Abstract
MicroRNAs (miRNAs) regulate most cellular functions, acting by posttranscriptionally repressing numerous eukaryotic mRNAs. They lead to translational repression, deadenylation and degradation of their target mRNAs. Yet, the relative contributions of these effects are controversial and little is known about the sequence of events occurring during the miRNA-induced response. Using stable human cell lines expressing inducible reporters, we found that translational repression is the dominant effect of miRNAs on newly synthesized targets. This step is followed by mRNA deadenylation and decay, which is the dominant effect at steady state. Our findings have important implications for understanding the mechanism of silencing and reconcile seemingly contradictory data.
Keywords: miRNA, translational control, mRNA decay, argonaute
Introduction
MicroRNAs (miRNAs) are ∼21-nt-long regulatory RNAs operating in most eukaryotes. They function in the form of a ribonucleoprotein complex termed miRISC, of which the Argonaute (AGO) and GW182 proteins are the key constituents. Bound to AGO, miRNAs serve as guides bringing miRISC to target mRNAs by hybridizing, generally through imperfect base pairing, with their 3′-untranslated regions (3′UTRs) [1, 2]. GW182 proteins (named TNRC6 in vertebrates), acting downstream of AGOs, recruit the CCR4–NOT deadenylation complex, resulting in the downregulation of mRNA function [3–5]. It is widely agreed that miRNAs induce the deadenylation and decay of target mRNAs. Yet, much evidence exists that miRNAs also induce translational repression independent of mRNA deadenylation and decay (reviewed in [2, 6]). Although considerable progress has been made towards the elucidation of the mechanism of miRNA action, the relative contributions of mRNA decay and translational inhibition to miRNA-mediated silencing remain controversial. In particular, recent genome-wide studies using microarrays, proteomics and/or ribosome profiling have indicated for a majority of mRNA targets that mRNA decay accounts for most of the observed repression of protein output [7–10]. Yet, in these studies, analyses were performed at steady state and, thus, could not assign a possible sequence of repressive events and may underestimate the contribution of translational repression. Indeed, it may be necessary to first inhibit translation of a target mRNA to allow its deadenylation and subsequent decay. Consistently, studies performed with cell extracts indicated that miRNA-induced translational repression of an artificial reporter may precede mRNA deadenylation [11]. Yet, it is not clear how far such a system, in which an in vitro transcribed RNA is added to a cell lysate, faithfully reproduces the steps leading to miRNA-mediated silencing. Two studies using transiently transfected mammalian cells showed that miRNAs induce rapid deadenylation of target mRNA reporters; but the contribution of translational inhibition to this process was not addressed [12, 13]. Moreover, miRNA-mediated repression may be affected by the nuclear history of target mRNAs [14], subcellular localization of the miRISC [15, 16], or the choice of cell transfection technique [17], suggesting that in vitro extracts and transiently transfected cells may not recapitulate all physiological effects of miRNAs.
In this study, we used stable HeLa cell lines expressing inducible miRNA-targeted reporters to monitor the contributions of translational repression and mRNA decay over time. We found that, at steady state, reporters fused to 3′-untranslated regions (3′UTRs) of endogenous miRNA targets may be repressed at both the translational and mRNA stability levels. We demonstrate that translational repression precedes mRNA decay and that initial steps of translational inhibition are not due to appreciable mRNA deadenylation.
