) comprising a total of 6634 individuals of European ancestry. According to the additive model of inheritance, the TRIB3 R84 variant has proved to be significantly associated with IGR, with an overall odds ratio (OR) for the additive model of inheritance (i.e. risk increase for each copy of the R84 variant) of 1.19 (95% confidence interval = 1.06–1.34). Most of the observed association was sustained by association with early IGR (i.e. diagnosed before age 45 yr), the additive OR being equal to 1.31 and reaching a robust level of statistical significance (P value = 0.00007) (87). This suggests that more than affecting the overall risk of abnormal glucose homeostasis, the TRIB3 R84 variant anticipates its appearance in predisposed individuals.
To further characterize the association between the TRIB3 R84 variant and impaired glucose homeostasis, OGTT-derived indexes accounting for insulin secretion and sensitivity were analyzed in 791 individuals (87) by the statistical methods known as the triangulation approach (92). The results of this study strongly suggest that reduced insulin secretion, rather than increased insulin resistance, is the main culprit for the elevated risk of IGR in individuals carrying the R84 variant (87). This finding is in keeping with the belief that, compared with insulin resistance, defective insulin secretion is a more important predictor of impaired glucose homeostasis (93, 94).
Consistent with the hypothesis that the TRIB3 R84 variant affects glucose homeostasis is the observation of a significant association between the variant and the presence of T2DM (additive OR of 1.4) in a sample of 1468 individual of mixed European descent from Boston (United States) and Krakow (Poland) (71). In contrast to what was observed among Europeans, no association between the variant and the presence of T2DM was observed in a small (n = 422) sample of Chinese individuals (89). This apparent discrepancy might be due to chance or, more likely, to the limited statistical power of this last study (i.e. false-negative result). Still, it cannot be excluded that differences in ethnicity, including genetic background and/or environmental factors such as dietary habits, may underlie the differences between results obtained in European (71, 87) and Chinese (89) populations. Finally, notwithstanding the robust statistical significance of the results, replicated in two different population samples (71, 87), it cannot definitely be ruled out that the association between the TRIB3 R84 variant and conditions of abnormal glucose homeostasis observed among Europeans is a false positive result. So far, the overall results obtained by association studies on candidate genes with T2DM have largely been frustrating, because of poor reproducibility. Thus, one needs to be very cautious in interpreting such data and before accepting them as fully established. We therefore believe that, before variability of the TRIB3 gene can be conclusively considered as playing a role in glucose metabolism modulation, these findings need to be replicated in additional large studies, amounting, if possible, to overall genome-wide significance.
In this specific context, it is unfortunate that the TRIB3 Q84R polymorphism (rs2295490) was not among the SNP examined by the recently published large genome-wide association studies (GWAS) on T2DM (95) and that no other SNP, among those tested, can function as a good proxy for it, because this means that data from these studies cannot be used to replicate the associations reported (71, 87).
3. In vivo data on cardiovascular disease
As previously discussed, several studies reported that the functional TRIB3 R84 variant is associated in vivo with insulin resistance and related abnormalities clustering within the metabolic syndrome (76, 87–89). In addition, studies on HUVEC have clearly described how TRIB3 R84 induces endothelial insulin resistance, resulting in reduced insulin-stimulated NO production (77) and a shift of cell membrane phenotype toward a proatherogenic profile (81). Given that both systemic (4) and endothelial (6, 17, 80) insulin resistance is supposed to be pathogenic for atherosclerosis, one might easily hypothesize that the TRIB3 R84 variant is associated with cardiovascular disease. Available data, albeit few, do suggest that this might indeed be the case. Prudente et al. (76) showed that among Italian patients with T2DM who survived myocardial infarction, age at myocardial ischemia was progressively younger in homozygous (RR individuals) and heterozygous (QR individuals) carriers of the R84 variant than among QQ carriers (i.e. 47 ± 11 vs. 55 ± 10 and 58 ± 9 yr). This association was independent of several possible confounders independently associated with age at myocardial infarction in that cohort.
Insulin resistance (96) and related abnormalities (97) are again considered pathogenic in the case of impaired glomerular filtration rate. In a cohort of 1012 patients with T2DM from Italy, the homozygous carriers of the TRIB3 R84 variant (i.e. RR individuals), compared with those carrying the QQ or the QR genotype considered together exhibited a significant 4.7-fold increased risk of chronic kidney disease (CKD), defined as glomerular filtration rate below 60 ml/min·1.73 m2 (98). In a different study carried out in 583 diabetic patients from the Boston area, a tendency toward greater risk of CKD for individuals carrying the R84 variant (i.e. QR or RR, according to a dominant genetic model) was also detected, but it did not attain statistical significance (OR = 1.44; 95% confidence interval = 0.97–2.15; P = 0.07) (98). The slight discrepancy between these two studies clearly calls for further investigation, possibly on a larger scale. Meanwhile, the notion that the TRIB3 R84 variant might be associated with CKD in T2DM patients deserves, with all due caution, to be earnestly considered. This is partly because the data on both myocardial infarction and reduced kidney function are somehow consistent, allowing one to speculate that the TRIB3 R84 variant predisposes to atherosclerosis.
