Abstract
CPAP—a gene mutated in primary microcephaly—is required for procentriole formation. Here we show that CPAP degradation and function is controlled by the poly(ADP-ribose) polymerase tankyrase 1. CPAP is PARsylated by tankyrase 1 in vitro and in vivo. Overexpression of tankyrase 1 leads to CPAP proteasomal degradation, preventing centriole duplication, whereas depletion of tankyrase 1 stabilizes CPAP in G1, generating elongated procentrioles and multipolarity. Tankyrase 1 localizes to centrosomes exclusively in G1, coinciding with CPAP degradation. Hence, tankyrase 1-mediated PARsylation regulates CPAP levels during the cell cycle to limit centriole elongation and ensure normal centrosome function.
Keywords: CPAP, tankyrase 1, centrosome
Introduction
The centrosome, the primary microtubule-organizing centre of the cell, is comprised of two centrioles surrounded by pericentriolar material that nucleates microtubules [1]. Centrosome duplication is a highly regulated process whose accuracy is essential for genome integrity [2]. The process involves the formation of two new centrioles (procentrioles) next to the two preexisting parental centrioles during S phase of the cell cycle. Studies in Caenorhabditis elegans and Drosophila melanogaster have defined a conserved pathway that relies on a key set of proteins for centriole duplication [3]. In some cases, human homologues have been identified and shown to have similar function. For example, SAS4 was shown to be required for procentriole formation through the deposition of centriolar microtubules in C. elegans [4, 5]. Centrosomal P4.1-associated protein (CPAP), a human SAS4-related protein, was found to be required for centrosome function in human cells [6, 7]. Depletion of CPAP in human cells prevented centrosome duplication, whereas overexpression of CPAP led to aberrant centriole elongation, supernumerary centrioles and spindle multipolarity [8–11]. A crucial role for CPAP and centriole maturation in humans is evidenced by its mutation in autosomal recessive primary microcephaly (MCPH) [12–14].
The aberrant centriole elongation and accompanying phenotypes observed on overexpression of CPAP suggest that centriole length must be restricted to ensure proper centrosome function and genome integrity. Recent studies indicated that CPAP protein levels were regulated across the cell cycle; as cells exited mitosis and entered G1, CPAP protein levels were reduced by proteasomal degradation [11]. The timing of degradation suggested that CPAP could be a target of the anaphase-promoting complex (APC)–Cdh1 complex, and (consistent with this notion) coimmunoprecipitation analysis revealed a CPAP–Cdh1 interaction [11]. However, the question remains how is centrosomal CPAP targeted for degradation at this specific point in the cell cycle.
Tankyrase 1 (TNKS1) is a multifunctional poly(ADP-ribose) polymerase (PARP) that uses NAD+ as substrate to transfer ADP-ribose polymers onto protein acceptors including itself [15, 16]. TNKS1-mediated PARsylation can influence protein degradation. PARsylation of TNKS1’s telomeric acceptor telomere-repeat-binding factor 1 (TRF1) led to loss of TRF1 from telomeres and subsequent ubiquitylation and proteasomal degradation [17]. TNKS1-mediated PARsylation of axin 1 and 2 (regulators of the Wnt signalling pathway) led to their degradation through the ubiquitin–proteasome pathway [18]. TNKS1 has a number of other binding partners that, though diverse, share a common RXXG/PDG (or degenerate)-binding motif [15, 19, 20]. Here we identify CPAP as an RXXPDG-containing TNKS1 binding partner. We show that TNKS1 PARsylates CPAP and regulates CPAP protein stability and function at centrosomes across the cell cycle.