Results and discussion
Stable cell lines expressing inducible miRNA targets
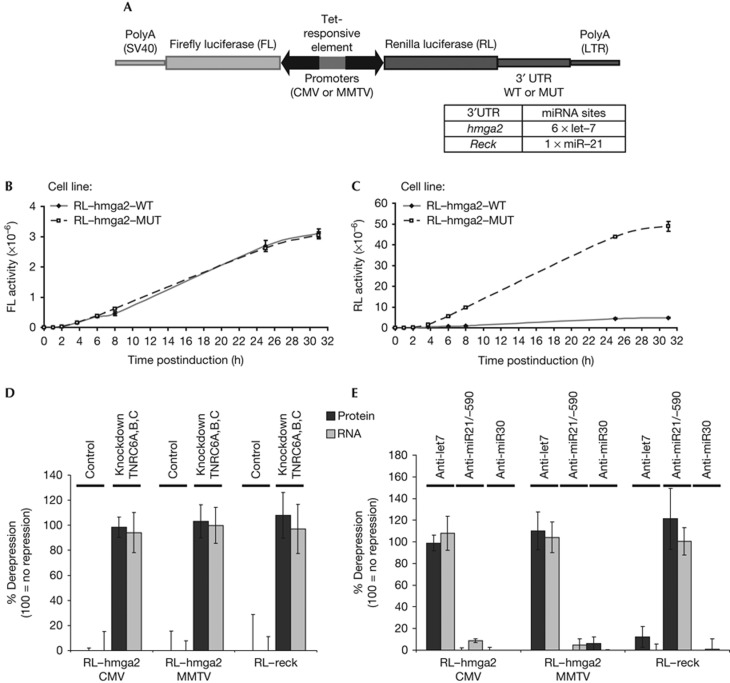
To address the sequence of events occurring during miRNA-mediated repression, we created stable HeLa cell lines in which expression of miRNA target reporters can be induced with doxycycline. The reporters, (i) a Renilla luciferase (RL) fused to 3′UTRs of validated human miRNA targets, representing a test mRNA, and (ii) a firefly luciferase (FL) used as a normalization control, were placed back-to-back under the control of the same tetracycline-responsive element (Fig 1A). We used the 3′UTRs of hmga2, a target of let7 miRNA containing six let7 binding sites [18], and of reck, a target of miR21 containing a single miRNA-binding site [19]. Cell lines were generated expressing reporters bearing either wild-type (WT) 3′UTR sequences or mutated versions thereof (MUT) containing mutations in the miRNA-binding sites. For the RL–hmga2 reporters, two sets of cell lines were generated: in one, expression was driven by cauliflower mosaic virus (CMV) promoters and, in a second, by ∼100-fold weaker mouse mammary tumour virus promoters. The WT and MUT pairs of different cell lines were directly comparable as the transgenes were directionally integrated as single copies at the same locus using a recently published strategy [20]. Indeed, on induction, expression of the control FL reporter was virtually identical in independent cell lines (Fig 1B; supplementary Fig S1 online). As expected, RL fused to WT 3′UTRs of miRNA targets showed a lower expression than mutants thereof (Fig 1C; supplementary Fig S1 online). Consistent with the effect being mediated by endogenous miRNAs, repression was relieved on knockdown of the three human TNRC6 proteins, which are necessary for miRNA-mediated silencing [1, 2] (Fig 1D), or on transfection of anti-miR oligonucleotides complementary to the targeting but not control miRNAs (Fig 1E).
Figure 1.
Construction and characterization of inducible cell lines. (A) Scheme of the genome-integrated reporters. RL miRNA-target reporters and a FL control reporter were cloned back-to-back under the control of the same tetracycline (tet)-responsive element. The indicated 3′UTRs in their wild-type (WT) form or containing mutated (MUT) miRNA-binding sites were fused to the RL coding sequence. Polyadenylation sites (SV40 or a 3′ long terminal repeat, LTR) are indicated. (B,C) Time course of RL and FL expression. CMV–RL–hmga2–WT and –MUT cell lines were seeded at the same density and induced for the indicated times. At each time point, the same number of cells was used for dual luciferase assay. Control FL reporters expressed in WT or MUT cell lines yielded similar readings (B), while the WT RL–hmga2 reporter was expressed at lower levels than the MUT RL–hmga2 reporter (C). Luciferase activity is expressed in arbitrary units. (D,E) Specificity of repression. Repression (ratio of normalized RL–hmga2–MUT over WT activities or mRNA levels) of the WT reporters was relieved on knockdown of the three TNRC6 proteins (D) or transfection of 2′-O-methyl oligonucleotides complementary to their targeting miRNA. As miR21 and miR590 share the same seed sequence, they were both targeted with specific anti-mirs (E). After 48 h transfection, cell lines were induced for 4 h before analysis. 0% indicates no effect on repression, 100%, no repression observed. Throughout this study, error bars represent 95% confidence intervals from at least three independent experiments done in duplicates. CMV, cauliflower mosaic virus; MMTV, mouse mammary tumour virus; 3′-UTR, 3′-untranslated region.