Because these data were gathered from patients with T2DM, they might not apply to a nondiabetic population. However, two additional studies, carried out in nondiabetic individuals, have explored the association between TRIB3 Q84R polymorphism and another proxy of atherosclerosis, such as the value of the carotid intima-media thickness (IMT). In 513 unrelated Chinese individuals, Gong et al. (88) found the R84 variant to be associated not only with insulin resistance and metabolic syndrome but also with carotid IMT, which increased from 0.75 to 0.76 and 0.95 mm in QQ, QR, and RR individuals, respectively. In a multivariate model, the TRIB3 R84 variant, together with age, waist-to-hip ratio, and systolic blood pressure, remained a significant independent risk factor for increased IMT (88). Data from an Italian cohort (81) are very much like those reported by Gong et al. (88) in Chinese individuals. Screening for the presence of the TRIB3 R84 variant, Formoso et al. (81) took 430 nondiabetic individuals of European ancestry whose carotid IMT was measured by high-resolution B-mode ultrasound. IMT progressively and significantly increased from 0.74 to 0.79 and 0.85 mm in Q84Q, Q84R, and R84R individuals, respectively. When a meta-analysis was performed, pooling data from the two aforementioned studies (a total of 943 individuals), a progressive increase in IMT going from Q84Q to Q84R and R84R individuals was confirmed, the IMT increase for one copy of the R84 variant (i.e. β-estimate) being 0.051 mm at a quite robust level of statistical significance (P = 0.000029) (81).
On the basis of the data discussed, it does appear that the TRIB3 R84 variant might contribute to the risk of developing atherosclerosis. Whether this is a consequence of the variant's deleterious effect on systemic insulin resistance and related traditional metabolic and hemodynamic cardiovascular risk factors (76, 87–89) or, on the contrary, due to a direct effect by the R84 variant on the endothelium, where it induces cellular insulin resistance (77) and proatherogenic changes (81), is not known. It is also possible, and probably most likely, that both mechanisms, which are not mutually exclusive, are actually operating in TRIB3 R84 carriers, causing increasing susceptibility to atherosclerosis and related clinical outcomes. In a broader perspective, the bulk of the available data point to TRIB3 overactivity (either because of overexpression or because of the R84 gain-of-function variant) as a possible shaper of individual susceptibility to cardiovascular disease.
Genetic variations of TRIB1 have been also associated with cardiovascular risk factors and events. Two GWAS have established that rs17321515 SNP at 8q24 near TRIB1 is associated with triglyceride concentration at a genome-wide level of statistical significance (99, 100).
Interestingly, in a large-scale gene-centric analysis, this same SNP was also associated with coronary artery disease (101). In addition, a GWAS involving nearly 100,000 individuals showed that SNP rs2954029 at the TRIB1 locus was associated with triglyceride, low-density lipoprotein cholesterol, and high-density lipoprotein cholesterol serum levels (102).
IV. Conclusions
As reviewed in this article, the last decade has witnessed mounting interest in TRIB3, a molecule that might indeed play a sizable role in several cellular functions. The studies prompted by this interest allow some conclusive remarks while at the same time leaving open some unanswered questions; both are briefly summarized below.
TRIB3 inhibits insulin signaling and action in various target cells and tissues. This effect is mainly due to the inhibition of insulin-induced Akt activation, although additional or alternative mechanisms cannot be excluded.
Many rodent models of insulin resistance indicate that TRIB3 expression levels are inversely related to whole-body insulin sensitivity, thus pointing to TRIB3 overexpression as a potential mechanism of insulin resistance.
Along the same line, the few data so far available in humans indicate that TRIB3 is overexpressed in patients with insulin resistance and/or T2DM; remarkably, in these patients, TRIB3 overexpression has been described in all analyzed tissues, including skeletal muscle, liver, adipose tissue, and pancreatic, insulin-secreting β-cells. Thus, this defect seems to be ubiquitous and to have the potential for playing a central role in modulating insulin sensitivity and glucose homeostasis in humans.
Data on nonsynonymous TRIB3 Q84R polymorphism lend further support to the notion that TRIB3 does play a role in modulating human insulin sensitivity and related clinical conditions. This gain-of-function amino acid substitution affects insulin signaling at the level of Akt activation and has been associated with abnormal glucose homeostasis and cardiovascular disease. Such epidemiological associations deserve to be confirmed in additional and broader studies before being accepted as fully established. In the meantime, in vitro and in vivo studies on the biological relevance of the TRIB3 R84 variant have provided a better understanding of insulin signaling and its part in modulating the physiology of several typical and nontypical insulin target tissues. These studies have also provided some clues as to novel molecular mechanisms potentially involved in the pathogenesis of highly prevalent diseases such as cardiovascular disease and T2DM.
Besides unraveling new pathogenic mechanisms underlying metabolic and cardiovascular abnormalities, studies on TRIB3 might turn out eventually to be relevant in the clinical setting, for both diagnosis and treatment. In the near future, evaluating TRIB3 expression level in target tissues as well as genotyping TRIB3 Q84R polymorphism may prove useful, in combination with other genetic and nongenetic markers, as a way of identifying individuals at high risk of metabolic disorders and atherosclerosis. In addition, data so far available clearly point to the modulation of TRIB3 activity (whether by inducing expression changes or by affecting its function) as an appealing target for treatments designed to reduce the burden of insulin resistance and related metabolic alterations. Additional studies addressing these issues would be both opportune and welcome.
All in all, the studies on TRIB3 reviewed in this article are part of a much broader scenario surrounding the role of several other players in abnormal insulin signaling. It is entirely possible that such studies might ultimately help pave the way for improving care delivery to a large number of people suffering from highly prevalent and costly illnesses such as T2DM and cardiovascular disease.
Acknowledgments
The funding for this manuscript is by the European Foundation for the Study of Diabetes (Sanofi-Aventis Partner Program 2010 to A.C.).