Results and discussion
TNKS1 binds to CPAP and influences its stability
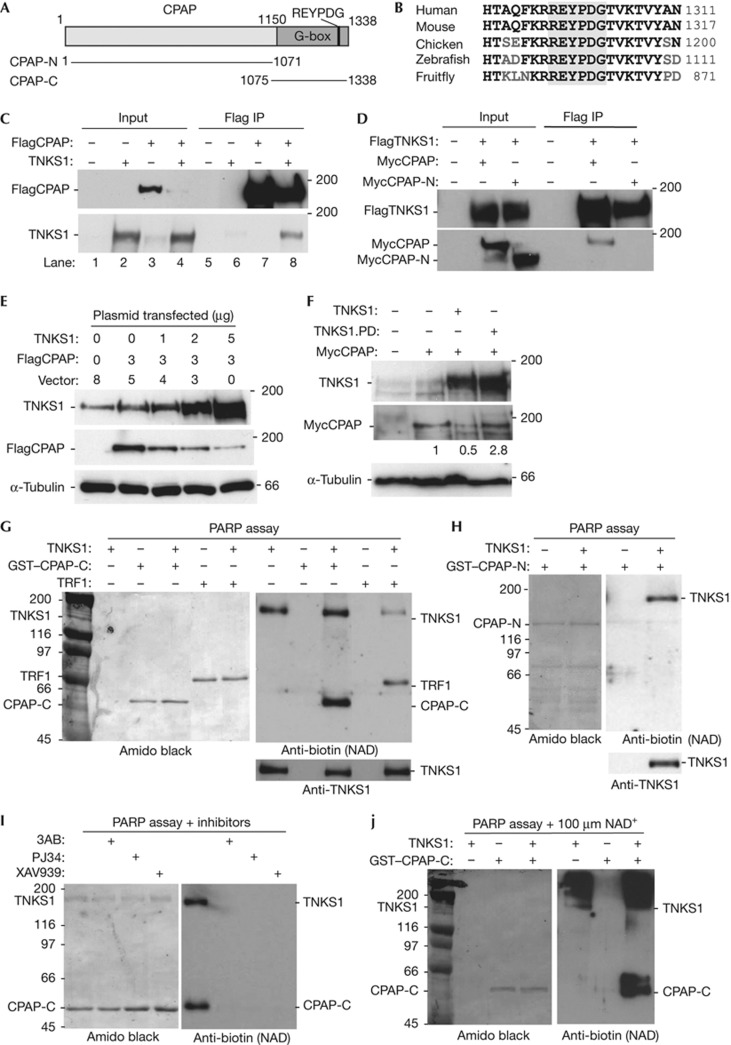
Human CPAP contains a highly conserved region of glycine repeats at its C terminus, termed the G-box, that houses a RXXPDG tankyrase-binding motif (REYPDG) (Fig 1A). The REYPDG domain shows 100% identity from fruitfly to human, suggesting an important function (Fig 1B). To determine whether CPAP interacts with TNKS1 in vivo, 293T cells were co-transfected with vectors expressing Flag epitope-tagged CPAP (FlagCPAP) and/or TNKS1. Immunoprecipitation with anti-Flag antibody showed that TNKS1 coimmunoprecipitated with FlagCPAP (Fig 1C). To determine whether the interaction was dependent on the TNKS1-binding motif, we generated a C-terminal deletion of the TNKS1-binding domain called CPAP-N (Fig 1A). Myc epitope-tagged CPAP (MycCPAP) or MycCPAP-N was co-transfected into 293T cells with Flag epitope-tagged TNKS1 (FlagTNKS1). As shown in Fig 1D, MycCPAP, but not MycCPAP-N, coimmunoprecipitated with FlagTNKS1.
Figure 1.
CPAP interacts with tankyrase 1 and is PARsylated in vitro. (A) Schematic representation of CPAP. The glycine-rich (G-box) is indicated. (B) Alignment of the tankyrase-binding REYPDG motif. Identical amino acids are in black. (C) Tankyrase 1 (TNKS1) binds to CPAP. Immunoblot analysis of proteins immunoprecipitated (IP) with anti-Flag beads from 293T cells transfected with FlagCPAP and/or TNKS1. (D) Tankyrase 1 binding to CPAP depends on the C-terminal domain of CPAP. Immunoblot analysis of proteins immunoprecipitated with anti-Flag beads from 293T cells transfected with Flag epitope-tagged TNKS1 (FlagTNKS1) and Myc epitope-tagged CPAP (MycCPAP) or MycCPAP-N. (E) Tankyrase 1 overexpression leads to loss of CPAP. Immunoblot analysis of extracts from 293T cells co-transfected with FlagCPAP and increasing amounts of TNKS1. (F) Tankyrase 1-induced loss of CPAP depends on tankyrase 1 poly(ADP-ribose) polymerase (PARP) catalytic activity. Immunoblot analysis of extracts from 293T cells co-transfected with MycCPAP and TNKS1 or TNKS1.PD (PARP-dead). Protein levels relative to α-tubulin and normalized to MycCPAP are indicated. (G–J) CPAP is PARsylated by tankyrase 1 in vitro. PARP assays containing recombinant tankyrase 1, 25 μM biotinylated NAD+, and the indicated recombinant proteins were analysed by immunblotting with anti-biotin. (G) CPAP-C is modified by tankyrase 1. (H) CPAP-N is not modified by tankyrase 1. (I) Tankyrase 1 modification of CPAP-C is inhibited by PARP inhibitors 3AB, PJ34, or XAV939. (J) PARsylation of CPAP-C by tankyrase 1 is demonstrated by addition of unlabelled NAD+ (100 μM). GST, Glutathione S-transferase; TRF1, telomere-repeat-binding factor 1.