Dynamics of repression of RL–hmga2 reporters
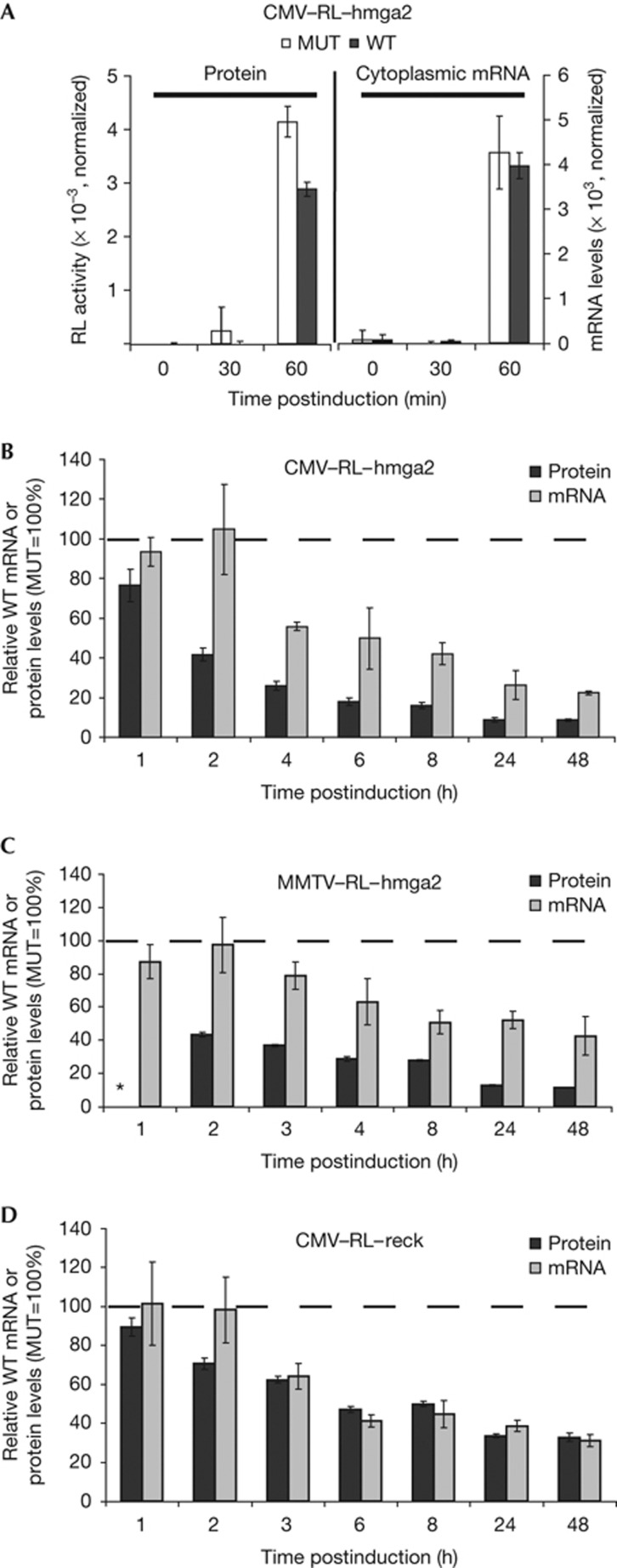
Using the cell lines, we analysed the dynamics of let7-mediated repression of RL–hmga2. First, we defined the earliest time point at which appreciable levels of luciferase activity could be detected. Monitoring expression of the reporters every 30 min postinduction, we could reliably measure RL and FL activities starting from 60 min postinduction (Fig 2A). This time point likely corresponded to early translation events following mRNA export, as it correlated with the appearance of reporter mRNA in the cytoplasmic fraction.
Figure 2.
