Abstract
Context:
Traditionally, factors predisposing to diseases are either genetic (“nature”) or environmental, also known as lifestyle-related (“nurture”). Papillary thyroid cancer is an example of a disease where the respective roles of these factors are surprisingly unclear.
Evidence Acquisition:
Original articles and reviews summarizing our current understanding of the role of microRNA in thyroid tumorigenesis are reviewed and evaluated.
Conclusion:
The genetic predisposition to papillary thyroid cancer appears to consist of a variety of gene mutations that are mostly either of low penetrance and common or of high penetrance but rare. Moreover, they likely interact with each other and with environmental factors. The culpable genes may not be of the traditional, protein-coding type. A limited number of noncoding candidate genes have indeed been described, and we propose here that the failure to find mutations in traditional protein-coding genes is not coincidental. Instead, a more likely hypothesis is that changes in the expression of multiple regulatory RNA genes, e.g. microRNAs, may be a major mechanism. Our review of the literature strongly supports this notion in that a polymorphism in one microRNAs (miR-146a) predisposes to thyroid carcinoma, whereas numerous other microRNAs are involved in signaling (mainly PTEN/PI3K/AKT and T3/THRB) that is central to thyroid carcinogenesis.
Most thyroid carcinomas originate from thyroid follicular cells and are categorized into well-differentiated papillary thyroid carcinoma (PTC; conventional or follicular variant) and follicular thyroid carcinoma (FTC; conventional or oncocytic type). Both PTC and FTC may progress to poorly differentiated carcinoma or may completely lose differentiation and proceed to anaplastic carcinoma. Follicular adenomas are benign thyroid tumors (conventional or oncocytic type). Medullary carcinoma arises from C cells and accounts for less than 5% of all thyroid cancers.
Somatic Genetic Changes in Thyroid Tumors
In the development of PTC, several genetic factors are involved, including the activation of the MAPK signaling pathway as a result of point mutations within BRAF (40% of PTC cases) or RAS (15% of PTC cases, exclusively the follicular variant) or RET/PTC rearrangement (18% of PTC cases). These mutually exclusive mutations are associated with distinct phenotypic and biological properties of the tumors (1). Contrary to PTC, FTC is known to harbor either RAS or phosphatase and tensin homolog (PTEN) mutations or PAX8/PPAR rearrangements, which are identified in up to 50–80% of FTC tumors (2). RAS mutations are also found in benign follicular adenomas, in which progression steps to follicular carcinoma are poorly understood (2).
Genetic Predisposition
Traditionally, factors predisposing to diseases are either genetic (“nature”) or environmental, also known as lifestyle-related (“nurture”). Papillary thyroid cancer is an example of a disease where the respective roles of these factors are surprisingly unclear. We shall briefly examine the evidence.
One of the most widely used methods of measuring genetic predisposition is the case control study. The family risk ratio is the ratio between the prevalence in the family members of two cohorts of probands, one with and the other without the disease phenotype. Case control studies require large series of well-studied patients and controls and disease databases such as cancer registries with good coverage. For cancer, two early studies are summarized in Table 1. Family risk ratios for first-degree relatives were calculated for various cancers. Although the ratio for all cancers was 2.2–3.5, such common cancers as breast, lung, colorectal, and prostate had ratios mostly in the 2–4 range (3, 4). Only a handful of cancers had values exceeding 4, and thyroid cancer stood out with the highest of all ratios, 8.5 in Utah and 12.4 in Sweden (5).
Table 1.
Familiality of selected cancers defined as family risk ratio derived from studies in Utah (3) and Sweden (4)
| Site | Family risk ratio |
Twin study (proportion of variance) | |
|---|---|---|---|
| Utah | Sweden | ||
| Thyroid | 8.48 | 12.42 | |
| Testis | 8.57 | 8.50 | |
| Multiple myeloma | 4.29 | 5.62 | |
| Prostate | 2.21 | 9.41 | 0.42 |
| Colorectum | 2.54 | 4.41 | 0.35 |
| Breast | 1.83 | 2.01 | 0.27 |
| Lung | 2.55 | 3.16 | 0.26 |
| All cancers | 2.15 | 3.53 | |
Heritability is defined as proportion of variance from a twin study (6). [Presentation of the data was adapted and modified from N. Risch, The genetic epidemiology of cancer: interpreting family and twin studies and their implications for molecular genetic approaches. Cancer Epidemiol Biomarkers Prev 10:733–741, 2001 (5), with permission. © American Association for Cancer Research.]
This does not automatically mean that the genetic component of the predisposition is exceptionally high in thyroid cancer. A main reason is that the method does not correct for the shared environment of family members. First-degree relatives in particular usually share not only the environment but also lifestyle. An independent method to correct for shared environment is the twin study. Here the co-occurrence of the phenotype in twins is considered separately for monozygotic and dizygotic twins. The basic assumption is that twins usually have the same environment, but monozygotic twins share all their genes whereas dizygotic twins share only 50%. Thus, the difference in concordance rates between mono- and dizygotic twins is a measure of the genetic predisposition. Twin studies require many twin pairs in which at least one member is affected. The largest twin study known to us that addresses cancer is by Lichtenstein et al. (6) from Scandinavia (Table 1). Sufficient numbers of twins to ensure strong results were only available for the most common cancers, resulting in proportions of variance between 0.42 and 0.26. For thyroid cancer, none of 39 monozygotic pairs and none of 63 dizygotic pairs were concordant for thyroid cancer. Although these numbers do not allow any statistical test, they do not support the notion of an overwhelming role of genes in the predisposition.
In the studies referred to above, all subtypes of thyroid cancer were considered. Although PTC accounts for 80–85% of all thyroid cancer, the medullary form constitutes some 5% of all, and about one third of medullary thyroid cancers are caused by germline mutations in the RET gene that are regularly inherited in a Mendelian fashion. Three studies address this question (7–9). The results suggest that the family risk ratio for well-differentiated thyroid cancer remains high even if medullary carcinoma is excluded. Further studies in a Swedish series showed the familiality of thyroid cancer to be by far the highest of all cancers (10). Finally, large series of cases and controls were studied in Iceland, where population records allowed the analysis of up to fifth-degree relatives. Again, thyroid cancer had one of the highest family risk ratios (11).
Combining the evidence from case control and twin studies, the conclusion is one of high familiality where both genes and environmental factors have a major impact.
Environmental Predisposing Factors
Ionizing radiation from fallout or ingestion of material contaminated by radioactive iodine isotopes is highly carcinogenic to the thyroid. For example, individuals exposed in the nuclear bombings in Hiroshima and Nagasaki showed clearly higher thyroid cancer incidence than controls. Likewise, the Chernobyl disaster led to an increased incidence of PTC, mainly or exclusively when the radiation occurred in infancy or childhood (12, 13). More pertinent to this discussion is lower level, chronic, or intermittent exposure. There is some evidence that the increased use of diagnostic methods such as computed tomography scans is in part responsible for the rising incidence of thyroid cancer (14).
