Abstract
MicroRNAs (miRNAs) associate with components of the RNA-induced silencing complex (RISC) to assemble on mRNA targets and regulate protein expression in higher eukaryotes. Here we describe a method for the intracellular single-molecule, high-resolution localization and counting (iSHiRLoC) of miRNAs. Microinjected, singly fluorophore-labelled, functional miRNAs were tracked within diffusing particles, a majority of which contained single such miRNA molecules. Mobility and mRNA-dependent assembly changes suggest the existence of two kinetically distinct pathways for miRNA assembly, revealing the dynamic nature of this important gene regulatory pathway. iSHiRLOC achieves an unprecedented resolution in the visualization of functional miRNAs, paving the way to understanding RNA silencing through single-molecule systems biology.
Keywords: Let-7 miRNA, microinjection, single-molecule fluorescence microscopy, single-particle tracking, stepwise photobleaching
Introduction
Gene regulation by microRNAs (miRNAs) is an evolutionarily conserved RNA silencing pathway, wherein ∼22-nt-short non-coding RNAs assemble with components of the RNA-induced silencing complex (RISC) so that their guide strand can bind to partially complementary sequences in mRNA 3′ untranslated regions to repress protein expression [1, 2]. To date, over 1,500 mammalian miRNAs have been identified that, collectively, are predicted to regulate over 60% of all protein coding genes [3]. The ubiquitous nature of miRNAs implicates their involvement in all aspects of multicellular life [1, 2], from general cellular processes such as cell differentiation and survival to pathologies such as cancer, thus creating an urgent need to understand the mechanism(s) by which miRNAs function. While standard ensemble assays, including intracellular fluorescence microscopy [4, 5], have revealed a wealth of information [1, 2], the mechanism of gene repression by miRNAs is still debated [6, 7]. We have developed an incisive tool to quantify the intracellular diffusion and assembly of single functional miRNAs, which are parameters fundamental to their biological function, in pursuit of their still elusive mode of action.
Single-molecule fluorescence microscopy has emerged as a powerful tool to quantify properties of biomolecules not accessible to conventional ensemble-averaging techniques [8, 9]. In particular, single-molecule fluorescence microscopy has been applied to living cells to assess diffusive properties by single-particle tracking (SPT) and the stoichiometry of molecular complexes by stepwise photobleaching [10]. The strategies available so far, however, either decorate the target molecule with multiple fluorescent probes, thereby creating high-molecular-weight appendages that may impede function, and/or have only been applied to proteins or large RNA–protein complexes [9, 11, 12]. Our intracellular single-molecule, high-resolution localization and counting (iSHiRLoC) method overcomes these caveats by combining microinjection (which defines an experimental start point) and low-background illumination with SPT and stepwise photobleaching to visualize the diffusive motions and distribution of singly fluorophore-labelled functional small RNAs inside cells. We observe two kinetically distinct pathways involving mRNA-dependent miRNA assembly processes, providing a direct look at the complexity of the intracellular processes involved in RNA silencing.
Results and discussion
iSHiRLoC: a biologically relevant method
To study intracellular miRNA transport and localization, we microinjected mature double-stranded let-7a-1 or artificial cxcr4 miRNA, labelled on the 3′-end of the guide strand with one Cy3 or Cy5 fluorophore, into human HeLa cervical cancer cells. Let-7a-1 miRNA is a member of the highly conserved, tumour suppressing let-7 family whose function is critical to cell proliferation [13]. Let-7a-1 and cxcr4 have ∼700 and ∼70 predicted mRNA targets in HeLa cells, respectively (supplementary Methods online), and both miRNAs have been reported to localize to processing bodies (PBs) [4, 14], subcellular ribonucleoprotein (RNP) complexes enriched in RNA-processing enzymes. Microinjection allowed us to control the number of molecules introduced into the cytoplasm (∼18,000) to be similar to the endogenous levels of let-7 miRNAs (supplementary Methods online), minimizing cell perturbation and facilitating single miRNA detection as distinct point-spread functions on imaging by highly inclined laminar optical sheet (HILO) microscopy [15]. We supplemented cell medium with an oxygen scavenger system to delay photobleaching (Methods). Microinjected cells divided normally after 24 h and remained viable for at least 32 h, attesting to the low invasiveness of iSHiRLoC.