Translational repression precedes mRNA decay. (A) Detection of the earliest measureable protein and cytoplasmic reporter mRNA. CMV–RL–hmga2–WT and –MUT cell lines were induced for the indicated times. Cytoplasmic extracts were then used for luciferase and RNA analysis. The earliest detectable cytoplasmic RL–hmga2 mRNA and RL protein appeared at 60-min postinduction. RNA levels are normalized to glyceraldehyde-3-phosphate dehydrogenase. RL activity was adjusted to FL values. Background values at t=0 were subtracted. (B–D) Relative expression of wild-type RL reporters mRNA and protein over time. CMV–RL–hmga2–WT and –MUT (B), MMTV–RL–hmga2–WT and –MUT (C), and CMV–RL–reck–WT and –MUT (D) cell lines were induced for the times indicated. RL activity and cytoplasmic mRNA level (normalized to corresponding FL values) of the WT reporters are expressed as percentages of values for the matching MUT reporter, which are set to 100% at each time point (broken line). In (C), the asterisk at 1 h indicates that protein levels were too low for reliable measurements. CMV, cauliflower mosaic virus; FL, firefly luciferase; MMTV, mouse mammary tumour virus; MUT, mutated; RL, Renilla luciferase; 3′-UTR, 3′-untranslated region; WT, wild type.
Next, expression of the reporters at both protein and cytoplasmic mRNA levels was analysed over 48 h, starting 1-h postinduction. RNA levels were analysed by reverse transcription quantitative PCR; linearity and accuracy of measurements were good enough to assess subtle changes (supplementary Fig S2 online). Activity and mRNA levels (both normalized to respective FL values) of the RL–hmga2–WT reporters were calculated relative to those of the matching MUT reporters, which were set at each time point to 100% (Fig 2, accumulation of the RL reporters over time is shown in supplementary Fig S3 online). This representation of the data makes it possible to directly assess at any given time point the contribution of mRNA decay to the observed repression at the protein level. Repression of the RL–hmga2 reporters, at both the protein and mRNA levels, increased over time (Fig 2B,C). Interestingly, at early time points up to 2-h postinduction, mRNA levels for the WT RL–hmga2 reporters matched those of the MUT reporters. In contrast, RL protein outputs were already markedly lower for the WT reporters. This indicates that miRNA-mediated repression starts shortly after mRNA export and first acts at the translational level, without inducing mRNA decay. Later, starting at 3-h postinduction, a marked effect on mRNA levels was also observed (Fig 2B,C), suggesting that miRNA-triggered mRNA decay occurs after translation has been inhibited. At these time points, protein levels were more repressed than mRNA levels, indicating that silencing of the reporters is due to a combination of translational inhibition and mRNA decay. Although at later time points the contribution of mRNA decay to the observed repression was higher for CMV-driven than mouse mammary tumour virus-driven transcripts, the effects were qualitatively similar and thus independent of the reporter expression level (Fig 2B,C).
Dynamics of RL–reck reporter repression
Similar kinetic analysis was performed for the RL–reck reporter (Fig 2D). As for the hmga2 reporters, no significant effect onRL–reck mRNA levels was observed within the first 2 h, while the protein output of the WT reporter was already diminished. At later time points, the relative levels of RL protein and mRNA decreased in parallel, indicating that, for this 3′UTR, mRNA decay is fully responsible for the lower protein output at steady state. Altogether, we show that miRNA-mediated repression is readily detectable early postinduction, with translational repression being the dominant mechanism for newly synthesized mRNA targets within the first hours of their lifetime. Thereafter, mRNA decay begins and becomes the main effect at steady state, explaining fully (RL–reck reporter) or partially (RL–hmga2 reporters) the miRNA-mediated repression we observe at equilibrium. Consequently, analyses of miRNA targets at steady state, 8- to 48-h posttransfection as performed in recent genome-wide studies [7–10], will underestimate the contribution of the translational component to the repression.