This work was partly supported by the Italian Ministry of Health (Ricerca Corrente 2011 and 2012 to S.P. and V.T.); by the European Foundation for the Study of Diabetes (Sanofi-Aventis Partner Program 2010 to A.C.); by the Italian Ministry for the University and Scientific Research (PRIN 2008 to A.C.); by the Italian Ministry for Agricultural, Nutritional Policies, and Forestry (Special Research Grant MiPAAF-CARONUT 2008 to A.P.); and by the European Union (FP6 EUGENE2 no. LSHM-CT-2004-512013 to G.S.).
Disclosure Summary: S.P., G.S, A.P., F.A., and V.T. have nothing to disclose.
Footnotes
- ACC
- Acetyl coenzyme A carboxylase
- APPL1
- adaptor protein, phosphotyrosine interaction, PH domain, and leucine zipper-containing 1
- ASO
- antisense oligonucleotide
- ATF4
- activating transcription factor-4
- CEBPα
- CCAAT/enhancer-binding protein α
- CHOP
- C/EBP homologous protein
- CKD
- chronic kidney disease
- COP1
- constitutive photomorphogenic protein 1
- DI
- disposition index
- eNOS
- endothelial nitric oxide synthase
- ENPP1
- ectonucleotide pyrophosphatase phosphodiesterase 1
- ER
- endoplasmic reticulum
- FoxO1
- forkhead box class O factor 1
- GLUT4
- glucose transporter 4
- GWAS
- genome-wide association studies
- HOMA-IR
- homeostasis model assessment of insulin resistance index
- HUVEC
- human vein endothelial cells
- IGR
- impaired glucose regulation
- IMT
- intima-media thickness
- IRS
- insulin receptor substrate
- JNK
- Jun N-terminal kinase
- LKB1
- liver kinase B1
- NO
- nitric oxide
- OGTT
- oral glucose tolerance test
- OR
- odds ratio
- PEPCK
- phosphoenolpyruvate carboxykinase
- PGC-1
- PPAR-γ coactivator-1
- PH
- pleckstrin homology
- PI3K
- phosphatidyl-inositol-3-kinase
- PIP3
- phosphatidylinositol 3,4,5-triphosphate
- PPAR-γ
- peroxisome proliferator-activated receptor-γ
- RAB3D
- Ras-like GTP-binding protein 3D
- SNAP25
- synaptosomal-associated protein 25
- SNP
- single-nucleotide polymorphism
- T2DM
- type 2 diabetes mellitus
- TRIB3
- tribbles homolog 3
- UPR
- unfolded protein response.
References
- 1. Yeni-Komshian H, Carantoni M, Abbasi F, Reaven GM. 2000. Relationship between several surrogate estimates of insulin resistance and quantification of insulin-mediated glucose disposal in 490 healthy nondiabetic volunteers. Diabetes Care 23:171–175 [DOI] [PubMed] [Google Scholar]
- 2. Warram JH, Martin BC, Krolewski AS, Soeldner JS, Kahn CR. 1990. Slow glucose removal rate and hyperinsulinemia precede the development of type II diabetes in the offspring of diabetic parents. Ann Intern Med 113:909–915 [DOI] [PubMed] [Google Scholar]
- 3. Lillioja S, Mott DM, Spraul M, Ferraro R, Foley JE, Ravussin E, Knowler WC, Bennett PH, Bogardus C. 1993. Insulin resistance and insulin secretory dysfunction as precursors of non-insulin-dependent diabetes mellitus. Prospective studies of Pima Indians. N Engl J Med 329:1988–1992 [DOI] [PubMed] [Google Scholar]
- 4. Reaven GM. 1988. Banting lecture 1988. Role of insulin resistance in human disease. Diabetes 37:1595–1607 [DOI] [PubMed] [Google Scholar]
- 5. Ferrannini E, Buzzigoli G, Bonadonna R, Giorico MA, Oleggini M, Graziadei L, Pedrinelli R, Brandi L, Bevilacqua S. 1987. Insulin resistance in essential hypertension. N Engl J Med 317:350–357 [DOI] [PubMed] [Google Scholar]
- 6. Muniyappa R, Montagnani M, Koh KK, Quon MJ. 2007. Cardiovascular actions of insulin. Endocr Rev 28:463–491 [DOI] [PubMed] [Google Scholar]
- 7. Hedblad B, Nilsson P, Engström G, Berglund G, Janzon L. 2002. Insulin resistance in non-diabetic subjects is associated with increased incidence of myocardial infarction and death. Diabet Med 19:470–475 [DOI] [PubMed] [Google Scholar]
- 8. Wilson PW, D'Agostino RB, Parise H, Sullivan L, Meigs JB. 2005. Metabolic syndrome as a precursor of cardiovascular disease and type 2 diabetes mellitus. Circulation 112:3066–3072 [DOI] [PubMed] [Google Scholar]
- 9. Wannamethee SG, Shaper AG, Lennon L, Morris RW. 2005. Metabolic syndrome vs Framingham Risk Score for prediction of coronary heart disease, stroke, and type 2 diabetes mellitus. Arch Intern Med 165:2644–2650 [DOI] [PubMed] [Google Scholar]
- 10. Goodarzi MO, Dumesic DA, Chazenbalk G, Azziz R. 2011. Polycystic ovary syndrome: etiology, pathogenesis and diagnosis. Nat Rev Endocrinol 7:219–231 [DOI] [PubMed] [Google Scholar]
- 11. Stefan N, Staiger H, Häring HU. 2011. Dissociation between fatty liver and insulin resistance: the role of adipose triacylglycerol lipase. Diabetologia 54:7–9 [DOI] [PubMed] [Google Scholar]
- 12. Ristow M. 2004. Neurodegenerative disorders associated with diabetes mellitus. J Mol Med (Berl) 82:510–529 [DOI] [PubMed] [Google Scholar]