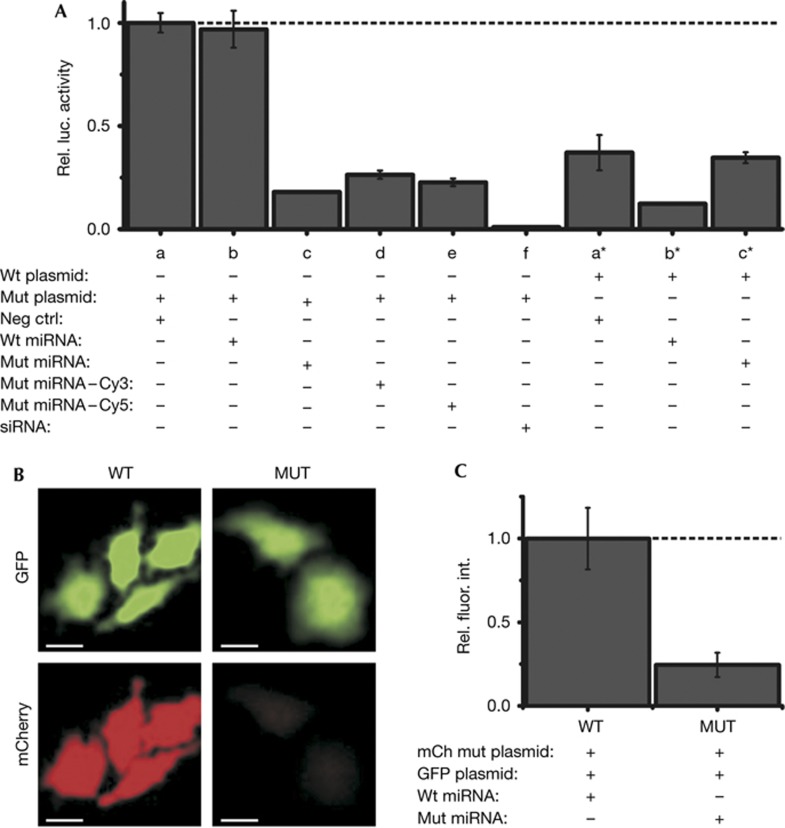
We further sought to ensure that microinjected, fluorophore-labelled miRNAs retain functionality. Transfection experiments showed that a luciferase reporter gene with seven let-7 target sequences was strongly repressed even without adding exogenous let-7a-1, most likely due to the fact that let-7 is one of most abundant miRNA families in HeLa cells. Accordingly, repression was moderately enhanced (only ∼2.8-fold) on co-transfection with let-7a-1 (Fig 1A; supplementary Methods online). To enhance the sensitivity of our assay, we included a mutant let-7a-1 that was previously shown to specifically repress a compensatory mutant of the target sequence [13]. As expected, the repression of mutant target by mutant let-7a-1 was strong (∼5.6-fold) and specific, consistent with previous observations [13]. In this more sensitive assay, we then showed that attaching either a single Cy3 or Cy5 to the 3′-end of the let-7a-1 guide strand does not significantly compromise repression (Fig 1A). Finally, we microinjected either mutant or wild-type let-7a-1 along with both an mCherry reporter plasmid containing seven mutant let-7 target sequences and a control green fluorescent protein (GFP) reporter plasmid, and found that the intended target protein was specifically and strongly repressed, supporting the biological relevance of microinjecting miRNAs as part of iSHiRLoC (Fig 1B,C; supplementary Methods online).
Figure 1.
Effect of fluorophore modification and microinjection on miRNA function. (A) Luciferase reporter assays of HeLa cells co-transfected with luciferase reporter plasmids bearing the wild-type (wt) or a mutant (mut) 3′ untranslated region of mouse HMGA2, and either a negative control siRNA (neg ctrl), wild-type let-7a-1 (wt miRNA) or mutant let-7a-1 (mut miRNA) miRNA. An siRNA, Siluc2, was used as a positive control for repression. Renilla luciferase activity was used for internal normalization of firefly luciferase activity within each sample. All samples were normalized with respect to negative control (a). Results presented are from four replicates. Error bars, standard deviations. (B) Representative images of GFP fluorescence (top) and mCherry fluorescence (bottom) in cells coinjected with an mCherry reporter plasmid, a GFP control plasmid and, either the wild-type (WT) or mutant let-7a-1 (MUT) miRNA are shown. Scale bar, 20 μm. (C) Quantification of mCherry fluorescence relative to GFP fluorescence from B, normalized with respect to the WT sample (n=3 independent trials, 50 cells per group). Error bars, s.e.m. GFP, green fluorescent protein; miRNA, microRNA; siRNA, small interfering RNA.