Although exposure to radioactive irradiation can usually be measured and monitored, other environmental factors are less well understood. Among these are iodine deficiency, benign thyroid disease such as Hashimoto's thyroiditis, and perhaps a diet low in fiber (15).
Occurrence in Families; Linkage, and Association Studies
Of all patients with PTC, up to 10% have a family history of PTC in a first- or second-degree relative. Given that PTC is not a common cancer [ninth most common cancer in the United States with 44,670 new patients estimated in 2010 (16)], 10% familiality is relatively high. Of note, in the great majority of cases, the “familial occurrence” is based on just one or two cases in addition to the proband. Pedigrees displaying Mendelian-type inheritance are conspicuously rare, and large pedigrees with more than five affected individuals are exceptional. Even in families involving typical Mendelian inheritance, skipping of generations (nonpenetrance) is common. In terms of the genetic predisposition to PTC, these facts predict that more than just a few genes are involved, that their penetrance is low, and that interaction with environmental factors plays a role.
Are there no high-penetrance Mendelian genes predisposing to PTC? One cannot exclude the existence of such genes, but the facts suggest that if they exist, they are rare. They might belong to the class termed “common disease, rare allele” by Bodmer and Bonilla (17). Such genes would not be detectable by present-day association studies [which so far are restricted to single nucleotide polymorphisms (SNP) that have a greater than 1% frequency of carriers in the population] and would only be detectable by linkage in large single families. Linkage analyses in PTC have resulted in numerous candidate loci (18), but although these have been carefully studied by positional cloning strategies, no convincing culpable gene has been found in this way.
Association studies might provide an answer but have so far not led to definitive results. Gudmundsson et al. (19) screened a small cohort of nonmedullary thyroid cancer cases from Iceland and validated the results in a small series from Spain and a larger series from Ohio. As a result, alleles at two loci (at 9q22 and 14q13) showed association with thyroid cancer. Both SNP were located far from known genes, and further studies into these have not been published. Because the odds ratios are modest (1.6 and 1.3, respectively) but the statistical evidence is strong, it is likely that the loci contain genes predisposing to thyroid cancer with low or ultra-low penetrance.
Conclusions
Taking all the above together, the genetic predisposition to PTC appears to consist of a variety of gene mutations that are mostly either of low penetrance and common or of high penetrance but rare. Moreover, they likely interact with each other and with environmental factors. The culpable genes may not be of the traditional, protein-coding type. A limited number of noncoding candidate genes have indeed been described, such as a large noncoding RNA gene in chromosome 8q24 (18) and microRNA gene miR-146a as described below in MicroRNA-associated genetic predisposition to PTC. Here we propose that the failure to find mutations in traditional protein coding genes is not coincidental. Instead, a more likely hypothesis is that changes in the expression of multiple noncoding RNA genes, e.g. miR, may be a major mechanism. Misexpression of miR (in both germline and tumor) probably primarily occurs as a result of a sequence change, as shown for miR-146a in PTC (20), but also as a result of DNA copy number alterations and epigenetic changes (21, 22). The sequence variation of miR-146a affects the processing and expression of mature microRNA and was linked to the risk of PTC (20). We describe some of the evidence pointing in this direction. In addition, microRNA expression might be induced by several other mechanisms, including BRAF mutation (miR-21) and immune stimulation (miR-155). It was shown that miR-146a is induced by nuclear factor κB upon ligation of toll-like receptors (TLR2, TLR4, or TLR5) as well as stimulation by TNF-α or IL-1β (23). A possible stimulus for an induction of the nuclear factor κB pathway in thyroid might be ionizing radiation, the strongest environmental risk factor for thyroid cancer.
MicroRNA
MicroRNA (miR) are small (19–25 nucleotides), noncoding RNA molecules that typically function as negative regulators of the expression of protein-encoding genes (24). It is speculated that miRs altogether regulate around 30% of the human genome, which highlights their potential importance as global regulators of gene expression. miRs regulate such major processes as development, apoptosis, cell proliferation, immune response, and hematopoiesis (25); they also may act as tumor suppressor genes and oncogenes (26, 27). Mature miRs target and inhibit translation or promote mRNA degradation by annealing to complementary sequences in mRNA 3′ untranslated regions. Watson-Crick complementarity between the target and the “seed” region comprising two to eight nucleotides of the mature miR is both necessary and sufficient for targeting and regulation of mRNA by miR. The sequence of this “seed” region is the basis of most genome-wide predictions of miR binding sites within miR-regulated genes (28). A SNP located in the crucial “seed” sequence affects its complementarity to potential target genes determining the functionality of a miR (29). Individual miR typically target dozens of mRNA, often encoding proteins with related functions (30). Therefore, although their inhibitory effects on individual mRNA are generally modest, their combined effects on multiple mRNA can induce strong biological responses (21, 31, 32).
Discovery of microRNA
Since microRNA were first identified in Caenorhabditis elegans (33), more than 1048 miR genes have been described in the human genome (34). The majority of currently known miR was identified by using traditional molecular methods, i.e. cloning and sequencing individual small RNA (35). Sequencing-based applications for identifying new miRs have been hindered by laborious cloning techniques and the expense of capillary DNA sequencing (36, 37). In contrast to capillary sequencing, recently available next-generation deep sequencing technologies offer inexpensive increases in throughput in addition to providing quantitative expression data (38). Novel miRs are deposited in miRBase, which acts as a public repository for microRNAs (39).
MicroRNA expression profiling in thyroid cancer
MicroRNAs were found to be involved in the development of several types of human cancers mainly by abnormal levels of expression of mature miR transcripts in tumors compared with the corresponding unaffected tissues (40). MicroRNA expression profiling of human tumors has identified signatures associated with diagnosis, staging, prognosis, and response to treatment (41, 42). The up-regulation of prooncogenic miRs (e.g. miR-155, miR-21) or down-regulation of miRs functioning as tumor suppressors (e.g. miR-15a, miR-16) have been shown to contribute to cancer development (43–45).
To profile global miR expression, a robust method for detection of the expression level of each miR in a single sample is required. The oligo DNA microarray was one of the first high-throughput platforms that allowed concurrent profiling of many miRs (46). Most profiling studies in thyroid cancers were performed by employing this method (45, 47–49). He et al. (45) detected preliminary evidence of a potential role for miRs in PTC. The authors noticed an overexpression of several miRs in tumor tissue compared with unaffected thyroid tissue. The list of up-regulated miRs (up to 19.3-fold) included miR-146, miR-221/222, miR-155, miR-34, and miR-181 and was soon confirmed (47). Tumors in which the up-regulation (11- to 19-fold) of miR-221, -222, and -146 was strong showed dramatic loss of KIT transcript and Kit protein (target shared by all three miRs). This down expression was associated with a SNP in each of the two recognition sequences in KIT mRNA for these miRs. The authors concluded that up-regulation of several miRs and altered inhibition of their targets are involved in PTC pathogenesis and that sequence changes in genes targeted by miRs can contribute to their regulation (45, 47). Different microRNAs are deregulated in FTC and anaplastic thyroid carcinoma (48, 49), proving that each type of thyroid cancer harbors specific signatures of microRNA expression. For useful tables summarizing the microRNA expression profiling in various thyroid samples, see Refs. 50–52.