- 13. Renehan A, Smith U, Kirkman MS. 2010. Linking diabetes and cancer: a consensus on complexity. Lancet 375:2201–2202 [DOI] [PubMed] [Google Scholar]
- 14. Sesti G, Federici M, Hribal ML, Lauro D, Sbraccia P, Lauro R. 2001. Defects of the insulin receptor substrate (IRS) system in human metabolic disorders. FASEB J 15:2099–2111 [DOI] [PubMed] [Google Scholar]
- 15. Cohen P. 2006. The twentieth century struggle to decipher insulin signalling. Nat Rev Mol Cell Biol 7:867–873 [DOI] [PubMed] [Google Scholar]
- 16. Taniguchi CM, Emanuelli B, Kahn CR. 2006. Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol 7:85–96 [DOI] [PubMed] [Google Scholar]
- 17. Yu Q, Gao F, Ma XL. 2011. Insulin says NO to cardiovascular disease. Cardiovasc Res 89:516–524 [DOI] [PubMed] [Google Scholar]
- 18. Elghazi L, Bernal-Mizrachi E. 2009. Akt and PTEN: β-cell mass and pancreas plasticity. Trends Endocrinol Metab 20:243–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goldfine ID, Maddux BA, Youngren JF, Reaven G, Accili D, Trischitta V, Vigneri R, Frittitta L. 2008. The role of membrane glycoprotein plasma cell antigen 1/ectonucleotide pyrophosphatase phosphodiesterase 1 in the pathogenesis of insulin resistance and related abnormalities. Endocr Rev 29:62–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Du K, Herzig S, Kulkarni RN, Montminy M. 2003. TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science 300:1574–1577 [DOI] [PubMed] [Google Scholar]
- 21. Goldstein BJ, Bittner-Kowalczyk A, White MF, Harbeck M. 2000. Tyrosine dephosphorylation and deactivation of insulin receptor substrate-1 by protein-tyrosine phosphatase 1B. Possible facilitation by the formation of a ternary complex with the Grb2 adaptor protein. J Biol Chem 275:4283–4289 [DOI] [PubMed] [Google Scholar]
- 22. D'Alessandris C, Lauro R, Presta I, Sesti G. 2007. C-reactive protein induces phosphorylation of insulin receptor substrate-1 on Ser307 and Ser 612 in L6 myocytes, thereby impairing the insulin signalling pathway that promotes glucose transport. Diabetologia 50:840–849 [DOI] [PubMed] [Google Scholar]
- 23. Schenk S, Saberi M, Olefsky JM. 2008. Insulin sensitivity: modulation by nutrients and inflammation. J Clin Invest 118:2992–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Andreozzi F, Laratta E, Procopio C, Hribal ML, Sciacqua A, Perticone M, Miele C, Perticone F, Sesti G. 2007. Interleukin-6 impairs the insulin signaling pathway, promoting production of nitric oxide in human umbilical vein endothelial cells. Mol Cell Biol 27:2372–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Clément S, Krause U, Desmedt F, Tanti JF, Behrends J, Pesesse X, Sasaki T, Penninger J, Doherty M, Malaisse W, Dumont JE, Le Marchand-Brustel Y, Erneux C, Hue L, Schurmans S. 2001. The lipid phosphatase SHIP2 controls insulin sensitivity. Nature 409:92–97 [DOI] [PubMed] [Google Scholar]
- 26. Gao T, Furnari F, Newton AC. 2005. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell 18:13–24 [DOI] [PubMed] [Google Scholar]
- 27. Andreozzi F, Procopio C, Greco A, Mannino GC, Miele C, Raciti GA, Iadicicco C, Beguinot F, Pontiroli AE, Hribal ML, Folli F, Sesti G. 2011. Increased levels of the Akt-specific phosphatase PH domain leucine-rich repeat protein phosphatase (PHLPP)-1 in obese participants are associated with insulin resistance. Diabetologia 54:1879–1887 [DOI] [PubMed] [Google Scholar]
- 28. Saltiel AR. 2003. Putting the brakes on insulin signaling. N Engl J Med 349:2560–2562 [DOI] [PubMed] [Google Scholar]
- 29. Prudente S, Scarpelli D, Chandalia M, Zhang YY, Morini E, Del Guerra S, Perticone F, Li R, Powers C, Andreozzi F, Marchetti P, Dallapiccola B, Abate N, Doria A, Sesti G, Trischitta V. 2009. The TRIB3 Q84R polymorphism and risk of early-onset type 2 diabetes. J Clin Endocrinol Metab 94:190–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mata J, Curado S, Ephrussi A, Rørth P. 2000. Tribbles coordinates mitosis and morphogenesis in Drosophila by regulating string/CDC25 proteolysis. Cell 101:511–522 [DOI] [PubMed] [Google Scholar]
- 31. Hanks SK, Hunter T. 1995. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J 9:576–596 [PubMed] [Google Scholar]
- 32. Ord D, Ord T. 2005. Characterization of human NIPK (TRB3, SKIP3) gene activation in stressful conditions. Biochem Biophys Res Commun 330:210–218 [DOI] [PubMed] [Google Scholar]
- 33. Hughes TA. 2006. Regulation of gene expression by alternative untranslated regions. Trends Genet 22:119–122 [DOI] [PubMed] [Google Scholar]
- 34. Yokoyama T, Nakamura T. 2011. Tribbles in disease: signaling pathways important for cellular function and neoplastic transformation. Cancer Sci 102:1115–1122 [DOI] [PubMed] [Google Scholar]