Assembled miRNAs diffuse within single particles
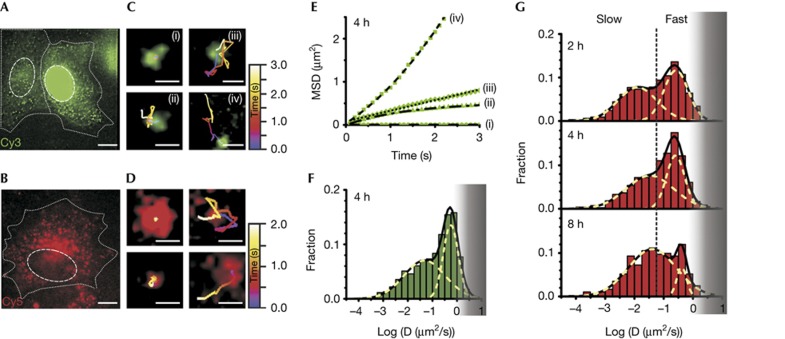
We next analysed SPT trajectories of miRNA particles 4 h after microinjecting cells with let-7a-1–Cy3 or let-7a-1–Cy5 (Fig 2A,B) and found individual particles to undergo (at least) four distinct diffusive patterns (Fig 2C–E; supplementary Videos 1 and 2 online). Control experiments showed that microinjection of PBS buffer alone or fluorescein-conjugated dextran, used as a marker for microinjection, contributed neither to the fluorescence signal (supplementary Video 3 online) nor impacted the function and mobility of miRNA particles (Figs 1B, 2F). Our SPT analysis reached a super-accuracy of ∼30 nm at 100 ms temporal resolution (supplementary Fig S1A online) with an observation window of, on average, ∼3 s for let-7a-1–Cy3 (and cxcr4–Cy3) and ∼1.5 s for let-7a-1–Cy5, before an individual particle went out of focus or photobleached (supplementary Fig S1B online). Only very few particles had large changes in intensity over their trajectory (supplementary Fig S1B online) that may report diffusion along the z axis. However, we cannot rule out that miRNA particles diffuse along the z axis between successive imaging frames, taken every 100 ms.
Figure 2.
Single-molecule high-resolution localization and tracking of miRNAs diffusing in living HeLa cells. Pseudo-coloured images of cells microinjected with (A) let-7a-1–Cy3 (left, cytoplasmic injection; right, nuclear injection, supplementary Video 1A online) imaged 4 h after microinjection and (B) let-7a-1–Cy5 (supplementary Video 1B online) imaged 2 h after microinjection, showing distinct particles containing miRNAs. Dashed and dotted lines indicate nuclear and cellular boundaries, respectively. Scale bar, 10 μm. Different types of diffusive motions exhibited by (C) let-7a-1–Cy3 (supplementary Video 2 online) and (D) let-7a-1–Cy5 miRNAs. Scale bars, 0.5 μm. (E) MSD plots of the let-7a-1–Cy3 particles shown in C. Data were fit with equations representing biased (iv), corralled (ii), fast (iii) and very slow (i) Brownian diffusion. Diffusion coefficients as derived from the fits are: D(i)=0.0001 μm2/s; D(ii)=0.06 μm2/s (corral radius=0.52 μm); D(iii)=0.062 μm2/s; and D(iv)=0.16 μm2/s (average velocity=0.46 μm/s). (F) Distribution of diffusion coefficients calculated from individual MSD plots of let-7a-1–Cy3 particles assuming Brownian diffusion (supplementary Methods online). Cells were imaged 4 h after microinjection (n=4 cells, supplementary Table S1 online). The grey shaded region represents diffusion coefficients of particles that are increasingly lost due to our limited time resolution of tracking. (G) Distribution of diffusion coefficients of let-7a-1–Cy5 at different time points after microinjection. Dotted lines represent demarcations of fast and slow particles to guide the eye, estimated based on segregation of the two Gaussian distributions 2 h after microinjection. Histograms represent data from multiple cells (n=4, 4 and 6 cells for data points corresponding to 2, 4 and 8 h, respectively, supplementary Table S1 online). Grey shaded region, as in F. MSD, mean squared displacement; miRNA, microRNA.