Diagnostic utility of microRNA
MiRs as a diagnostic tool might be used: 1) to detect an increased risk of acquiring cancer by studying disease-associated variations of miR sequences (like miR-146a in thyroid cancer); 2) to diagnose early-stage cancer by determining miR expression profiles in the blood [like miR-141 in prostate cancer (53)]; 3) to distinguish tumor from normal tissue in fine-biopsy specimens by miR expression profiling (54); and 4) to predict clinical outcome [like in leukemia (42)].
In the thyroid field, one of the main diagnostic problems is the assessment of thyroid nodules, which are palpable in 5–7% of adults. Thyroid fine-needle aspiration biopsy is a widespread method for preoperative evaluation of thyroid nodules, although in up to 20% of cases a precise diagnosis cannot be obtained by cytological methods. Most of these patients undergo surgery, and less than 20% of surgically removed nodules are found to be malignant. Whether determining the expression profiles of selected miRs might improve the diagnostic accuracy of fine-needle aspiration biopsy is presently an open question. Both promising (55–57) and negative (52) studies have been published. Larger prospective clinical studies are still awaited. In a retrospective study, it was shown that some miRs (miR-221/222 and miR-146b) were associated with clinically aggressive PTC tumors (58).
Variations in microRNA sequences
Several SNP have been detected within the precursors of canonical miRs deposited in miRBase (59, 60). At first, most of them seemed to be examples of “innocent” variation; however, some soon proved to be of fundamental functional importance contributing to cancer susceptibility. That SNP might affect miR maturation was first shown for a viral miR (61) and for human miR-125a (62). To what extent sequence variation in miR genes is associated with tumorigenesis is a matter of intensive investigation (63, 64).
MicroRNA-associated genetic predisposition to PTC
Endocrinologists were among the first to demonstrate a role for a miR polymorphism in the predisposition to cancer (20). In an effort to elucidate the putative effects of overexpressed miRs in PTC, the authors sequenced the genomic DNA of 15 PTC patients and noted a common G/C polymorphism in miR-146a. The SNP resides at position +60 relative to the first nucleotide of pre-miR-146a, placing it in the passenger strand. The rarer C allele causes mispairing within the hairpin and a lowering of the predicted minimum free energy (65). The expression of the pre-miR and mature miR was 2-fold lower in the C allele compared with the G allele, indicating that this single nucleotide difference significantly alters the amount of mature miR produced in an allele-specific way (20). It was also shown that the G/C polymorphism affects both the efficiency of pri-miR processing and protein binding to the pre-miR product. This likely affects the stability and/or efficiency of pre-miR export to the cytoplasm. The reduction in miR expression led to less efficient inhibition of its target genes, attesting to the importance of the SNP to the miR function potentially leading to tumorigenesis (20).
In an association study of 608 PTC patients and 901 controls, a marked difference in genotype distribution of the miR-146a SNP was found (P = 2.2 × 10−6), the heterozygous state being associated with risk of PTC (GC vs. GG + CC, odds ratio = 1.62; P = 7.2 · 10−6). Moreover, 14 of 300 (4.7%) tumors had undergone somatic mutations of the SNP sequence from GG or CC toward heterozygosity, underscoring the role of sequence variations in shaping the miR function in thyroid cancer (20).
SNP-dependent novel miRs
A fascinating role of miR genetic variations is that they can create new functional mature miRs by changing the structure of precursors and/or modifying the sequence of the “seed regions” responsible for targeting mRNA (66). The study of miR-146a showed not only that novel mature miR (marked with an asterisk) are produced from the passenger strand but also that the presence of the SNP generates two isoforms of the alternative miR: miR-146a*G from the allele carrying G, and miR-146a*C from the allele carrying C, each with its distinct set of target genes (29). The SNP located in the crucial seed sequence of miR-146a* determines its complementarity to potential target genes affecting the functionality of both isoforms (28). In a microarray expression study, the transcriptomes of individual genotypes differed profoundly, with the modulated genes mainly involved in regulation of apoptosis. This can lead to exaggerated DNA-damage response in heterozygotes, potentially explaining the predisposition to thyroid cancer (29, 67). The impact of the passenger strand microRNAs on regulatory networks is now extensively studied (68, 69).
Subsequent results implicating inherited SNP were reported in breast cancer (70, 71), hepatocellular carcinoma (72), and chronic lymphocytic leukemia (64). Polymorphisms in microRNA and their target sites became a “gold mine” for molecular pathology and pharmacogenomics (63, 73–75).
Next-generation sequencing of the microRNA transcriptome
The oligo DNA microarray presently used for miR expression profiling relies on probes designed to detect each of the currently known reference miR sequences listed in miRBase (39). The main weakness of this method is that it does not allow unknown miRs and their expression to be detected. Next-generation deep sequencing platforms detect new miRs and at the same time measure their expression (76). Based on next-generation deep sequencing data, it was recently reported that for many miRs the most abundant transcript differs from the canonical sequence in miRBase (77, 78). Notably, the data showed that choosing a different isomiR sequence for measuring miR expression level could affect the ability to detect differential miR expression. The read count for the most abundant isomiR, rather than the miRBase reference sequence, provides the most robust approach to compare miR expression between libraries (79). For thyroid carcinomas, the most abundant isoforms have yet to be reliably determined for all miRs.
In addition to single nucleotide substitution/insertion/deletions, the next-generation deep sequencing offers the potential to detect other variations such as RNA editing (80) and 3′ nucleotide additions (79). In the few hitherto published studies of deep miR sequencing, miRs frequently exhibited variation from their “reference” sequences, producing multiple mature variants, as well as miR* from the passenger strand. Although it is possible that isomiR share a common set of targets with the canonical miR, it is likely that some of them target totally different sets of genes similar to the two isomiRs derived from the passenger strand of pre-miR-146a. Those isomiRs resulting from variation at the 5′ end may be of particular interest when they have a seed sequence different from the reference miR indicating their potential to target different transcripts. The functional implications of the more widespread 3′ modifications and RNA editing are unclear so far (66).