We noticed that the level of FlagCPAP was dramatically reduced upon co-transfection with TNKS1 (Fig 1C, compare lanes 3 and 4). Indeed, co-transfection of increasing amounts of TNKS1 with FlagCPAP led to a dose-dependant reduction in FlagCPAP (Fig 1E). This reduction was dependent on TNKS1’s catalytic PARP domain; cotransfection of MycCPAP with wild-type (WT), but not a PARP dead (PD) allele of tankyrase 1 [6], induced loss of CPAP (Fig 1F).
CPAP is PARsylated by TNKS1 in vitro
To determine whether TNKS1 PARsylates CPAP, we performed in vitro PARP assays with purified recombinant proteins. Glutathione S-transferase (GST)–CPAP-C purified from Escherichia coli was incubated with baculovirus-derived TNKS1 and biotinylated NAD+ substrate. Proteins were fractionated by SDS–polyacrylamide gel electrophoresis, transferred to nitrocellulose, stained with amido black to visualize the proteins (Fig 1G, left panel), and probed with anti-biotin to visualize ADP-ribosylated proteins (Fig 1G, right panel). TNKS1 modified itself and CPAP-C. As a control we show a similar reaction for the known acceptor TRF1. PARsylation was dependent on TNKS1 binding, as GST–CPAP-N was not modified by TNKS1 (Fig 1H). Modification of CPAP-C by TNKS1 was inhibited by the general PARP inhibitors 3AB and PJ34, and by the tankyrase-specific PARP inhibitor XAV939, confirming that the modification was the result of PARP activity (Fig 1I). When the in vitro PARP assays were performed in the presence of excess unlabelled NAD+ substrate, biotin-labelled TNKS1 and CPAP were both converted into slower migrating diffuse bands, indicative of PARsylation (Fig 1J).
CPAP is ubiquitylated and PARsylated by TNKS1 in vivo
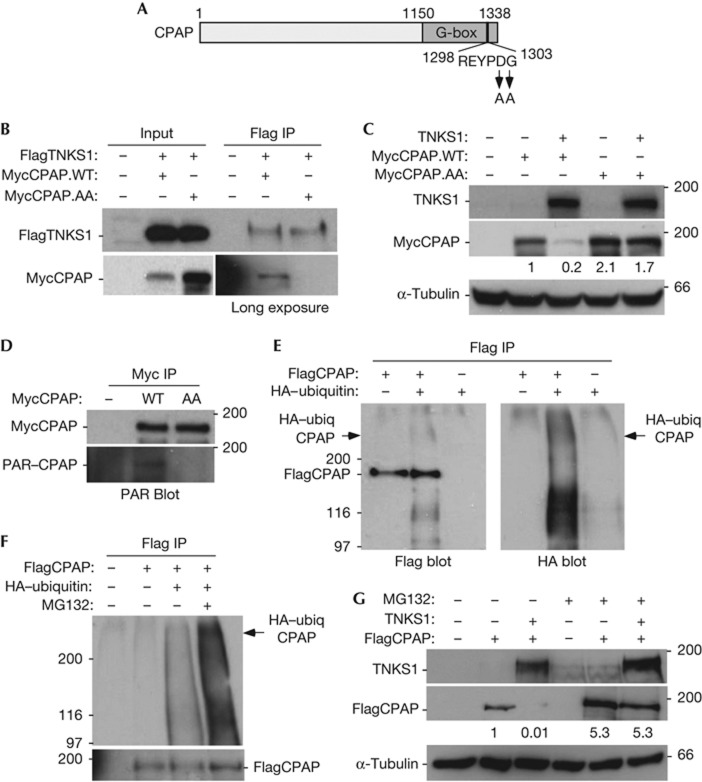
To determine whether the REYPDG motif was required for TNKS1 binding to CPAP, we generated a double-point mutation converting DG to AA (CPAP.AA) (Fig 2A). As shown in Fig 2B, MycCPAP.WT (but not MycCPAP.AA) was coimmunoprecipitated by FlagTNKS1. Co-transfection of TNKS1 led to loss of CPAP.WT, but not CPAP.AA (Fig 2C), consistent with the notion that TNKS1 is responsible for the observed reduction in CPAP protein levels. We showed in Fig 1F that the reduction in CPAP levels in vivo depended on the PARP activity of TNKS1 and that TNKS1 PARsylated CPAP in vitro (Fig 1G,J). To determine whether CPAP was PARsylated in vivo, MycCPAP (WT or AA) was immunoprecipitated with anti-Myc beads and immunoblotted with anti-PAR antibody. As shown in Fig 2D, CPAP was PARsylated in vivo, dependent on its TNKS1-binding site. We next sought to determine whether CPAP was ubiquitylated under the same conditions that we observed its PARsylation and degradation. FlagCPAP was