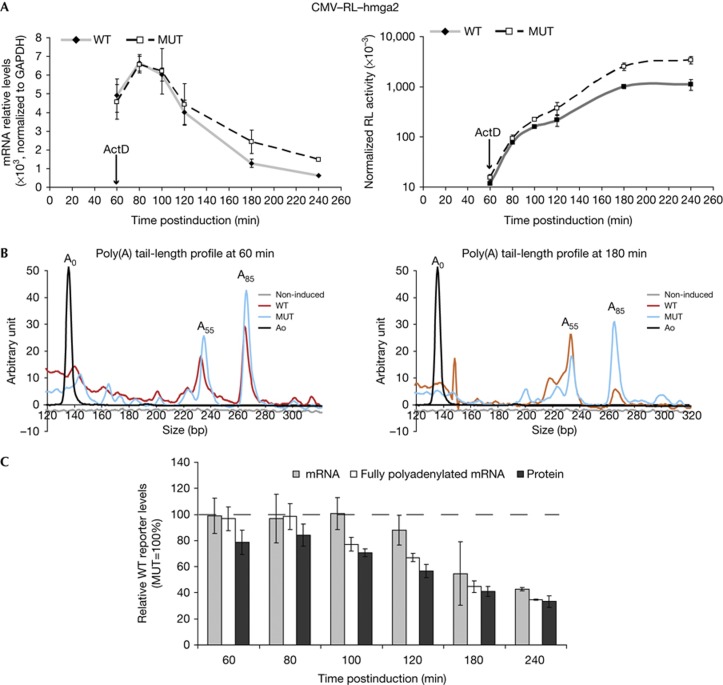
Translational inhibition precedes poly(A) tail shortening
MiRNA-induced repression of protein expression observed early postinduction, when mRNA levels are unaffected, may be explained by an effect on translation itself or, alternatively, by mRNA deadenylation, which may lead to decreased translation efficiency [21]. To distinguish the two possibilities, we analysed the length of the reporters poly(A) tails over time. To this end, we used a transcription pulse-chase approach to monitor a population of mRNAs undergoing largely synchronous expression and decay. HeLa cell lines expressing the CMV–RL–hmga2 reporters were induced for 1 h, transcription was then stopped by adding actinomycin D (ActD), and expression of reporters was analysed over a 3-h period. As shown in Fig 3A, whereas decreased protein expression from the WT reporter was observed already at the earliest time points, cytoplasmic mRNA levels of WT and MUT reporters were similar over the first 100 min following the pulse of transcription. Thereafter, the WT RL–hmga2 mRNA decayed more rapidly than its MUT version.
Figure 3.
Translational repression precedes mRNA deadenylation. (A) Expression of RL reporters after a 1-h transcription pulse. CMV–RL–hmga2–WT and –MUT cell lines were induced for 1 h. Transcription was then stopped with actinomycin D (ActD, arrow) and reporters analysed at the indicated time points for mRNA (left, normalized to GAPDH) and RL activity (right, adjusted for FL values, log10 scale). (B) Estimation of poly(A) tail lengths by PCR analysis after polyG/I extension. Ao was obtained with reporter-specific primers amplifying a region upstream of the polyadenylation site. To measure poly(A) tail lengths, the reverse specific primer was replaced by a primer annealing to the poly(A)-polyG/I junction. Sizing of the PCR products was performed on a microfluidic chip. Poly(A) tail lengths estimated at different time points are indicated. (C) Relative levels of WT RL protein, mRNA, and fully polyadenylated mRNA after a 1-h transcription pulse. At each time point, RL activity and cytoplasmic mRNA level of the RL–hmga2–WT reporter are expressed as percentages of values for the matching MUT reporter. In addition, cytoplasmic RNA was fractionated according to poly(A) tail length (see text and supplementary Fig S4 online). The amount of mRNA present in the fraction containing reporters with the longest poly(A) tails (fraction E4 in supplementary Fig S4 online, termed ‘fully polyadenylated mRNA’) was calculated for both WT and MUT reporters. The percentage of fully polyadenylated WT reporter, relative to MUT reporter is indicated. The broken line at 100% indicates levels of the matching MUT (non-repressed) reporter (set to 100% at each time point). CMV, cauliflower mosaic virus; FL, firefly luciferase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MUT, mutated; RL, Renilla luciferase; WT, wild type.