To further clarify the role of miRs in thyroid tumorigenesis, it is crucial to generate a complete dataset of variations in miRs sequences and to profile the expression of miR isoforms in thyroid cancer tissues. The introduction of the next-generation deep sequencing technology facilitates the study of the RNA transcriptome of thyroid cancer on the genome-wide level. Both sequence variation and expression levels of small RNA, including microRNAs, can be analyzed simultaneously. This approach will lead to the creation of strong comprehensive datasets that will be important assets when miRs are developed to serve as diagnostic, preventive, and therapeutic tools.
MicroRNA and PTEN/PI3K/AKT signaling
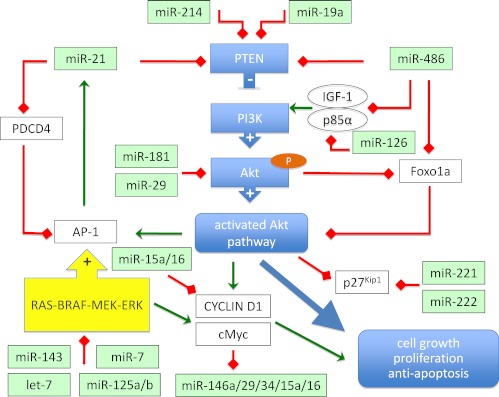
AKT (protein kinase B) is a central signaling molecule in the phosphatidyl inositol 3-kinase (PI3K) pathway that is frequently activated in thyroid cancer, especially in the follicular subtype (Fig. 1). AKT activation regulates apoptosis, proliferation, and migration in many cell systems and appears to play an important role in tumor formation and progression of thyroid carcinomas (81). Enhanced AKT activity results from increased PI3K activity or reduced activity of the PTEN. These events are associated with advanced tumor size and invasion of both PTC and FTC (81).
Fig. 1.
Regulation of PTEN/PI3K/AKT pathway by microRNAs. Arrows represent positive (green) or negative (red) regulation of the genes.
It was demonstrated that sporadic FTC are characterized by reduced expression of PTEN mRNA and protein and increased levels of total AKT activity in comparison with normal thyroid tissue (82, 83). The AKT pathway is also activated in PTC cases with RET/PTC rearrangements or RAS mutations, but not in BRAF-mutation-positive cases. Enhanced AKT activation is also present in all thyroid tumors of Cowden syndrome, an autosomal-dominant multiple hamartoma syndrome in which more than 50% of patients develop thyroid neoplasia.
PTEN expression is repressed by activator protein-1 (AP-1), a known regulator of cell proliferation, apoptosis, tumor invasiveness, and angiogenesis, and that process is proposed to be mediated by miR-21 (84, 85). MicroRNA-21 is a highly relevant tumor-associated microRNA that is overexpressed in almost all tumors analyzed so far, including PTC (45, 47, 86). It was recently shown that miR-21 is induced by AP-1 (85, 87). In a rat thyroid cell system, inhibition of miR-21 increased the expression of PTEN, which led to decreased tumor cell proliferation, migration, and invasion. The opposite effect was observed when miR-21 expression was enhanced by transfection with the precursor of miR-21; expression of PTEN and PTEN-dependent genes like matrix metalloproteases 2 and 9 was diminished, and tumor cell proliferation, migration, and invasion was enhanced (84, 85). Interestingly, miR-21 is significantly overexpressed in PTEN mutation-positive patients with Cowden syndrome (88).
It is reported that in addition, miR-486 is involved in enhancing PI3K/Akt signaling by repressing the expression of PTEN and Foxo1a, both negative components of the pathway (89). In PTC tumors, miR-486 is 5.1-fold down-regulated compared with unaffected tissue (our unpublished data), which suggests that miR-486 may have a dual, positive and negative impact on the Akt-signaling pathway. Indeed, miR-486 is predicted to target positive regulators of the Akt-signaling pathway, including IGF-I and p85α. Negative regulation of IGF-I and p85α by miR-486 would be predicted to counteract the positive influence on the PI3K/Akt-signaling pathway via inhibition of PTEN and Foxo1a (89).
Activated AKT is exported from the nucleus via interactions with chromosome region maintenance protein-1; the process is inhibited by AKT coactivating protein T-cell leukemia/lymphoma 1 (Tcl1) (90). Subcellular localization of AKT isoforms is a potentially important determinant of biological effects, as noted in the case of T-cell leukemias caused by TCL1 gene rearrangements leading to nuclear AKT accumulation and malignant transformation (90). Interestingly, the expression of Tcl1 is regulated by miR-29 and miR-181, the latter being 2-fold overexpressed in papillary thyroid tumors (45, 91). Cytoplasmic relocalization is also crucial for p27(Kip1), a key regulator of the cell cycle, that is inhibited by an activated AKT pathway (92–94). As shown recently p27(Kip1) is also inhibited by miR-221/222. These two miR are up-regulated (5-fold) in PTC, which leads to reduction in the protein level of p27(Kip1) in PTC tumors (95). Rapid activation of PI3K/AKT signaling by thyroid hormones creates a link between PTEN/PI3K/AKT signaling and the thyroid hormone receptor β (96).
MicroRNA and the thyroid hormone receptor β (THRB) tumor suppressor gene
The role of the THRB in tumorigenesis has been studied extensively. A mutant knock-in mouse (TRβPV) with a truncated THRB receptor has been generated (97). As TRβPV/PV mice age, they spontaneously develop thyroid carcinomas with a pathological progression similar to human follicular thyroid cancer (98). Growth stimulated by TSH, elevated in TRβPV mice, is a prerequisite but not sufficient factor for cancer to occur (99). Enhanced AKT activation was shown in primary and metastatic tumor tissue of TRβPV/PV animals (100). Hemizygous mice with one mutated THRB allele in the absence of the wild-type allele (TRβPV/− mouse) also develop thyroid carcinoma. Thus, a mutation of one THRB allele in the absence of the wild-type allele is sufficient to induce thyroid carcinoma. Loss of function of THRB alleles might occur as a result of somatic mutation or hypermethylation, as reported in liver cancer (101), kidney cancer (102), breast cancer (103), and thyroid cancer (104). A recent study showed that up-regulation of miR might well be an important mechanism of silencing of THRB (105). Interestingly, the 3′ untranslated region of THRB contains 14 binding sites for the top seven miR up-regulated in PTC (Fig. 2). Four of them (miR-221, miR-146a, miR-21, and miR-181a) were experimentally tested and proved to inhibit THRB mRNA and protein leading to the inhibition of T3-THRB-dependent genes (105). The THRB gene also is inhibited by miR-204 (106), which is highly deregulated in PTC (4-fold down-regulation). Down-regulation of miR-204 as opposed to up-regulation of other THRB-regulating miR suggests a highly complex T3-THRB-microRNA regulatory circuit in PTC.
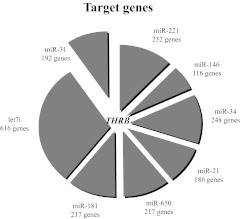
Fig. 2.