co-transfected with haemagglutinin (HA) epitope-tagged ubiquitin into 293T cells, immunoprecipitated with anti-Flag beads and analysed by immunoblotting with anti-Flag or anti-HA antibody. As shown in Fig 2E, a smear of high molecular weight bands migrating slower than CPAP (indicated by the arrowhead) corresponding to HA-ubiquitylated FlagCPAP was detected. We additionally observed lower molecular weight HA-ubiquitylated CPAP, as has been observed previously [11], that likely corresponds to degraded CPAP. Ubiquitylated CPAP was degraded by the proteasome as demonstrated by its stabilization upon treatment with the proteasome inhibitor MG132 (Fig 2F). Finally, we show that TNKS1-induced loss of CPAP was rescued by treatment with MG132 (Fig 2G). Together, these data indicate that TNKS1 PARsylates CPAP and promotes CPAP degradation in vivo.
Figure 2.
CPAP is PARsylated in vivo dependent on its tankyrase 1-binding motif. (A) Schematic representation of CPAP with the double-point mutation indicated. (B) Tankyrase 1 (TNKS1) binding to CPAP depends on the REYPDG TNKS1-binding site. Immunoblot analysis of proteins immunoprecipitated (IP) with anti-Flag beads from 293T cells transfected with Flag epitope-tagged TNKS1 (FlagTNKS1) and Myc epitope-tagged CPAP (MycCPAP; wild type (WT) or AA). (C) Tankyrase 1 overexpression leads to loss of CPAP dependent on the REYPDG TNKS1-binding site. Immunoblot analysis of extracts from 293T cells co-transfected with TNKS1 and MycCPAP (WT or AA). Protein levels relative to α-tubulin and normalized to MycCPAP are indicated. (D) CPAP is PARsylated in vivo dependent on its REYPDG TNKS1-binding site. Immunoblot analysis of proteins immunoprecipitated with anti-Myc beads from 293T cells transfected with MycCPAP (WT or AA) and probed with anti-Myc or anti-PAR antibodies. (E) CPAP is ubiquitylated in vivo. Immunoblot analysis of proteins immunoprecipitated with anti-Flag beads from 293T cells co-transfected with FlagCPAP and/or haemagglutinin (HA)-ubiquitin and probed with anti-Flag antibody (left panel) or anti-HA antibody (right panel). (F) Ubiquitylated CPAP is degraded by the proteasome. Immunoblot analysis of proteins immunoprecipitated with anti-Flag beads from 293T cells co-transfected with FlagCPAP and/or HA-ubiquitin, treated with (+) or without (−) MG132, and probed with anti-HA antibody (top panel) or anti-Flag antibody (bottom panel). (G) TNKS1-induced degradation of CPAP depends on the proteasome. Immunoblot analysis of extracts from 293T cells co-transfected with TNKS1 and/or Flag epitope-tagged CPAP (FlagCPAP), and treated with (+) or without (−) MG132. Protein levels relative to α-tubulin and normalized to FlagCPAP are indicated.