Poly(A) tail length of the WT and MUT reporters was qualitatively estimated using a polyG/I extension procedure followed by PCR analysis [4, 22]. PCR products indicative of poly(A) tail lengths were analysed with a high-resolution microfluidic chip (Fig 3B). Poly(A) tail-length profiles for WT and MUT reporters were similar at 60-min postinduction, with two major peaks corresponding to A85 and A55 (most probably representing fully polyadenylated reporter and a deadenylation intermediate), indicating that initial repression of the WT reporter is not due to stimulated deadenylation. By contrast, at 180 min, the peak corresponding to A85 was significantly decreased for the WT reporter, indicating accelerated deadenylation. To quantitatively estimate the amounts of fully polyadenylated RL–hmga2 reporters over time, cytoplasmic RNA purified from the cell lines was fractionated on an oligo(dT) matrix using a decreasing salt gradient (supplementary Fig S4 online), following an established procedure [23]. No apparent difference in poly(A) tail-length distribution was observed between WT and MUT reporters at early times up to 80- min postinduction, when reporter mRNA levels were also unchanged (Fig 3C). At later time points, the fraction of fully polyadenylated WT reporter gradually decreased, suggesting that deadenylation and ensuing mRNA decay potentiate the repressive effect of miRNA.
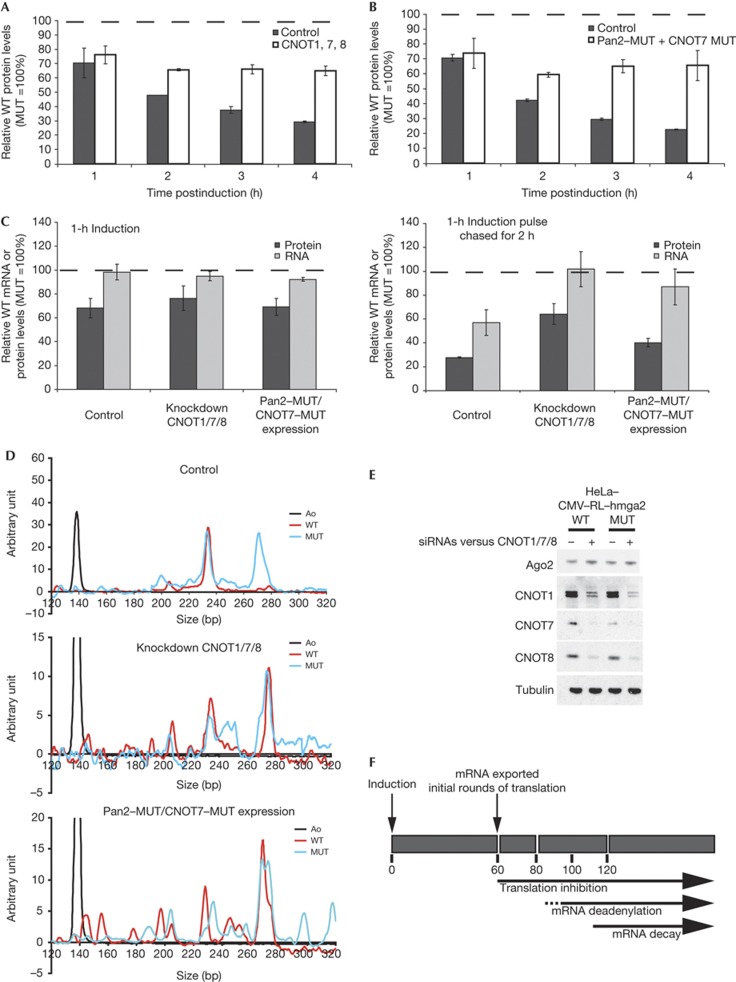
Analyses were also performed under deadenylation-impaired conditions, either by simultaneous knockdown of CNOT1 (protein responsible for the recruitment of the CCR4–NOT deadenylation complex by miRISC [3–5]), and the deadenylases CNOT7 and CNOT8, or overexpression of dominant-negative mutants of PAN2 and CNOT7, which were shown to efficiently block miRNA-mediated deadenylation of reporters [12]. Neither of the treatments affected the initial ∼30% repression of the hmga2–WT reporter at 1-h postinduction. Strikingly, while repression increased over time in the control experiments, it stayed unchanged at the initial 30% level when deadenylation was impaired (Fig 4A,B). Using the transcription pulse-chase approach, we confirmed that, on blockage of deadenylation, mRNA levels (Fig 4C) and poly(A) tail length (Fig 4D) of the WT reporter did not decrease over time when compared to the MUT counterpart. This indicates that the knockdown efficiencies (Fig 4E) and effects of the dominant-negative mutants were strong enough to efficiently impair deadenylation and thus suppress the mRNA decay component of the miRNA-mediated repression. Taken together, the data strongly indicate that the initial miRNA-mediated translation inhibition precedes mRNA deadenylation and is independent of it.