Time-averaged mean squared displacement (MSD) analysis of individual SPT trajectories allowed us to characterize the distinct diffusive patterns as biased, corralled, fast or very slow Brownian diffusion, leading to a wide range of diffusion coefficients (Fig 2E,F; supplementary Fig S1C–E online, supplementary Video 2 online, and supplementary Methods online). Analysis of a large number of trajectories (supplementary Table S1 online) 4 h after microinjection revealed two distinct Gaussian distributions of microscopic diffusion constants, with average diffusion coefficients of ∼0.26 and ∼0.034 μm2/s for both let-7a-1–Cy3 (Fig 2F) and let-7a-1–Cy5 miRNA (Fig 2G), smaller than those reported for RISC by fluorescence correlation spectroscopy, ∼5.4 μm2/s [5]. We did not observe more rapidly diffusing particles, possibly due to limits in our time resolution (100 ms camera integration time) and/or as they may not be visible for all of nine frames, our threshold for diffusion coefficient calculation. The main particles we did observe had diffusion coefficients that resemble those of messenger RNPs (mRNPs) [16] and PBs (supplementary Fig S2 online) [17], key mechanistic intermediates of mRNA repression by miRNAs [1, 2] that have much higher molecular masses than RISC. These assignments were further supported by the observed colocalization of microinjected miRNAs both with mRNPs containing matching miRNA target sites and with PBs (Fig 3). Taken together, our microinjected miRNAs are functionally fully active (Fig 1), and are associated with mRNAs and PBs (Fig 3), strongly suggesting that the observed particles must contain miRNAs bound to and actively repressing target mRNAs.
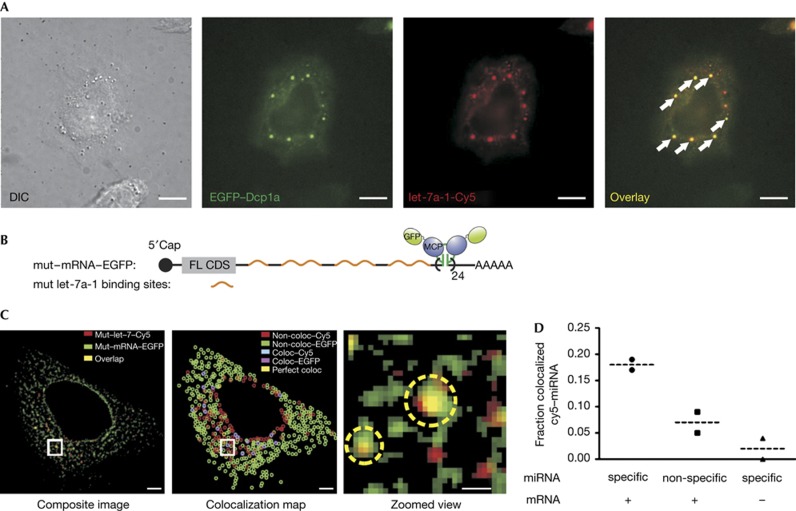
Figure 3.
Colocalization of microinjected miRNAs with PBs and cognate mRNA targets. (A) miRNA–PB colocalization. Differential interference contrast (left) and pseudo-coloured images depicting the colocalization (overlay, right) of ectopically expressed EGFP-tagged Dcp1a, a component of PBs (green), and microinjected let-7a-1–Cy5 miRNA (red). Wide-field epi-illumination was used to visualize these cells. Arrows show representative foci containing both Dcp1a and miRNA. Scale bar, 10 μm. (B) Schematic of an EGFP-labelled mRNA (top) based on the MS2–MS2 coat protein (MCP) mRNA-labelling system [16]. (C) miRNA–mRNA colocalization. Left, composite image of GFP (green) and Cy5 (red) channels from a representative cell expressing the mutant–mRNA–EGFP and mutant let-7a-1–Cy5 miRNA. Yellow particles indicate colocalization. Scale bar, 5 μm. Centre, colocalization map representing independent miRNA (red) and mRNA (GFP) particles, and either perfectly colocalized (yellow) or adjacent miRNA–mRNA localizations with an interparticle centre–centre distance less than the lateral resolution limit of the image (cyan and magenta, supplementary Methods online). Right, zoomed view of the region in the preceding images marked by a white box. Yellow circles indicate colocalized spots; scale bar, 1 μm. (D) Fraction of specific or nonspecific miRNA colocalizing with mRNA–EGFP. Cells were injected with the MS2/MCP plasmids and either mutant (specific; circle) or wild-type (nonspecific; square) let-7a-1. To test for the contribution of background fluorescence, control cells were injected with mutant let-7a-1 and MCP plasmid without the MS2–mRNA expression plasmid (specific; triangle). Data points within each group and their associated means (dashed lines) are shown (n=2 cells for each group). EGFP, enhanced green fluorescent protein; miRNA, microRNA; PB, processing body.