- 35. Hegedus Z, Czibula A, Kiss-Toth E. 2007. Tribbles: a family of kinase-like proteins with potent signalling regulatory function. Cell Signal 19:238–250 [DOI] [PubMed] [Google Scholar]
- 36. Matsushima R, Harada N, Webster NJ, Tsutsumi YM, Nakaya Y. 2006. Effect of TRB3 on insulin and nutrient-stimulated hepatic p70 S6 kinase activity. J Biol Chem 281:29719–29729 [DOI] [PubMed] [Google Scholar]
- 37. Liu J, Wu X, Franklin JL, Messina JL, Hill HS, Moellering DR, Walton RG, Martin M, Garvey WT. 2010. Mammalian Tribbles homolog 3 impairs insulin action in skeletal muscle: role in glucose-induced insulin resistance. Am J Physiol Endocrinol Metab 298:E565–E576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Koh HJ, Arnolds DE, Fujii N, Tran TT, Rogers MJ, Jessen N, Li Y, Liew CW, Ho RC, Hirshman MF, Kulkarni RN, Kahn CR, Goodyear LJ. 2006. Skeletal muscle-selective knockout of LKB1 increases insulin sensitivity, improves glucose homeostasis, and decreases TRB3. Mol Cell Biol 26:8217–8227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cheng KK, Iglesias MA, Lam KS, Wang Y, Sweeney G, Zhu W, Vanhoutte PM, Kraegen EW, Xu A. 2009. APPL1 potentiates insulin-mediated inhibition of hepatic glucose production and alleviates diabetes via Akt activation in mice. Cell Metab 9:417–427 [DOI] [PubMed] [Google Scholar]
- 40. Kiss-Toth E, Bagstaff SM, Sung HY, Jozsa V, Dempsey C, Caunt JC, Oxley KM, Wyllie DH, Polgar T, Harte M, O'neill LA, Qwarnstrom EE, Dower SK. 2004. Human tribbles, a protein family controlling mitogen-activated protein kinase cascades. J Biol Chem 279:42703–42708 [DOI] [PubMed] [Google Scholar]
- 41. Bi XP, Tan HW, Xing SS, Wang ZH, Tang MX, Zhang Y, Zhang W. 2008. Overexpression of TRB3 gene in adipose tissue of rats with high fructose-induced metabolic syndrome. Endocr J 55:747–752 [DOI] [PubMed] [Google Scholar]
- 42. Wang YG, Shi M, Wang T, Shi T, Wei J, Wang N, Chen XM. 2009. Signal transduction mechanism of TRB3 in rats with non-alcoholic fatty liver disease. World J Gastroenterol 15:2329–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Okamoto H, Latres E, Liu R, Thabet K, Murphy A, Valenzeula D, Yancopoulos GD, Stitt TN, Glass DJ, Sleeman MW. 2007. Genetic deletion of Trb3, the mammalian Drosophila tribbles homolog, displays normal hepatic insulin signaling and glucose homeostasis. Diabetes 56:1350–1356 [DOI] [PubMed] [Google Scholar]
- 44. Weismann D, Erion DM, Ignatova-Todorava I, Nagai Y, Stark R, Hsiao JJ, Flannery C, Birkenfeld AL, May T, Kahn M, Zhang D, Yu XX, Murray SF, Bhanot S, Monia BP, Cline GW, Shulman GI, Samuel VT. 2011. Knockdown of the gene encoding Drosophila tribbles homologue 3 (Trib3) improves insulin sensitivity through peroxisome proliferator-activated receptor-γ (PPAR-γ) activation in a rat model of insulin resistance. Diabetologia 54:935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Matos A, Ropelle ER, Pauli JR, Frederico MJ, de Pinho RA, Velloso LA, De Souza CT. 2010. Acute exercise reverses TRB3 expression in the skeletal muscle and ameliorates whole body insulin sensitivity in diabetic mice. Acta Physiol (Oxf) 198:61–69 [DOI] [PubMed] [Google Scholar]
- 46. Lima AF, Ropelle ER, Pauli JR, Cintra DE, Frederico MJ, Pinho RA, Velloso LA, De Souza CT. 2009. Acute exercise reduces insulin resistance-induced TRB3 expression and amelioration of the hepatic production of glucose in the liver of diabetic mice. J Cell Physiol 221:92–97 [DOI] [PubMed] [Google Scholar]
- 47. Koo SH, Satoh H, Herzig S, Lee CH, Hedrick S, Kulkarni R, Evans RM, Olefsky J, Montminy M. 2004. PGC-1 promotes insulin resistance in liver through PPAR-α-dependent induction of TRB-3. Nat Med 10:530–534 [DOI] [PubMed] [Google Scholar]
- 48. Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC. 2003. β-Cell deficit and increased β-cell apoptosis in humans with type 2 diabetes. Diabetes 52:102–110 [DOI] [PubMed] [Google Scholar]
- 49. Robertson RP. 2009. β-Cell deterioration during diabetes: what's in the gun? Trends Endocrinol Metab 20:388–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Humphrey RK, Newcomb CJ, Yu SM, Hao E, Yu D, Krajewski S, Du K, Jhala US. 2010. Mixed lineage kinase-3 stabilizes and functionally cooperates with TRIBBLES-3 to compromise mitochondrial integrity in cytokine-induced death of pancreatic β-cells. J Biol Chem 285:22426–22436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bezy O, Vernochet C, Gesta S, Farmer SR, Kahn CR. 2007. TRB3 blocks adipocyte differentiation through the inhibition of C/EBPβ transcriptional activity. Mol Cell Biol 27:6818–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Qi L, Heredia JE, Altarejos JY, Screaton R, Goebel N, Niessen S, Macleod IX, Liew CW, Kulkarni RN, Bain J, Newgard C, Nelson M, Evans RM, Yates J, Montminy M. 2006. TRB3 links the E3 ubiquitin ligase COP1 to lipid metabolism. Science 312:1763–1766 [DOI] [PubMed] [Google Scholar]