TNKS1 overexpression prevents centrosome duplication
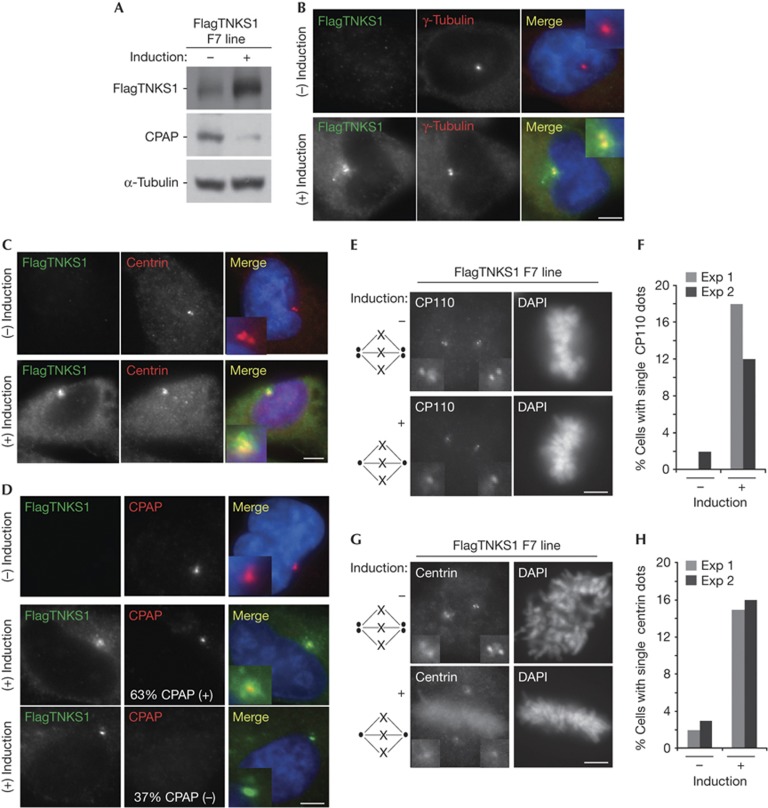
To gain insight into the functional consequences of TNKS1-induced loss of CPAP, we expressed FlagTNKS1 stably in HTC75 cells, a human fibrosarcoma cell line (HT1080) that contains a tetracycline-controlled gene expression system. Immunoblot analysis of cells grown with and without induction shows induced expression of FlagTNKS1 and concomitant loss of CPAP (Fig 3A). Immunofluorescence analysis showed that induced FlagTNKS1 accumulated at interphase centrosomes, as measured by costaining with γ-tubulin (Fig 3B) or centrin (Fig 3C). Costaining for FlagTNKS1 and CPAP showed that they colocalized (Fig 3D, middle panels). However, in approximately one-third of FlagTNKS1-expressing cells CPAP was lost (Fig 3D, bottom panels), consistent with the reduction of CPAP protein by immunoblot (Fig 3A).
Figure 3.
Tankyrase 1 overexpression leads to loss of CPAP and prevents centrosome duplication. (A) Immunoblot analysis of inducible Flag epitope-tagged Tankyrase 1 (FlagTNKS1) expression in a HTC75 clonal cell line (F7). (B–C) FlagTNKS1 colocalizes with centrosomal markers. Immunofluorescence analysis of inducible FlagTNKS1 expression in F7 cells fixed with methanol and stained with anti-Flag (green) and (B) anti-γ-tubulin (red) or (C) anti-centrin (red). DAPI (4,6-diamidino-2-phenylindole) staining is in blue and merge is yellow. (D) FlagTNKS1 induces loss of CPAP at centrosomes in a fraction of cells. Immunofluorescence analysis of inducible FlagTNKS1 expression in F7 cells fixed with methanol and stained with anti-Flag (green) and anti-CPAP (red). DAPI staining is in blue and merge is yellow. Overall, 63% of cells show colocalization of FlagTNKS1 with CPAP, but in 37% of cells expressing FlagTNKS1 CPAP is lost from centrosomes (n=100 cells). (E–H) Centrosome duplication is inhibited in tankyrase 1-overexpressing cells. (E,G) Immunofluorescence analysis of mitotic F7 cells fixed with methanol and stained with (E) anti-CP110 or (G) anti-centrin and DAPI. (F,H) Graphical representation of the frequency of mitotic cells in E and G, respectively, from two independent experiments, where n=50–100 cells each. Scale bar, 5 μm (B,C,D,E,G).
Previous studies showed that short interfering RNA-mediated CPAP depletion prevented centriole duplication in cycling human cells. We hypothesized that as TNKS1 overexpression leads to loss of CPAP, it should mimic the effect of CPAP depletion by short interfering RNA. Normal centriole duplication can be readily scored as two centrioles at each spindle pole in a mitotic cell. Whereas, cells depleted of CPAP show single centrioles at each pole. We stained cells with the centrosome marker CP110 and scored mitotic cells containing a single centriole at each spindle pole. As shown in Fig 3E,F, induction of FlagTNKS1 led to a dramatic increase in cells containing unduplicated centrioles. Similar results were obtained using centrin as a marker (Fig 3G,H).