miRNP mobility increases over an 8 h period
Next, we asked whether the diffusive properties of miRNA-containing particles changed over time. Such changes may be expected for RISC-mediated loading of miRNAs onto mRNA targets and subsequent processing of the resulting mRNPs. To this end, we incubated let-7a-1–Cy5-microinjected cells for 1, 2, 4, 8 and 32 h prior to imaging. After 1 and 32 h, high background and large interframe displacements of individual particles, respectively, precluded accurate tracking. This observation, together with further analysis (supplementary Fig S3 online), is consistent with the presence of fast diffusing free miRNAs and smaller miRNA–protein complexes, respectively, that become blurred at our 100-ms time resolution. Notably, after 32 h the 3′ fluorophore-labelled miRNAs, whose 3′ ends are blocked against polyuridylation—a key step in the miRNA degradation pathway [18], remained detectable as intracellular particles of relatively slow diffusion, suggesting that they were neither (fully) degraded nor exported. For the intermediate time points (2, 4 and 8 h), we again clearly discerned diffusing particles within two distinct Gaussian distributions, whose average Brownian diffusion constants systematically increased over time (Fig 2G). This increase in mobility over time was not caused by an increase in the fraction of particles undergoing directed motion (supplementary Fig S4 online), suggesting that, on average, the molecular weights of the RNPs decrease in the 2–8 h timeframe. We note that the shifted distributions measured at later time points became increasingly curtailed above a diffusion coefficient of ∼1 μm2/s (Fig 2F,G; supplementary Fig S1E online) due to our limited time resolution, somewhat diminishing the observed effect by narrowing the fast diffusing population.
miRNA molecule counting by stepwise photobleaching
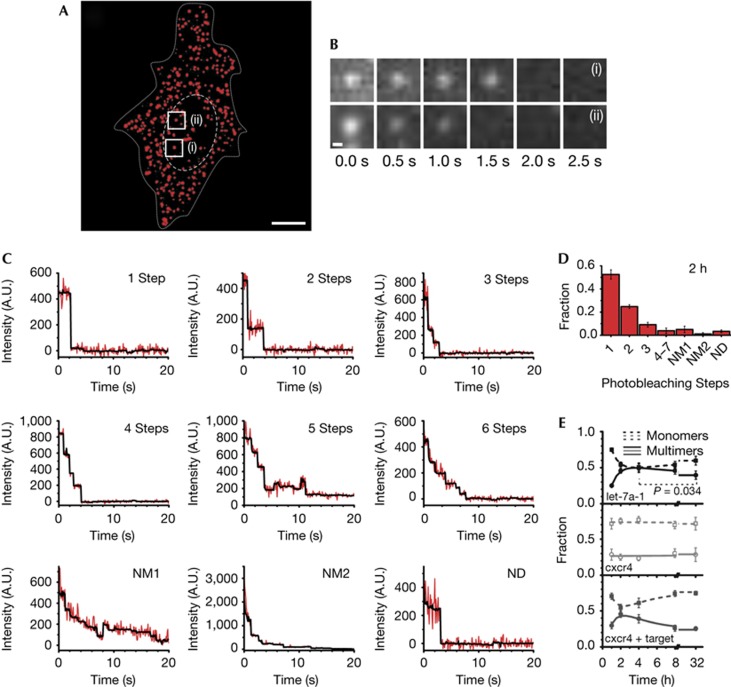
The goal of iSHiRLoC is to measure both diffusion properties and assembly states of individual small RNA containing particles. To assess assembly, we used the relatively rapid stepwise photobleaching [10] of single Cy5 fluorophores to quantify the number of miRNA molecules per particle. HeLa cells microinjected with either let-7a-1–Cy5 or cxcr4–Cy5 were formaldehyde-fixed and continuously illuminated until all fluorescent particles had photobleached (Fig 4A,B; supplementary Video 4 online). By counting the number of photobleaching steps for many particles (Fig 4C; supplementary Fig S5A online and supplementary Table S1 online), we were able to deduce the distribution of their assembly states (Fig 4D). While close to 50% of all particles contained single fluorophore-labelled miRNAs 2 h after microinjection, a significant fraction held up to seven labelled miRNAs (Fig 4C,D; supplementary Fig S5A online). To test for bias from ‘dark’ (prebleached) Cy5, we microinjected a control DNA labelled with an average of 3.45 Cy5 fluorophores per molecule and counted a majority of three and four photobleaching steps in each particle (supplementary Fig S5B–D online), as expected if prebleaching is negligible. In addition, we observed a strong overlap in the distribution of fluorescence intensities between particles in fixed and living cells 2 h after microinjection (supplementary Fig S5E online), when most miRNAs are assembled into RNPs, suggesting that our counting results in fixed cells closely reflect the miRNA assembly states in living cells.