- 53. Naiki T, Saijou E, Miyaoka Y, Sekine K, Miyajima A. 2007. TRB2, a mouse Tribbles ortholog, suppresses adipocyte differentiation by inhibiting AKT and C/EBPβ. J Biol Chem 282:24075–24082 [DOI] [PubMed] [Google Scholar]
- 54. Takahashi Y, Ohoka N, Hayashi H, Sato R. 2008. TRB3 suppresses adipocyte differentiation by negatively regulating PPARγ transcriptional activity. J Lipid Res 49:880–892 [DOI] [PubMed] [Google Scholar]
- 55. Burkhardt R, Toh SA, Lagor WR, Birkeland A, Levin M, Li X, Robblee M, Fedorov VD, Yamamoto M, Satoh T, Akira S, Kathiresan S, Breslow JL, Rader DJ. 2010. Trib1 is a lipid- and myocardial infarction-associated gene that regulates hepatic lipogenesis and VLDL production in mice. J Clin Invest 120:4410–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kato S, Du K. 2007. TRB3 modulates C2C12 differentiation by interfering with Akt activation. Biochem Biophys Res Commum 353:933–938 [DOI] [PubMed] [Google Scholar]
- 57. Hotamisligil GS. 2010. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 140:900–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mayumi-Matsuda K, Kojima S, Suzuki H, Sakata T. 1999. Identification of a novel kinase-like gene induced during neuronal cell death. Biochem Biophys Res Commun 258:260–264 [DOI] [PubMed] [Google Scholar]
- 59. Ord D, Ord T. 2003. Mouse NIPK interacts with ATF4 and affects its transcriptional activity. Exp Cell Res 286:308–320 [DOI] [PubMed] [Google Scholar]
- 60. Wu M, Xu LG, Zhai Z, Shu HB. 2003. SINK is a p65-interacting negative regulator of NF-κB-dependent transcription. J Biol Chem 278:27072–27079 [DOI] [PubMed] [Google Scholar]
- 61. He L, Marecki JC, Serrero G, Simmen FA, Ronis MJ, Badger TM. 2007. Dose-dependent effects of alcohol on insulin signaling: partial explanation for biphasic alcohol impact on human health. Mol Endocrinol 21:2541–2550 [DOI] [PubMed] [Google Scholar]
- 62. Hartley T, Siva M, Lai E, Teodoro T, Zhang L, Volchuk A. 2010. A Endoplasmic reticulum stress response in an INS-1 pancreatic β-cell line with inducible expression of a folding-deficient proinsulin. BMC Cell Biol 11:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nicoletti-Carvalho JE, Nogueira TC, Gorjão R, Bromati CR, Yamanaka TS, Boschero AC, Velloso LA, Curi R, Anhê GF, Bordin S. 2010. UPR-mediated TRIB3 expression correlates with reduced AKT phosphorylation and inability of interleukin 6 to overcome palmitate-induced apoptosis in RINm5F cells. J Endocrinol 206:183–193 [DOI] [PubMed] [Google Scholar]
- 64. Yacoub Wasef SZ, Robinson KA, Berkaw MN, Buse MG. 2006. Glucose, dexamethasone, and the unfolded protein response regulate TRB3 mRNA expression in 3T3-L1 adipocytes and L6 myotubes. Am J Physiol Endocrinol Metab 291:E1274–E1280 [DOI] [PubMed] [Google Scholar]
- 65. Ohoka N, Yoshii S, Hattori T, Onozaki K, Hayashi H. 2005. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J 24:1243–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jousse C, Deval C, Maurin AC, Parry L, Chérasse Y, Chaveroux C, Lefloch R, Lenormand P, Bruhat A, Fafournoux P. 2007. TRB3 inhibits the transcriptional activation of stress-regulated genes by a negative feedback on the ATF4 pathway. J Biol Chem 282:15851–15861 [DOI] [PubMed] [Google Scholar]
- 67. Avery J, Etzion S, DeBosch BJ, Jin X, Lupu TS, Beitinjaneh B, Grand J, Kovacs A, Sambandam N, Muslin AJ. 2010. TRB3 function in cardiac endoplasmic reticulum stress. Circ Res 106:1516–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Eizirik DL, Cardozo AK, Cnop M. 2008. The role for endoplasmic reticulum stress in diabetes mellitus. Endocr Rev 29:42–61 [DOI] [PubMed] [Google Scholar]
- 69. Bromati CR, Lellis-Santos C, Yamanaka TS, Nogueira TC, Leonelli M, Caperuto LC, Gorjão R, Leite AR, Anhê GF, Bordin S. 2011. UPR induces transient burst of apoptosis in islets of early lactating rats through reduced AKT phosphorylation via ATF4/CHOP stimulation of TRB3 expression. Am J Physiol Regul Integr Comp Physiol 300:R92–R100 [DOI] [PubMed] [Google Scholar]
- 70. Qian B, Wang H, Men X, Zhang W, Cai H, Xu S, Xu Y, Ye L, Wollheim CB, Lou J. 2008. TRIB3 [corrected] is implicated in glucotoxicity- and endoplasmic reticulum-stress-induced β-cell apoptosis. J Endocrinol [Errata (2009) 203:399, 200:243] 199:407–416 [DOI] [PubMed] [Google Scholar]