Target genes of microRNA deregulated in PTC tumors. The top seven microRNA up-regulated in PTC contain binding sites for the same one gene, namely THRB, as predicted by TargetScan software (P = 0.0000002). The size of every sector of the circle graph reflects the number of genes predicted to be regulated by a given microRNA. [Reproduced from K. Jazdzewski et al.: Thyroid hormone receptor β (THRB) is a major target gene for microRNAs deregulated in papillary thyroid carcinoma (PTC). J Clin Endocrinol Metab 96:E546–E553, 2011 (105), with permission. © The Endocrine Society.]
Summary
We outline in the introductory section why the strong genetic predisposition displayed by thyroid cancer may be due to low- or medium-penetrance gene mutations and predict that RNA genes such as miR may play a major role. Our review of the literature strongly supports this notion in that a polymorphism in one miR (miR-146a) predisposes to thyroid carcinoma, whereas numerous other miR are involved in signaling (mainly PTEN, PI3K, AKT) that is central to thyroid carcinogenesis. For thyroid carcinomas, the most abundant sequence variants have yet to be determined for all miR, and next-generation deep sequencing platforms seem to be a reliable method for that purpose.
Acknowledgments
This work was supported by National Cancer Institute Grants P30 CA16058 (to the Ohio State University Comprehensive Cancer Center) and P01 CA124570 (to Matthew D. Ringel, principal investigator, and A.d.l.C, leader of a project); Polish Ministry of Science and Higher Education Grants NN401 584838 and 0150/P01/2010/70 (to K.J.); and the Foundation for Polish Science (to K.J.).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AP-1
- Activator protein-1
- FTC
- follicular thyroid carcinoma
- PI3K
- phosphatidyl inositol 3-kinase
- PTC
- papillary thyroid carcinoma
- PTEN
- phosphatase and tensin homolog
- SNP
- single nucleotide polymorphism
- THRB
- thyroid hormone receptor β.
References
- 1. Adeniran AJ, Zhu Z, Gandhi M, Steward DL, Fidler JP, Giordano TJ, Biddinger PW, Nikiforov YE. 2006. Correlation between genetic alterations and microscopic features, clinical manifestations, and prognostic characteristics of thyroid papillary carcinomas. Am J Surg Pathol 30:216–222 [DOI] [PubMed] [Google Scholar]
- 2. Nikiforova MN, Lynch RA, Biddinger PW, Alexander EK, Dorn GW, 2nd, Tallini G, Kroll TG, Nikiforov YE. 2003. RAS point mutations and PAX8-PPAR γ rearrangement in thyroid tumors: evidence for distinct molecular pathways in thyroid follicular carcinoma. J Clin Endocrinol Metab 88:2318–2326 [DOI] [PubMed] [Google Scholar]
- 3. Goldgar DE, Easton DF, Cannon-Albright LA, Skolnick MH. 1994. Systematic population-based assessment of cancer risk in first-degree relatives of cancer probands. J Natl Cancer Inst 86:1600–1608 [DOI] [PubMed] [Google Scholar]
- 4. Dong C, Hemminki K. 2001. Modification of cancer risks in offspring by sibling and parental cancers from 2,112,616 nuclear families. Int J Cancer 92:144–150 [PubMed] [Google Scholar]
- 5. Risch N. 2001. The genetic epidemiology of cancer: interpreting family and twin studies and their implications for molecular genetic approaches. Cancer Epidemiol Biomarkers Prev 10:733–741 [PubMed] [Google Scholar]
- 6. Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. 2000. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 343:78–85 [DOI] [PubMed] [Google Scholar]
- 7. Hemminki K, Dong C. 2001. Population-based study of familial medullary thyroid cancer. Fam Cancer 1:45–49 [DOI] [PubMed] [Google Scholar]
- 8. Frich L, Glattre E, Akslen LA. 2001. Familial occurrence of nonmedullary thyroid cancer: a population-based study of 5673 first-degree relatives of thyroid cancer patients from Norway. Cancer Epidemiol Biomarkers Prev 10:113–117 [PubMed] [Google Scholar]
- 9. Hrafnkelsson J, Tulinius H, Jonasson JG, Olafsdottir G, Sigvaldason H. 1989. Papillary thyroid carcinoma in Iceland. A study of the occurrence in families and the coexistence of other primary tumours. Acta Oncol 28:785–788 [DOI] [PubMed] [Google Scholar]
- 10. Czene K, Lichtenstein P, Hemminki K. 2002. Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish Family-Cancer Database. Int J Cancer 99:260–266 [DOI] [PubMed] [Google Scholar]
- 11. Amundadottir LT, Thorvaldsson S, Gudbjartsson DF, Sulem P, Kristjansson K, Arnason S, Gulcher JR, Bjornsson J, Kong A, Thorsteinsdottir U, Stefansson K. 2004. Cancer as a complex phenotype: pattern of cancer distribution within and beyond the nuclear family. PLoS Med 1:e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boice JD., Jr 2006. Thyroid disease 60 years after Hiroshima and 20 years after Chernobyl. JAMA 295:1060–1062 [DOI] [PubMed] [Google Scholar]
- 13. Rahu M, Rahu K, Auvinen A, Tekkel M, Stengrevics A, Hakulinen T, Boice JD, Jr, Inskip PD. 2006. Cancer risk among Chernobyl cleanup workers in Estonia and Latvia, 1986–1998. Int J Cancer 119:162–168 [DOI] [PubMed] [Google Scholar]
- 14. Schonfeld SJ, Lee C, Berrington de González A. 2011. Medical exposure to radiation and thyroid cancer. Clin Oncol (R Coll Radiol) 23:244–250 [DOI] [PubMed] [Google Scholar]
- 15. Dal Maso L, Bosetti C, La Vecchia C, Franceschi S. 2009. Risk factors for thyroid cancer: an epidemiological review focused on nutritional factors. Cancer Causes Control 20:75–86 [DOI] [PubMed] [Google Scholar]
- 16. Jemal A, Siegel R, Xu J, Ward E. 2010. Cancer statistics, 2010. CA Cancer J Clin 60:277–300 [DOI] [PubMed] [Google Scholar]