Figure 4.
Translational repression is independent of deadenylation. (A,B) Relative expression of RL–hmga2–WT protein over time. CMV–RL–hmga2–WT and –MUT cell lines were induced for the times indicated under control or deadenylation-impaired conditions. WT RL activity is presented as in Fig 2. (C) Expression of RL reporters after a 1 h transcription pulse. CMV–RL–hmga2–WT and –MUT cell lines were induced for 1 h. Transcription was then stopped with actinomycin D and reporters analysed at the indicated time points. (D) Poly(A) tail-length profiles of the RL reporters after a 1 h transcription pulse, chased for 2 h as in (C). (E) Analysis of knockdown efficiencies. Cell lines were transfected with siRNAs against CNOT1, 7, 8. Cytoplasmic extracts were analysed by western blot. The control proteins Ago2 and tubulin were not affected. (F) Sequence of events leading to miRNA-mediated repression as observed at steady state. The dashed line indicates that mRNA deadenylation may already start at 80 min postinduction. CMV, cauliflower mosaic virus; MUT, mutated; RL, Renilla luciferase; siRNAs, small interfering RNAs; WT, wild type.
A unifying model for miRNA-mediated repression
While genome-wide studies proposed that miRNA-mediated repression mainly leads to mRNA decay at steady state [7–10], other reports describe situations in which it can be rapidly reversed in response to different cellular cues [24–26]. Reversible silencing may be of particular importance in cells such as neurons, where localized translation at dendritic spines responds to synaptic stimulation [27]; and requires that target mRNAs are repressed translationally, without major mRNA decay as reported [24–26]. This apparent discrepancy may be explained by mechanistic differences between different cell types and/or miRNAs investigated, or by the presence of regulatory factors that modulate the miRNA effects in a target-specific way.
Our present work, combined with published data, supports the second possibility with a model that reconciles seemingly contradictory observations on the outcome of miRNA-mediated silencing (Fig 4F). We propose that the initial effect of miRNAs is inhibition of translation, most probably at the initiation step (reviewed in [2, 6]), without mRNA decay (step 1). This is followed by increased mRNA deadenylation (step 2), possibly as a consequence of the initial translation inhibition that may render the poly(A) tail more accessible. Alternatively, deadenylation may happen independently of the initial translational block but at a slower rate. The stimulated deadenylation, catalysed by PAN2–PAN3 and CCR4–NOT complexes [12] that are directly recruited by the miRISC [3–5], may potentiate the effect on translational inhibition and lead to decay of target mRNAs (step 3) through the recruitment of decapping machinery [12, 28]. This model, similar to another proposed recently [6], involves three distinct and successive major steps, and thus implies the possibility of reversibility or stalling at the transition from one step to the next. In most cases, miRNA-mediated repression may proceed through the three steps and lead to mRNA decay. In examples when miRNA-silencing is reversible or no pronounced mRNA decay is observed, repression may be stalled at step 1 and relieved in response to an appropriate stimulus. Interestingly, it was found recently that the CCR4–NOT complex may be involved not only in the deadenylation but also in the translational inhibition of miRNA targets, as it represses poly(A)-free RNA reporters without concomitant decay [3, 4]. This complex might thus be a key player in regulating transition from step 1 (translation inhibition) to step 2 (deadenylation) of our model. In future, it will be important to identify signals in the mRNA 3′UTRs and factors that may regulate transition from one silencing stage to another, and thus control the final outcome of miRNA-mediated repression of different mRNAs.
During revision of our manuscript, two papers reported that also in zebrafish and Drosophila S2 cells, translational repression precedes mRNA deadenylation and decay [29, 30].