Figure 4.
Counting of single miRNA molecules in fixed HeLa cells. (A) A pseudo-coloured (background corrected) image of a formaldehyde-fixed HeLa cell (supplementary Video 4 online) showing the intracellular distribution of miRNAs 2 h after microinjection. Dashed and dotted lines indicate nuclear and cellular boundaries, respectively. Scale bar, 10 μm. (B) Sets of frames showing photobleaching of two different miRNA particles over the indicated time. The locations of the particles are labelled in A. Scale bar, 300 nm. (C) Representative stepwise photobleaching traces of individual miRNA particles within the cell shown in A. For some traces, the number of steps could not accurately be determined. These traces were further classified as either non-determinable multimer with indistinct steps (NM1); non-determinable multimer with initial peak intensity exceeding that expected from up to 7 fluorophores based on an average intensity of ∼250 arbitrary units (A.U.) per fluorophore bleaching step (NM2); or non-determinable as either monomer or multimer due to large fluctuations in intensity and rapid (<3 frames) photobleaching (ND). (D) Distribution of miRNA photobleaching steps observed 2 h after microinjection (n=4, supplementary Table S1 online). Error bars, s.d. (E) Change in the fraction of monomers (particles undergoing a single step of photobleaching, dotted line) and multimers (particles that bleach in two or more steps, solid line) over time for let-7a-1 (black), cxcr4 (grey) and cxcr4 miRNAs comicroinjected with specific mRNA targets (dark grey, supplementary Fig S7A,B online), respectively. Each data point is derived from multiple cells (nlet-7a-1=4, 4, 3, 4, 8 cells; ncxcr4=5, 5, 5, 7, 9 cells; and ncxcr4+mRNA=4, 4, 4, 6 and 8 for 1, 2, 4, 8 and 32 h, respectively; supplementary Table S1 online). Error bars, s.d. The relative changes marked by asterisk are statistically significant with greater than 95% confidence, based on a paired t-test (P=0.034). miRNA, microRNA.
Time- and mRNA-dependent changes in miRNA assembly
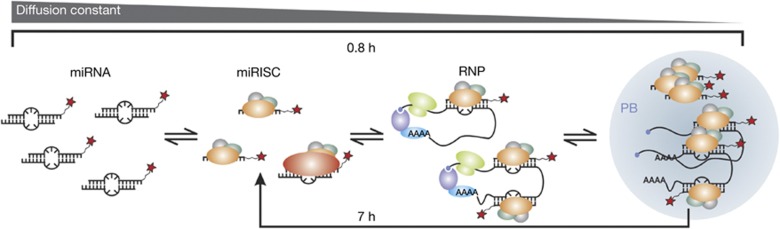
We monitored miRNA assembly over time at the same time points as in our diffusion measurements. Similar to our observations in living cells, high background made it impossible to accurately localize single-miRNA particles in cells that were fixed only 20 min after microinjection (supplementary Fig S6A online). At later time points (1–32 h), we found two distinct assembly phases for let-7a-1 miRNA that fit well with a double-exponential function, suggesting the existence of two kinetically distinct processes (Fig 4E; supplementary Fig S6 online). The faster phase is characterized by a rate constant of 1.2±0.2 h−1, during which the number of RNPs containing more than one miRNA increases; we note that this time evolution resembles that of the initial assembly of miRNAs into RNPs observed over the earliest (1–2 h) live cell time points. The slower phase with a rate constant of 0.14±0.08 h−1 leads to a modest increase in RNPs containing just a single miRNA and matches the timing of the increase in mobility of miRNA-containing particles in live cells (Fig 2G; supplementary Fig S3B online) as well as that of miRNA-induced mRNA target degradation [6]. On the basis of these observations, we propose the following time-dependent model for miRNA assembly: At initial time points, such as 20 min and up to 1 h (supplementary Figs S3A,S6A online) after microinjection, a significant fraction of our miRNAs remains free, thus contributing to a large background in our live and fixed cell experiments. Between 1 and 2 h, most miRNAs have assembled into miRISC–mRNA complexes, thereby depleting the free miRNA population. This assembly leads to relatively slowly diffusing complexes in living cells and a corresponding decrease in the population of