TNKS1 depletion impacts centrosome function
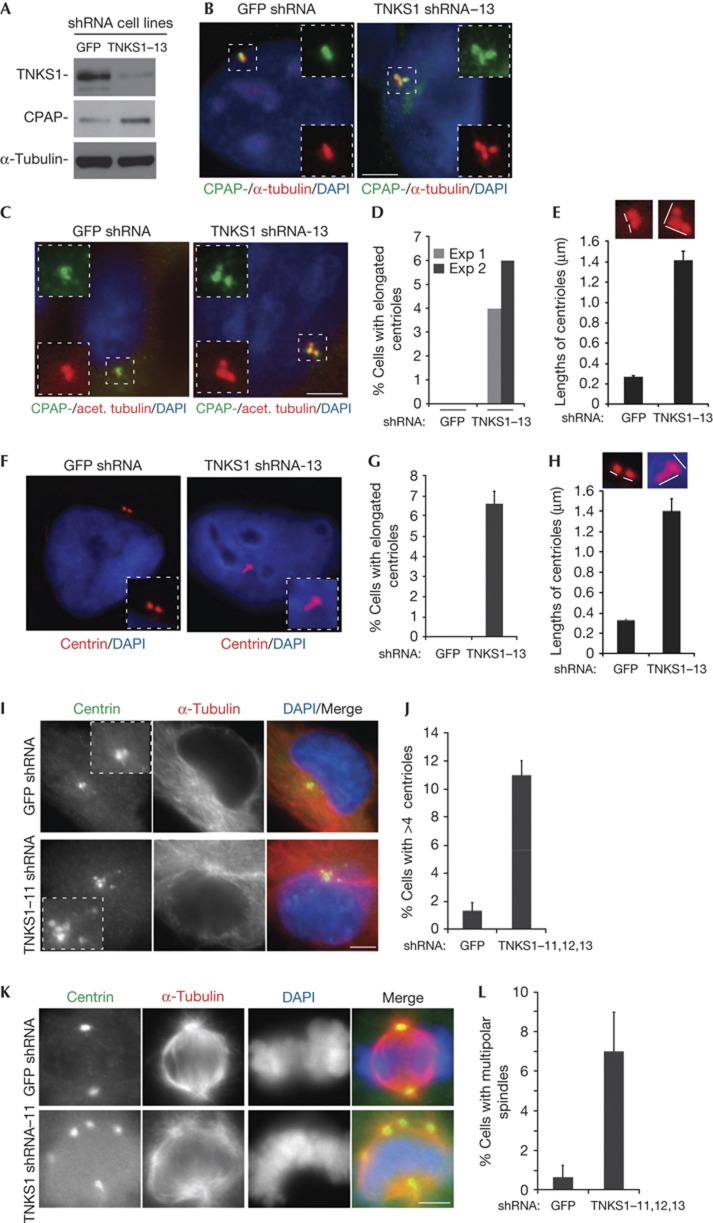
To determine whether endogenous CPAP levels are influence by TNKS1, we analysed stable TNKS1 knockdown HTC75 cell lines generated by infection with lentiviruses expressing TNKS1 or green fluorescent protein (GFP) short hairpin RNA (shRNA). As shown in Fig 4A, immunoblot analysis of line TNKS1–13 showed depletion of TNKS1 and concomitant increase in CPAP protein (compared with the GFP control line), indicating that TNKS1 is required to maintain WT levels of CPAP. To determine whether the increased CPAP protein levels influenced CPAP function at centrosomes, we considered phenotypes induced by CPAP overexpression. Previous studies showed that overexpression of CPAP in human cells led to abnormally elongated centrioles, supernumerary procentrioles and multipolar spindles. First we analysed cells for centriolar elongation. Costaining of TNKS1–13 cells with CPAP and α-tubulin revealed long threads containing CPAP (Fig 4B). Moreover, these CPAP containing long threads costained with acetylated tubulin (Fig 4C), a hallmark of stable microtubules and a characteristic of CPAP-induced centriolar elongations. We detected these threads in TNKS1-depleted cells, but not in GFP control (Fig 4D,E). Similar results were obtained using centrin as a marker (Fig 4F,G). Previous studies showed that CPAP overexpression led to supernumerary centrosomes and as a result, multipolar spindles. Consistent with this, we observed centrosome amplification (measured as greater than four centrioles per interphase cell; Fig 4I,J) and multipolar spindles (Fig 4K,L) in three different TNKS1 shRNA lines (TNKS1–11, 12 and 13). We note that the multipolarity could also be due to other aspects of TNKS1 function at spindle poles. Together these data indicate that in the absence of TNKS1, CPAP levels are overexpressed sufficiently to impair normal centrosome function.
Figure 4.