- 71. Liew CW, Bochenski J, Kawamori D, Hu J, Leech CA, Wanic K, Malecki M, Warram JH, Qi L, Krolewski AS, Kulkarni RN. 2010. The pseudokinase tribbles homolog 3 interacts with ATF4 to negatively regulate insulin exocytosis in human and mouse β-cells. J Clin Invest 120:2876–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ding J, Kato S, Du K. 2008. PI3K activates negative and positive signals to regulate TRB3 expression in hepatic cells. Exp Cell Res 314:1566–1574 [DOI] [PubMed] [Google Scholar]
- 73. Du K, Ding J. 2009. Insulin regulates TRB3 and other stress-responsive gene expression through induction of C/EBPβ. Mol Endocrinol 23:475–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Matsumoto M, Han S, Kitamura T, Accili D. 2006. Dual role of transcription factor FoxO1 in controlling hepatic insulin sensitivity and lipid metabolism. J Clin Invest 116:2464–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Oberkofler H, Pfeifenberger A, Soyal S, Felder T, Hahne P, Miller K, Krempler F, Patsch W. 2010. Aberrant hepatic TRIB3 gene expression in insulin-resistant obese humans. Diabetologia 53:1971–1975 [DOI] [PubMed] [Google Scholar]
- 76. Prudente S, Hribal ML, Flex E, Turchi F, Morini E, De Cosmo S, Bacci S, Tassi V, Cardellini M, Lauro R, Sesti G, Dallapiccola B, Trischitta V. 2005. The functional Q84R polymorphism of mammalian Tribbles homolog TRB3 is associated with insulin resistance and related cardiovascular risk in Caucasians from Italy. Diabetes 54:2807–2811 [DOI] [PubMed] [Google Scholar]
- 77. Andreozzi F, Formoso G, Prudente S, Hribal ML, Pandolfi A, Bellacchio E, Di Silvestre S, Trischitta V, Consoli A, Sesti G. 2008. TRIB3 R84 variant is associated with impaired insulin-mediated nitric oxide production in human endothelial cells. Arterioscler Thromb Vasc Biol 28:1355–1360 [DOI] [PubMed] [Google Scholar]
- 78. Federici M, Pandolfi A, De Filippis EA, Pellegrini G, Menghini R, Lauro D, Cardellini M, Romano M, Sesti G, Lauro R, Consoli A. 2004. G972R IRS-1 variant impairs insulin regulation of endothelial nitric oxide synthase in cultured human endothelial cells. Circulation 109:399–405 [DOI] [PubMed] [Google Scholar]
- 79. Bacci S, Di Paola R, Menzaghi C, Di Fulvio P, Di Silvestre S, Pellegrini F, Baratta R, Marucci A, Mastroianno S, Fini G, Formoso G, Consoli A, Perticone F, Frittitta L, Pandolfi A, Trischitta V. 2009. ENPP1 Q121 variant, increased pulse pressure and reduced insulin signaling, and nitric oxide synthase activity in endothelial cells. Arterioscler Thromb Vasc Biol 29:1678–1683 [DOI] [PubMed] [Google Scholar]
- 80. Rask-Madsen C, Li Q, Freund B, Feather D, Abramov R, Wu IH, Chen K, Yamamoto-Hiraoka J, Goldenbogen J, Sotiropoulos KB, Clermont A, Geraldes P, Dall'Osso C, Wagers AJ, Huang PL, Rekhter M, Scalia R, Kahn CR, King GL. 2010. Loss of insulin signaling in vascular endothelial cells accelerates atherosclerosis in apolipoprotein E null mice. Cell Metab 11:379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Formoso G, Di Tomo P, Andreozzi F, Succurro E, Di Silvestre S, Prudente S, Perticone F, Trischitta V, Sesti G, Pandolfi A, Consoli A. 2011. The TRIB3 R84 variant is associated with increased carotid intima-media thickness in vivo and with enhanced MAPK signalling in human endothelial cells. Cardiovasc Res 89:184–192 [DOI] [PubMed] [Google Scholar]
- 82. Porzio O, Federici M, Hribal ML, Lauro D, Accili D, Lauro R, Borboni P, Sesti G. 1999. The Gly972→Arg amino acid polymorphism in IRS-1 impairs insulin secretion in pancreatic β-cells. J Clin Invest 104:357–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Federici M, Hribal ML, Ranalli M, Marselli L, Porzio O, Lauro D, Borboni P, Lauro R, Marchetti P, Melino G, Sesti G. 2001. The common Arg972 polymorphism in insulin receptor substrate-1 causes apoptosis of human pancreatic islets. FASEB J 15:22–24 [DOI] [PubMed] [Google Scholar]
- 84. Marchetti P, Lupi R, Federici M, Marselli L, Masini M, Boggi U, Del Guerra S, Patanè G, Piro S, Anello M, Bergamini E, Purrello F, Lauro R, Mosca F, Sesti G, Del Prato S. 2002. Insulin secretory function is impaired in isolated human islets carrying the Gly(972)→Arg IRS-1 polymorphism. Diabetes 51:1419–1424 [DOI] [PubMed] [Google Scholar]
- 85. Di Paola R, Caporarello N, Marucci A, Dimatteo C, Iadicicco C, Del Guerra S, Prudente S, Sudano D, Miele C, Parrino C, Piro S, Beguinot F, Marchetti P, Trischitta V, Frittitta L. 2011. ENPP1 affects insulin action and secretion: evidences from in vitro studies. PloS One 6:e19462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kulkarni RN, Brüning JC, Winnay JN, Postic C, Magnuson MA, Kahn CR. 1999. Tissue-specific knockout of the insulin receptor in pancreatic β-cells creates an insulin secretory defect similar to that in type 2 diabetes. Cell 96:329–339 [DOI] [PubMed] [Google Scholar]