- 17. Bodmer W, Bonilla C. 2008. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet 40:695–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. He H, Nagy R, Liyanarachchi S, Jiao H, Li W, Suster S, Kere J, de la Chapelle A. 2009. A susceptibility locus for papillary thyroid carcinoma on chromosome 8q24. Cancer Res 69:625–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gudmundsson J, Sulem P, Gudbjartsson DF, Jonasson JG, Sigurdsson A, Bergthorsson JT, He H, Blondal T, Geller F, Jakobsdottir M, Magnusdottir DN, Matthiasdottir S, Stacey SN, Skarphedinsson OB, Helgadottir H, Li W, Nagy R, Aguillo E, Faure E, Prats E, Saez B, Martinez M, Eyjolfsson GI, Bjornsdottir US, Holm H, Kristjansson K, Frigge ML, Kristvinsson H, Gulcher JR, Jonsson T, Rafnar T, Hjartarsson H, Mayordomo JI, de la Chapelle A, Hrafnkelsson J, Thorsteinsdottir U, Kong A, Stefansson K. 2009. Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nat Genet 41:460–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. 2008. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci USA 105:7269–7274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Volinia S, Galasso M, Costinean S, Tagliavini L, Gamberoni G, Drusco A, Marchesini J, Mascellani N, Sana ME, Abu Jarour R, Desponts C, Teitell M, Baffa R, Aqeilan R, Iorio MV, Taccioli C, Garzon R, Di Leva G, Fabbri M, Catozzi M, Previati M, Ambs S, Palumbo T, Garofalo M, Veronese A, Bottoni A, Gasparini P, Harris CC, Visone R, Pekarsky Y, de la Chapelle A, Bloomston M, Dillhoff M, Rassenti LZ, Kipps TJ, Huebner K, Pichiorri F, Lenze D, Cairo S, Buendia MA, Pineau P, Dejean A, Zanesi N, Rossi S, Calin GA, Liu CG, Palatini J, Negrini M, Vecchione A, Rosenberg A, Croce CM. 2010. Reprogramming of miRNA networks in cancer and leukemia. Genome Res 20:589–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fabbri M, Calin GA. 2010. Epigenetics and miRNAs in human cancer. Adv Genet 70:87–99 [DOI] [PubMed] [Google Scholar]
- 23. Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. 2008. MicroRNAs: new regulators of immune cell development and function. Nat Immunol 9:839–845 [DOI] [PubMed] [Google Scholar]
- 24. Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Croce CM, Calin GA. 2005. miRNAs, cancer, and stem cell division. Cell 122:6–7 [DOI] [PubMed] [Google Scholar]
- 26. Zhang B, Pan X, Cobb GP, Anderson TA. 2007. microRNAs as oncogenes and tumor suppressors. Dev Biol 302:1–12 [DOI] [PubMed] [Google Scholar]
- 27. Esquela-Kerscher A, Slack FJ. 2006. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 6:259–269 [DOI] [PubMed] [Google Scholar]
- 28. Nielsen CB, Shomron N, Sandberg R, Hornstein E, Kitzman J, Burge CB. 2007. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA 13:1894–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jazdzewski K, Liyanarachchi S, Swierniak M, Pachucki J, Ringel MD, Jarzab B, de la Chapelle A. 2009. Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc Natl Acad Sci USA 106:1502–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Santarpia L, Nicoloso M, Calin GA. 2010. MicroRNAs: a complex regulatory network drives the acquisition of malignant cell phenotype. Endocr Relat Cancer 17:F51–F75 [DOI] [PubMed] [Google Scholar]
- 31. Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433:769–773 [DOI] [PubMed] [Google Scholar]
- 32. Negrini M, Nicoloso MS, Calin GA. 2009. MicroRNAs and cancer—new paradigms in molecular oncology. Curr Opin Cell Biol 21:470–479 [DOI] [PubMed] [Google Scholar]
- 33. Lee RC, Feinbaum RL, Ambros V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854 [DOI] [PubMed] [Google Scholar]
- 34. Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. 2008. miRBase: tools for microRNA genomics. Nucleic Acids Res 36:D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meyers BC, Souret FF, Lu C, Green PJ. 2006. Sweating the small stuff: microRNA discovery in plants. Curr Opin Biotechnol 17:139–146 [DOI] [PubMed] [Google Scholar]
- 36. Pfeffer S, Lagos-Quintana M, Tuschl T. 2005. Cloning of small RNA molecules. Curr Protoc Mol Biol Chapter 26:Unit 26.24 [DOI] [PubMed] [Google Scholar]
- 37. Cummins JM, Velculescu VE. 2006. Implications of micro-RNA profiling for cancer diagnosis. Oncogene 25:6220–6227 [DOI] [PubMed] [Google Scholar]
- 38. Shendure J, Ji H. 2008. Next-generation DNA sequencing. Nat Biotechnol 26:1135–1145 [DOI] [PubMed] [Google Scholar]
- 39. Griffiths-Jones S. 2006. miRBase: the microRNA sequence database. Methods Mol Biol 342:129–138 [DOI] [PubMed] [Google Scholar]
- 40. Barbarotto E, Schmittgen TD, Calin GA. 2008. MicroRNAs and cancer: profile, profile, profile. Int J Cancer 122:969–977 [DOI] [PubMed] [Google Scholar]
- 41. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. 2005. MicroRNA expression profiles classify human cancers. Nature 435:834–838 [DOI] [PubMed] [Google Scholar]
- 42. Marcucci G, Radmacher MD, Maharry K, Mrózek K, Ruppert AS, Paschka P, Vukosavljevic T, Whitman SP, Baldus CD, Langer C, Liu CG, Carroll AJ, Powell BL, Garzon R, Croce CM, Kolitz JE, Caligiuri MA, Larson RA, Bloomfield CD. 2008. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med 358:1919–1928 [DOI] [PubMed] [Google Scholar]
- 43. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. 2002. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 99:15524–15529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. 2005. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA 102:3627–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, Kloos RT, Croce CM, de la Chapelle A. 2005. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA 102:19075–19080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu CG, Spizzo R, Calin GA, Croce CM. 2008. Expression profiling of microRNA using oligo DNA arrays. Methods 44:22–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pallante P, Visone R, Ferracin M, Ferraro A, Berlingieri MT, Troncone G, Chiappetta G, Liu CG, Santoro M, Negrini M, Croce CM, Fusco A. 2006. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer 13:497–508 [DOI] [PubMed] [Google Scholar]
- 48. Visone R, Pallante P, Vecchione A, Cirombella R, Ferracin M, Ferraro A, Volinia S, Coluzzi S, Leone V, Borbone E, Liu CG, Petrocca F, Troncone G, Calin GA, Scarpa A, Colato C, Tallini G, Santoro M, Croce CM, Fusco A. 2007. Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene 26:7590–7595 [DOI] [PubMed] [Google Scholar]
- 49. Weber F, Teresi RE, Broelsch CE, Frilling A, Eng C. 2006. A limited set of human microRNA is deregulated in follicular thyroid carcinoma. J Clin Endocrinol Metab 91:3584–3591 [DOI] [PubMed] [Google Scholar]
- 50. Pallante P, Visone R, Croce CM, Fusco A. 2010. Deregulation of microRNA expression in follicular-cell-derived human thyroid carcinomas. Endocr Relat Cancer 17:F91–104 [DOI] [PubMed] [Google Scholar]
- 51. Menon MP, Khan A. 2009. Micro-RNAs in thyroid neoplasms: molecular, diagnostic and therapeutic implications. J Clin Pathol 62:978–985 [DOI] [PubMed] [Google Scholar]