Tankyrase 1 depletion leads to an increase in CPAP protein and impacts centrosome function. (A) Immunoblot analysis of HTC75 stable cell lines expressing green fluorescent protein (GFP) or Tankyrase 1 (TNKS1)–13 short hairpin RNA (shRNA). (B–H) Depletion of tankyrase 1 leads to centriolar elongation. (B,C,F) Immunofluorescence analysis of shRNA cell lines cold treated for 1 h at 4 °C, pre-extracted with Triton X-100 (0.5%) for 1 min, fixed in methanol, and stained with antibodies to (B,C) CPAP (green) and (B) α-tubulin (red) or (C) acetylated tubulin (red) or (F) centrin (red). DAPI (4,6-diamidino-2-phenylindole) staining is in blue and merge is yellow. (D,G) Graphical representation of the frequency of cells with elongated centrioles from (D) two experiments, where n=100 cells each, or (G) from three experiments, where n=100 cells each; data represent the mean±s.d. (E,H) Graphical representation of the lengths of centrioles±s.e.m. Labelled with (E) acetylated tubulin (n=20 centrioles) or with centrin (n=40 centrioles). Depletion of tankyrase 1 leads to (I,J) centriole amplification and to (K and L) multipolarity. (I,K) Immunofluorescence analysis of GFP and TNKS1–11 shRNA cell lines fixed in methanol and stained with antibodies to centrin (green) and α-tubulin (red). DAPI staining is in blue and merge is yellow. (J,L) Graphical representation of the frequency of cells with (J) greater than 4 centrioles or (L) with multipolar spindles. Data represent the mean±s.d. from three experiments using TNKS1–11, –12, and –13, where n=100 cells each. Scale bar, 5 μm.
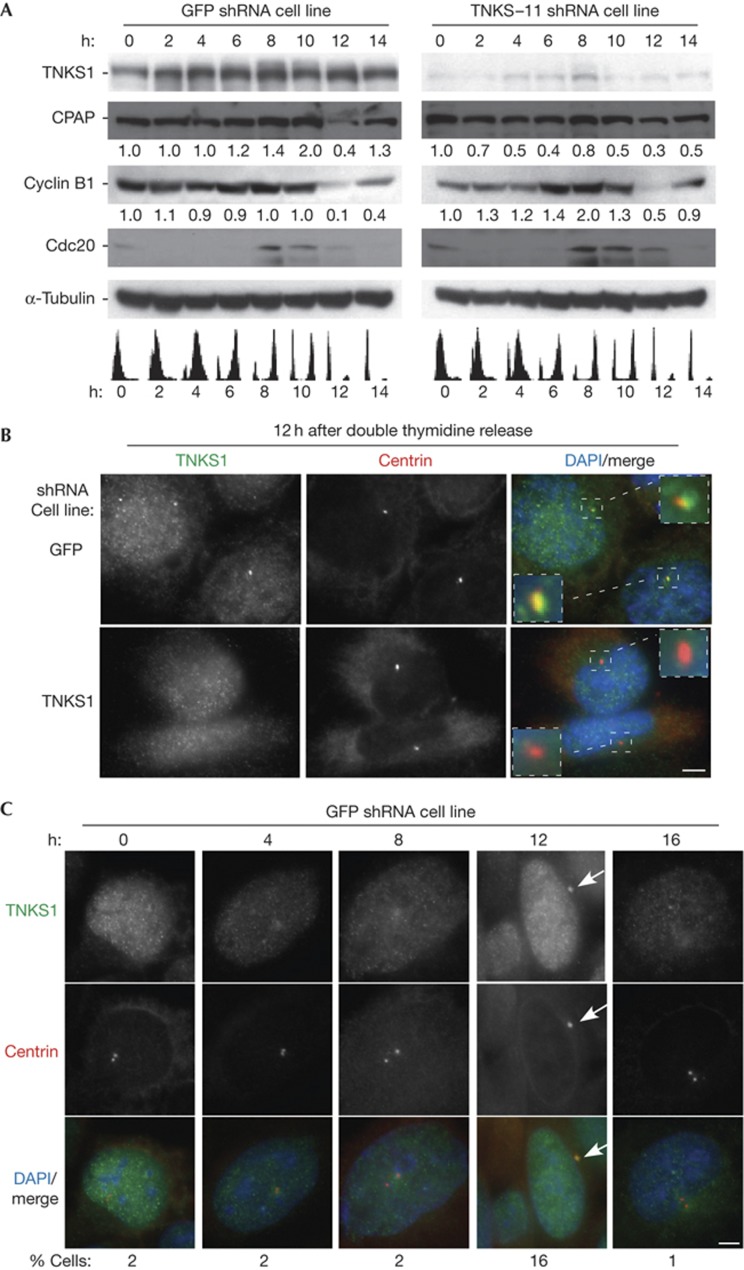
TNKS1 induces loss of CPAP in G1
Previous studies showed that CPAP was degraded in G1 phase of the cell cycle, likely mediated by the APC–Cdh1 E3 ligase [11]. We thus asked if CPAP was specifically stabilized in this stage of the cell cycle in the absence of TNKS1. TNKS1 or GFP shRNA cells were synchronized by a double thymidine block, released, and collected every 2 h. The cell-cycle stage was analysed by fluorescence-activated cell sorting, and the protein content at each stage by immunoblotting (Fig 5A). As shown previously, in GFP shRNA control cells CPAP was degraded at 12 h (early G1) (Fig 5A, left panel). However, in TNKS1-depleted cells CPAP was stabilized at 12 h (Fig 5A, right panel). This stabilization was not due to a defect in the E3 ligase, as cyclin B1 (the target of the APC–Cdc20 and APC–Cdh1) and Cdc20 (the target of the APC–Cdh1) were degraded normally at 12 h in the absence of TNKS1.