single miRNAs containing particles in fixed cells. Still later, miRNAs mediate mRNA degradation through association with PBs, eventually releasing miRISC complexes to possibly elicit further rounds of repression, which is represented by a relative increase in the mobility of miRNA particles in living cells and an increase in the population of monomeric miRNAs in fixed cells (Fig 5). Consistent with this model, cells microinjected with the artificial cxcr4 miRNA, which is predicted to find 10-fold fewer mRNA target molecules in a HeLa cell (supplementary Methods online), do not show these time-dependent changes in the fractions of single and multiple miRNA-containing particles (Fig 4E; as the decrease in monomeric miRNAs is expected to be ∼10-fold smaller for cxcr4 than let-7a-1, it becomes indiscernible in our experiments with an estimated standard deviation about the mean of about 5–10%). By contrast, when cxcr4 was comicroinjected with ∼3,000 specific mRNA target molecules, miRNA assembly adopted again two kinetically distinct phases (with rate constants of 1.14±0.12 and 0.30±0.07 h−1; Fig 4E and supplementary Fig S7A,B online). Additionally, and as expected from our model, miRNAs shifted towards multimeric assemblies when we found them colocalized with target mRNA, but were more monomeric in the absence of a colocalized target (supplementary Fig S7C online). Taken together, we find strong evidence for mRNA-dependent miRNA assembly.
Figure 5.
Model illustrating intracellular miRNA assembly as derived from our data. miRNAs assemble into miRISC complexes that bind to translationally active mRNA targets to form large RNPs, associate with PBs, and are finally released on PB-mediated mRNA degradation to potentially engage in multiple turnover of targets. The indicated timeframes were derived from the kinetics measured in the current study for let-7a-1. miRISC, microRNA-induced silencing complex; miRNA, microRNA; PB, processing body; RNP, ribonucleoprotein.
In summary, in iSHiRLoC we have developed a broadly applicable method that visualizes both diffusive properties and assembly states of functional small non-coding RNAs in cultured cells. We find that miRNAs show different types of Brownian diffusion in live HeLa cells with (at least) two widely ranging Gaussian distributions of diffusion coefficients, representing two complexes of distinct molecular mass, consistent with mRNPs and PBs. Stepwise photobleaching revealed that the largest fraction of these particles contains single fluorophore-labelled miRNA molecules. A still significant fraction of particles, however, contains multiple miRNA molecules, strongly invoking the formation of higher-order miRNA complexes either assembled on mRNA targets or associated with PBs. Time-dependent changes in diffusion and assembly of miRNAs were observed, supporting the model in Fig 5, featuring two kinetically distinct processes wherein miRNAs first assemble into large RNPs (such as a miRISC bound to an mRNA or PB), then are released from these complexes. In the future, iSHiRLoC can be extended by multicolour imaging [9] to cotrack a miRNA with its corresponding target mRNA, RISC or PB proteins and map the dynamic interaction network of RNA silencing, with the ultimate goal of understanding its mechanism through single-molecule systems biology.
Methods
Repression assays. Dual luciferase assays were performed on HeLa cells co-transfected (Lipofectamine 2000, Invitrogen) with the indicated plasmid and mature double-stranded miRNA 24 h after transfection. For microinjection repression assays, 0.1 μg/μl of both the reporter (mCherry) and control (GFP) plasmids were microinjected with 1.5 μM miRNA and 0.025% Alexa647 dextran (Invitrogen) in PBS. Cells were imaged 24 h after injection. Fluorescence signals were quantified using ImageJ software (NIH).