- 52. Sheu SY, Grabellus F, Schwertheim S, Worm K, Broecker-Preuss M, Schmid KW. 2010. Differential miRNA expression profiles in variants of papillary thyroid carcinoma and encapsulated follicular thyroid tumours. Br J Cancer 102:376–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. 2008. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 105:10513–10518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Calin GA, Croce CM. 2006. MicroRNA signatures in human cancers. Nat Rev Cancer 6:857–866 [DOI] [PubMed] [Google Scholar]
- 55. Mazeh H, Mizrahi I, Halle D, Ilyayev N, Stojadinovic A, Trink B, Mitrani-Rosenbaum S, Roistacher M, Ariel I, Eid A, Freund HR, Nissan A. 2011. Development of a microRNA-based molecular assay for the detection of papillary thyroid carcinoma in aspiration biopsy samples. Thyroid 21:111–118 [DOI] [PubMed] [Google Scholar]
- 56. Chen YT, Kitabayashi N, Zhou XK, Fahey TJ, 3rd, Scognamiglio T. 2008. MicroRNA analysis as a potential diagnostic tool for papillary thyroid carcinoma. Mod Pathol 21:1139–1146 [DOI] [PubMed] [Google Scholar]
- 57. Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE. 2008. MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab 93:1600–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chou CK, Chen RF, Chou FF, Chang HW, Chen YJ, Lee YF, Yang KD, Cheng JT, Huang CC, Liu RT. 2010. miR-146b is highly expressed in adult papillary thyroid carcinomas with high risk features including extrathyroidal invasion and the BRAF(V600E) mutation. Thyroid 20:489–494 [DOI] [PubMed] [Google Scholar]
- 59. Iwai N, Naraba H. 2005. Polymorphisms in human pre-miRNAs. Biochem Biophys Res Commun 331:1439–1444 [DOI] [PubMed] [Google Scholar]
- 60. Saunders MA, Liang H, Li WH. 2007. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci USA 104:3300–3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gottwein E, Cai X, Cullen BR. 2006. A novel assay for viral microRNA function identifies a single nucleotide polymorphism that affects drosha processing. J Virol 80:5321–5326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Duan R, Pak C, Jin P. 2007. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet 16:1124–1131 [DOI] [PubMed] [Google Scholar]
- 63. Nicoloso MS, Sun H, Spizzo R, Kim H, Wickramasinghe P, Shimizu M, Wojcik SE, Ferdin J, Kunej T, Xiao L, Manoukian S, Secreto G, Ravagnani F, Wang X, Radice P, Croce CM, Davuluri RV, Calin GA. 2010. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res 70:2789–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wojcik SE, Rossi S, Shimizu M, Nicoloso MS, Cimmino A, Alder H, Herlea V, Rassenti LZ, Rai KR, Kipps TJ, Keating MJ, Croce CM, Calin GA. 2010. Non-coding RNA sequence variations in human chronic lymphocytic leukemia and colorectal cancer. Carcinogenesis 31:208–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Krol J, Sobczak K, Wilczynska U, Drath M, Jasinska A, Kaczynska D, Krzyzosiak WJ. 2004. Structural features of microRNA (miRNA) precursors and their relevance to miRNA biogenesis and small interfering RNA/short hairpin RNA design. J Biol Chem 279:42230–42239 [DOI] [PubMed] [Google Scholar]
- 66. Starega-Roslan J, Krol J, Koscianska E, Kozlowski P, Szlachcic WJ, Sobczak K, Krzyzosiak WJ. 2011. Structural basis of microRNA length variety. Nucleic Acids Res 39:257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jazdzewski K, de la Chapelle A. 2009. Genomic sequence matters: a SNP in microRNA-146a can turn anti-apoptotic. Cell Cycle 8:1642–1643 [PMC free article] [PubMed] [Google Scholar]
- 68. Yang JS, Phillips MD, Betel D, Mu P, Ventura A, Siepel AC, Chen KC, Lai EC. 2011. Widespread regulatory activity of vertebrate microRNA species. RNA 17:312–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Guo L, Lu Z. 2010. The fate of miRNA strand through evolutionary analysis: implication for degradation as merely carrier strand or potential regulatory molecule? PLoS ONE 5:e11387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hu Z, Liang J, Wang Z, Tian T, Zhou X, Chen J, Miao R, Wang Y, Wang X, Shen H. 2009. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum Mutat 30:79–84 [DOI] [PubMed] [Google Scholar]
- 71. Shen J, Ambrosone CB, DiCioccio RA, Odunsi K, Lele SB, Zhao H. 2008. A functional polymorphism in the miR-146a gene and age of familial breast/ovarian cancer diagnosis. Carcinogenesis 29:1963–1966 [DOI] [PubMed] [Google Scholar]
- 72. Xu T, Zhu Y, Wei QK, Yuan Y, Zhou F, Ge YY, Yang JR, Su H, Zhuang SM. 2008. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis 29:2126–2131 [DOI] [PubMed] [Google Scholar]
- 73. Chen K, Song F, Calin GA, Wei Q, Hao X, Zhang W. 2008. Polymorphisms in microRNA targets: a gold mine for molecular epidemiology. Carcinogenesis 29:1306–1311 [DOI] [PubMed] [Google Scholar]
- 74. Mishra PJ, Mishra PJ, Banerjee D, Bertino JR. 2008. MiRSNPs or MiR-polymorphisms, new players in microRNA mediated regulation of the cell: introducing microRNA pharmacogenomics. Cell Cycle 7:853–858 [DOI] [PubMed] [Google Scholar]
- 75. Sun G, Yan J, Noltner K, Feng J, Li H, Sarkis DA, Sommer SS, Rossi JJ. 2009. SNPs in human miRNA genes affect biogenesis and function. RNA 15:1640–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Morozova O, Marra MA. 2008. Applications of next-generation sequencing technologies in functional genomics. Genomics 92:255–264 [DOI] [PubMed] [Google Scholar]
- 77. Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, Eaves CJ, Marra MA. 2008. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 18:610–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li SC, Chan WC, Ho MR, Tsai KW, Hu LY, Lai CH, Hsu CN, Hwang PP, Lin WC. 2010. Discovery and characterization of medaka miRNA genes by next generation sequencing platform. BMC Genomics 11(Suppl 4):S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129:1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. 2007. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep 8:763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Shinohara M, Chung YJ, Saji M, Ringel MD. 2007. AKT in thyroid tumorigenesis and progression. Endocrinology 148:942–947 [DOI] [PubMed] [Google Scholar]