- 87. Prudente S, Baratta R, Andreozzi F, Morini E, Farina MG, Nigro A, Copetti M, Pellegrini F, Succurro E, Di Pietrantonio L, Brufani C, Barbetti F, Dallapiccola B, Sesti G, Trischitta V, Frittitta L. 2010. TRIB3 R84 variant affects glucose homeostasis by altering the interplay between insulin sensitivity and secretion. Diabetologia 53:1354–1361 [DOI] [PubMed] [Google Scholar]
- 88. Gong HP, Wang ZH, Jiang H, Fang NN, Li JS, Shang YY, Zhang Y, Zhong M, Zhang W. 2009. TRIB3 functional Q84R polymorphism is a risk factor for metabolic syndrome and carotid atherosclerosis. Diabetes Care 32:1311–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Shi Z, Liu J, Guo Q, Ma X, Shen L, Xu S, Gao H, Yuan X, Zhang J. 2009. Association of TRB3 gene Q84R polymorphism with type 2 diabetes mellitus in Chinese population. Endocrine 35:414–419 [DOI] [PubMed] [Google Scholar]
- 90. Stumvoll M, Fritsche A, Volk A, Stefan N, Madaus A, Maerker E, Teigeler A, Koch M, Machicao F, Häring H. 2001. The Gly972Arg polymorphism in the insulin receptor substrate-1 gene contributes to the variation in insulin secretion in normal glucose-tolerant humans. Diabetes 50:882–885 [DOI] [PubMed] [Google Scholar]
- 91. Baratta R, Rossetti P, Prudente S, Barbetti F, Sudano D, Nigro A, Farina MG, Pellegrini F, Trischitta V, Frittitta L. 2008. Role of the ENPP1 K121Q polymorphism in glucose homeostasis. Diabetes 57:3360–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Freathy RM, Timpson NJ, Lawlor DA, Pouta A, Ben-Shlomo Y, Ruokonen A, Ebrahim S, Shields B, Zeggini E, Weedon MN, Lindgren CM, Lango H, Melzer D, Ferrucci L, Paolisso G, Neville MJ, Karpe F, Palmer CN, Morris AD, Elliott P, Jarvelin MR, Smith GD, McCarthy MI, Hattersley AT, Frayling TM. 2008. Common variation in the FTO gene alters diabetes-related metabolic traits to the extent expected given its effect on BMI. Diabetes 57:1419–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lyssenko V, Almgren P, Anevski D, Perfekt R, Lahti K, Nissén M, Isomaa B, Forsen B, Homström N, Saloranta C, Taskinen MR, Groop L, Tuomi T. 2005. Predictors of and longitudinal changes in insulin sensitivity and secretion preceding onset of type 2 diabetes. Diabetes 54:166–174 [DOI] [PubMed] [Google Scholar]
- 94. Abdul-Ghani MA, Williams K, DeFronzo RA, Stern M. 2007. What is the best predictor of future type 2 diabetes? Diabetes Care 30:1544–1548 [DOI] [PubMed] [Google Scholar]
- 95. Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, Zeggini E, Huth C, Aulchenko YS, Thorleifsson G, McCulloch LJ, Ferreira T, Grallert H, Amin N, Wu G, Willer CJ, Raychaudhuri S, McCarroll SA, Langenberg C, Hofmann OM, Dupuis J, Qi L, Segrè AV, van Hoek M, Navarro P, et al. 2010. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 42:579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Nerpin E, Risérus U, Ingelsson E, Sundström J, Jobs M, Larsson A, Basu S, Arnlöv J. 2008. Insulin sensitivity measured with euglycemic clamp is independently associated with glomerular filtration rate in a community-based cohort. Diabetes Care 31:1550–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Chen J, Muntner P, Hamm LL, Jones DW, Batuman V, Fonseca V, Whelton PK, He J. 2004. The metabolic syndrome and chronic kidney disease in U.S. adults. Ann Int Med 140:167–174 [DOI] [PubMed] [Google Scholar]
- 98. De Cosmo S, Prudente S, Andreozzi F, Morini E, Rauseo A, Scarpelli D, Zhang YY, Xu R, Perticone F, Dallapiccola B, Sesti G, Doria A, Trischitta V. 2007. Glutamine to arginine substitution at amino acid 84 of mammalian tribbles homolog TRIB3 and CKD in whites with type 2 diabetes. Am J Kidney Dis 50:688–689 [DOI] [PubMed] [Google Scholar]
- 99. Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, Wahlstrand B, Hedner T, Corella D, Tai ES, Ordovas JM, Berglund G, Vartiainen E, Jousilahti P, Hedblad B, Taskinen MR, Newton-Cheh C, Salomaa V, Peltonen L, Groop L, Altshuler DM, Orho-Melander M. 2008. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet 40:189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, et al. 2008. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet 40:161–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. IBC 50K CAD Consortium 2011. Large-scale gene-centric analysis identifies novel variants for coronary artery disease. PLoS Genet 7:e1002260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP, Penninx BW, Janssens AC, Wilson JF, Spector T, Martin NG, Pedersen NL, Kyvik KO, Kaprio J, Hofman A, Freimer NB, Jarvelin MR, Gyllensten U, Campbell H, Rudan I, Johansson A, Marroni F, Hayward C, Vitart V, Jonasson I, et al. 2009. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet 41:47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]