Methods
Cell culture and reporter assays. HeLa cell lines were generated and characterized as described in [20]. Cytoplasmic extracts were prepared, as in [12], using cytoplasmic lysis buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA and 0.5% NP-40). Luciferase activities and mRNA levels were determined by standard procedures using the Dual luciferase reporter assay system (Promega) and Transcriptor first-strand cDNA synthesis kit (Roche) followed by quantitative PCR. Procedures are detailed in the supplementary information online.
Poly(A) tail-length measurement. Fractionation of mRNAs bearing poly(A) tails of different lengths was performed according to [23] and is detailed in the supplementary information online. RNA from all collected fractions was precipitated with 10 μg of yeast tRNA and 15 μg glycoblue (Ambion). Samples were analysed by reverse transcription quantitative PCR using random hexamers for the cDNA synthesis step.
Supplementary Material
Acknowledgments
We thank G. Meister, A. Krichevsky, A.B. Shyu and K. Schönig for sharing reagents. We thank P. King for editing the manuscript, and M. Chekulaeva for discussions. J.B. is the recipient of postdoctoral fellowships from the German Research Foundation (DFG) and the Peter and Traudl Engelhorn Foundation. The Friedrich Miescher Institute is supported by the Novartis Research Foundation.
Author contributions: J.B. designed and performed experiments, analysed the data, wrote the manuscript. C.A.-R. performed experiments and analysed the data. W.F. analysed the data, wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bushati N, Cohen SM (2007) microRNA functions. Annu Rev Cell Dev Biol 23: 175–205 [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W (2010) Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 79: 351–379 [DOI] [PubMed] [Google Scholar]
- Braun JE, Huntzinger E, Fauser M, Izaurralde E (2011) GW182 Proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell 44: 120–133 [DOI] [PubMed] [Google Scholar]
- Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, Filipowicz W (2011) miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat Struct Mol Biol 18: 1218–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR et al. (2011) miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat Struct Mol Biol 18: 1211–1217 [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R (2011) A parsimonious model for gene regulation by miRNAs. Science 331: 550–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP (2008) The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO (2009) Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol 7: e1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR et al. (2009) Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell 35: 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Zheng D, Xia Z, Shyu AB (2009) Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat Struct Mol Biol 16: 1160–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG (2006) MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA 103: 4034–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong YW et al. (2008) The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene. Proc Natl Acad Sci USA 105: 8866–8871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O (2009) Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol 11: 1143–1149 [DOI] [PubMed] [Google Scholar]
- Lee YS et al. (2009) Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol 11: 1150–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA (2007) Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5' UTR as in the 3' UTR. Proc Natl Acad Sci USA 104: 9667–9672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP (2007) Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 315: 1576–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, Krichevsky AM (2008) MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol 28: 5369–5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidenfeld I, Gossen M, Low R, Kentner D, Berger S, Gorlich D, Bartsch D, Bujard H, Schonig K (2009) Inducible expression of coding and inhibitory RNAs from retargetable genomic loci. Nucleic Acids Res 37: e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm AC, Wickens M (2008) Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol 9: 337–344 [DOI] [PubMed] [Google Scholar]
- Kusov YY, Shatirishvili G, Dzagurov G, Gauss-Muller V (2001) A new G-tailing method for the determination of the poly(A) tail length applied to hepatitis A virus RNA. Nucleic Acids Res 29: E57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HA, Bushell M, Hill K, Gant TW, Willis AE, Jones P, de Moor CH (2007) A novel method for poly(A) fractionation reveals a large population of mRNAs with a short poly(A) tail in mammalian cells. Nucleic Acids Res 35: e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W (2006) Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125: 1111–1124 [DOI] [PubMed] [Google Scholar]
- Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, Laur O, Warren ST, Bassell GJ (2011) Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell 42: 673–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME (2006) A brain-specific microRNA regulates dendritic spine development. Nature 439: 283–289 [DOI] [PubMed] [Google Scholar]
- Schratt G (2009) microRNAs at the synapse. Nat Rev Neurosci 10: 842–849 [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E (2006) mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 20: 1885–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ (2012) Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336: 233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336: 237–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.