- 82. Ringel MD, Hayre N, Saito J, Saunier B, Schuppert F, Burch H, Bernet V, Burman KD, Kohn LD, Saji M. 2001. Overexpression and overactivation of Akt in thyroid carcinoma. Cancer Res 61:6105–6111 [PubMed] [Google Scholar]
- 83. Bruni P, Boccia A, Baldassarre G, Trapasso F, Santoro M, Chiappetta G, Fusco A, Viglietto G. 2000. PTEN expression is reduced in a subset of sporadic thyroid carcinomas: evidence that PTEN-growth suppressing activity in thyroid cancer cells mediated by p27kip1. Oncogene 19:3146–3155 [DOI] [PubMed] [Google Scholar]
- 84. Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. 2007. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133:647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Talotta F, Cimmino A, Matarazzo MR, Casalino L, De Vita G, D'Esposito M, Di Lauro R, Verde P. 2009. An autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1 activity in RAS transformation. Oncogene 28:73–84 [DOI] [PubMed] [Google Scholar]
- 86. Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. 2006. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 103:2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, Iba H. 2008. miR-21 gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol 378:492–504 [DOI] [PubMed] [Google Scholar]
- 88. Pezzolesi MG, Platzer P, Waite KA, Eng C. 2008. Differential expression of PTEN-targeting microRNAs miR-19a and miR-21 in Cowden syndrome. Am J Hum Genet 82:1141–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Small EM, O'Rourke JR, Moresi V, Sutherland LB, McAnally J, Gerard RD, Richardson JA, Olson EN. 2010. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc Natl Acad Sci USA 107:4218–4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Pekarsky Y, Koval A, Hallas C, Bichi R, Tresini M, Malstrom S, Russo G, Tsichlis P, Croce CM. 2000. Tcl1 enhances Akt kinase activity and mediates its nuclear translocation. Proc Natl Acad Sci USA 97:3028–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V, Volinia S, Alder H, Liu CG, Rassenti L, Calin GA, Hagan JP, Kipps T, Croce CM. 2006. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res 66:11590–11593 [DOI] [PubMed] [Google Scholar]
- 92. Viglietto G, Motti ML, Bruni P, Melillo RM, D'Alessio A, Califano D, Vinci F, Chiappetta G, Tsichlis P, Bellacosa A, Fusco A, Santoro M. 2002. Cytoplasmic relocalization and inhibition of the cyclin-dependent kinase inhibitor p27(Kip1) by PKB/Akt-mediated phosphorylation in breast cancer. Nat Med 8:1136–1144 [DOI] [PubMed] [Google Scholar]
- 93. Liang J, Zubovitz J, Petrocelli T, Kotchetkov R, Connor MK, Han K, Lee JH, Ciarallo S, Catzavelos C, Beniston R, Franssen E, Slingerland JM. 2002. PKB/Akt phosphorylates p27, impairs nuclear import of p27 and opposes p27-mediated G1 arrest. Nat Med 8:1153–1160 [DOI] [PubMed] [Google Scholar]
- 94. Shin I, Yakes FM, Rojo F, Shin NY, Bakin AV, Baselga J, Arteaga CL. 2002. PKB/Akt mediates cell-cycle progression by phosphorylation of p27(Kip1) at threonine 157 and modulation of its cellular localization. Nat Med 8:1145–1152 [DOI] [PubMed] [Google Scholar]
- 95. Visone R, Russo L, Pallante P, De Martino I, Ferraro A, Leone V, Borbone E, Petrocca F, Alder H, Croce CM, Fusco A. 2007. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer 14:791–798 [DOI] [PubMed] [Google Scholar]
- 96. Cao X, Kambe F, Moeller LC, Refetoff S, Seo H. 2005. Thyroid hormone induces rapid activation of Akt/protein kinase B-mammalian target of rapamycin-p70S6K cascade through phosphatidylinositol 3-kinase in human fibroblasts. Mol Endocrinol 19:102–112 [DOI] [PubMed] [Google Scholar]
- 97. Cheng SY. 2000. Multiple mechanisms for regulation of the transcriptional activity of thyroid hormone receptors. Rev Endocr Metab Disord 1:9–18 [DOI] [PubMed] [Google Scholar]
- 98. Kaneshige M, Kaneshige K, Zhu X, Dace A, Garrett L, Carter TA, Kazlauskaite R, Pankratz DG, Wynshaw-Boris A, Refetoff S, Weintraub B, Willingham MC, Barlow C, Cheng S. 2000. Mice with a targeted mutation in the thyroid hormone β receptor gene exhibit impaired growth and resistance to thyroid hormone. Proc Natl Acad Sci USA 97:13209–13214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lu C, Zhao L, Ying H, Willingham MC, Cheng SY. 2010. Growth activation alone is not sufficient to cause metastatic thyroid cancer in a mouse model of follicular thyroid carcinoma. Endocrinology 151:1929–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kim CS, Vasko VV, Kato Y, Kruhlak M, Saji M, Cheng SY, Ringel MD. 2005. AKT activation promotes metastasis in a mouse model of follicular thyroid carcinoma. Endocrinology 146:4456–4463 [DOI] [PubMed] [Google Scholar]
- 101. Lin KH, Shieh HY, Chen SL, Hsu HC. 1999. Expression of mutant thyroid hormone nuclear receptors in human hepatocellular carcinoma cells. Mol Carcinog 26:53–61 [DOI] [PubMed] [Google Scholar]
- 102. Kamiya Y, Puzianowska-Kuznicka M, McPhie P, Nauman J, Cheng SY, Nauman A. 2002. Expression of mutant thyroid hormone nuclear receptors is associated with human renal clear cell carcinoma. Carcinogenesis 23:25–33 [DOI] [PubMed] [Google Scholar]
- 103. Li Z, Meng ZH, Chandrasekaran R, Kuo WL, Collins CC, Gray JW, Dairkee SH. 2002. Biallelic inactivation of the thyroid hormone receptor β1 gene in early stage breast cancer. Cancer Res 62:1939–1943 [PubMed] [Google Scholar]
- 104. Puzianowska-Kuznicka M, Krystyniak A, Madej A, Cheng SY, Nauman J. 2002. Functionally impaired TR mutants are present in thyroid papillary cancer. J Clin Endocrinol Metab 87:1120–1128 [DOI] [PubMed] [Google Scholar]
- 105. Jazdzewski K, Boguslawska J, Jendrzejewski J, Liyanarachchi S, Pachucki J, Wardyn KA, Nauman A, de la Chapelle A. 2011. Thyroid hormone receptor β (THRB) is a major target gene for microRNAs deregulated in papillary thyroid carcinoma (PTC). J Clin Endocrinol Metab 96:E546–E553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Master A, Wójcicka A, Piekiełko-Witkowska A, Bogusławska J, Popławski P, Tañski Z, Darras VM, Williams GR, Nauman A. 2010. Untranslated regions of thyroid hormone receptor β1 mRNA are impaired in human clear cell renal cell carcinoma. Biochim Biophys Acta 1802:995–1005 [DOI] [PubMed] [Google Scholar]