Microinjection and intracellular single-molecule imaging. HeLa cells (1 to 1.25 × 105) were seeded onto delta-T dishes (Bioptechs) 1 day before microinjection. Regular medium was replaced with phenol red-free medium 4 h prior to microinjection and, immediately before microinjection, with a minimal HEPES buffered saline (HBS) medium without serum and vitamins, but containing 20 mM HEPES-KOH, pH 7.4, 135 mM NaCl, 5 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2 and 5.6 mM glucose. The micropipette (Femtotip, Eppendorf) was loaded with 0.5–2 μM fluorophore-labelled double-stranded miRNA (sometimes supplemented with 0.05% (w/v) 10 kDa Fluorescein dextran marker, Invitrogen) in PBS and injected using a Femtojet pump and Injectman NI2 micromanipulator (Eppendorf) at 100 hPa for 0.5 s with 20–40 hPa compensation pressure. Injected cells were incubated at 37 °C in phenol red-free DMEM containing 2% (v/v) fetal bovine serum and in the presence of a 5% CO2 atmosphere for the indicated amounts of time prior to imaging. For live cell imaging, microinjected cells were washed several times with HBS and imaged in HBS supplemented with an oxygen scavenging antioxidant mix (OSS) consisting of 0.3 U/ml Oxyfluor (Oxyrase), 20 mM sodium succinate, 2 mg/ml ascorbic acid and 200 μM trolox (supplementary Methods online), at 37 °C. For fixed cell imaging, cells were washed with PBS, fixed using warm 4% (w/v) paraformaldehyde in PBS for 20 min, washed again with PBS and imaged in PBS supplemented with OSS. We used HILO illumination to image cells at × 120 magnification with 100 ms camera exposure using a cell-TIRF system on an Olympus IX81 microscope (supplementary Methods online).
Data analysis. Single particles were tracked using Imaris (Bitplane). In-house MATLAB routines were used to calculate MSDs and diffusion coefficients. The number of photobleaching steps was determined using a custom LabView (National Instruments) code (supplementary Methods online).
Supplementary Material
Acknowledgments
We thank D. Bartel, C. Mayr, R. Tsien, N. Kedersha and R. Singer for generous gifts of plasmids containing the 3′ untranslated region of HMGA2, mCherry ORF, Dcp1a ORF and the MS2 system of plasmids, A. Manzo for MSD software development, H. Ding, A. Gafni and D. Steel for sharing their intensity analysis routine, the Microscopy and Image Analysis Laboratory at the University of Michigan for access to Imaris, X.S. Xie for important input at the onset of the project, and A. Mapp for access to her plate reader. This work was supported by National Institutes of Health (NIH) grant GM081025 to N.G.W. and an NIH Cellular Biotechnology training grant fellowship to J.R.A.
Author contributions: S.P. performed all experiments and analysis, except for the fluorescence repression and miRNA–mRNA colocalization experiments. J.R.A. performed the latter two assays and assisted with the bioinformatic analysis. The manuscript was prepared jointly by S.P., J.R.A. and N.G.W.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ (2011) Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet 12: 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kouwenhove M, Kedde M, Agami R (2011) MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer 11: 644–656 [DOI] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W (2005) Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 309: 1573–1576 [DOI] [PubMed] [Google Scholar]
- Ohrt T, Mutze J, Staroske W, Weinmann L, Hock J, Crell K, Meister G, Schwille P (2008) Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy reveal the cytoplasmic origination of loaded nuclear RISC in vivo in human cells. Nucleic Acids Res 36: 6439–6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R (2011) A parsimonious model for gene regulation by miRNAs. Science 331: 550–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter NG, Huang CY, Manzo AJ, Sobhy MA (2008) Do-it-yourself guide: how to use the modern single-molecule toolkit. Nat Methods 5: 475–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald D, Singer RH, Rout M (2011) Nuclear export dynamics of RNA-protein complexes. Nature 475: 333–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leake MC, Chandler JH, Wadhams GH, Bai F, Berry RM, Armitage JP (2006) Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443: 355–358 [DOI] [PubMed] [Google Scholar]
- Babcock HP, Chen C, Zhuang X (2004) Using single-particle tracking to study nuclear trafficking of viral genes. Biophys J 87: 2749–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzkovitz S, van Oudenaarden A (2011) Validating transcripts with probes and imaging technology. Nat Methods 8: S12–S19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP (2007) Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 315: 1576–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Calabrese JM, Sharp PA (2006) Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci USA 103: 18125–18130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga M, Imamoto N, Sakata-Sogawa K (2008) Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods 5: 159–161 [DOI] [PubMed] [Google Scholar]
- Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E (2003) Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol 13: 161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizer A, Brody Y, Ler LW, Sonenberg N, Singer RH, Shav-Tal Y (2008) The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol Biol Cell 19: 4154–4166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom KH, Heo I, Lee J, Hohng S, Kim VN, Joo C (2011) Single-molecule approach to immunoprecipitated protein complexes: insights into miRNA uridylation. EMBO Rep 12: 690–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.