Figure 5.
Tankyrase 1 is required for degradation of CPAP in G1 phase of the cell cycle. (A) Immunoblot analysis of staged cell cycle extracts from green fluorescent protein (GFP; left panel) or Tankyrase 1 (TNKS1)–11 (right panel) stable HTC75 short hairpin RNA (shRNA) cell lines. Cells were synchronized by a double thymidine block and released for 0 h (G1/S), 2 h (early S), 4 h (mid S), 6 h (late S), 8 h (G2), 10 h (M), 12 h (early G1), and 14 h (mid G1). Fluorescence-activated cell sorting analysis is indicated below. Protein levels relative to α-tubulin and normalized to the 0 h time point, are indicated beneath the blots. (B) Tankyrase 1 is detected at interphase centrosomes at 12 h (early G1) in control GFP (top panels) but not TNKS1 (bottom panels) shRNA cell lines. Immunofluorescence analysis of cells at 12 h fixed with methanol and stained with anti-TNKS1 763 (green) or anti-centrin (red) antibodies. DAPI (4,6-diamidino-2-phenylindole) staining is in blue and merge is yellow. (C) Tankyrase 1 localizes to centrosomes exclusively in G1. Immunofluorescence analysis of GFP shRNA cells at 0, 4, 8, 12, and 16 h fixed with methanol and stained with anti-TNKS1 763 (green) or anti-centrin (red) antibodies. DAPI staining is in blue and merge is yellow. The frequency of cells where tankyrase 1 localizes to the interphase centrosome is indicated at the bottom (n=100 cells). Scale bar, 5 μm.
Immunofluorescence analysis of synchronized cells detected TNKS1 at interphase centrosomes at 12 h in GFP shRNA control cells (Fig 5B, upper panels). The specificity of the signal was confirmed by the absence of TNKS1 at centrosomes in the TNKS1 shRNA cells (Fig 5B, lower panels). Immunofluorescence analysis across the cell cycle showed that TNKS1 was detected at interphase centrosomes at 12 h in 16% compared with 1%–2% of cells at all other stages of the cell cycle (Fig 5C). Together, these data indicate that TNKS1 localizes to centrosomes in G1, coinciding precisely with the cell cycle–regulated timing of CPAP degradation.
Conclusion
The results described here identify CPAP, a protein crucial for centrosome function, as a new acceptor of PARsylation by TNKS1. Our studies suggest that TNKS1 localizes to centrosomes promoting degradation of CPAP in early G1, thereby preventing over-elongation of centrioles and ensuring proper centrosome function. At least two other centrosomal proteins Cep76 [22] and Cep170 [23] contain RXXPDG TNKS1 binding sites and a number of other centrosomal proteins contain degenerate motifs, raising the possibility that TNKS could regulate stability of additional centrosomal proteins. Previous studies have demonstrated localization of other PARPs (PARP1 [24] and PARP3 [25]) to interphase centrosomes and PARP1 was found to be required for maintenance of proper centrosome number [26]. Hence, PARsylation (through multiple PARPs and targets) may have a general role in centrosome function.
Methods
PARP assays. Samples containing recombinant baculovirus-derived TNKS1 (0.2 μg), 25 μM biotinylated NAD+ (Trevigen), and recombinant E. coli–derived GST–CPAP-C or GST–CPAP-N (2 μg) or baculovirus-derived TRF1 (2 μg) were performed as described previously [16]. See supplementary Methods online for further detail.
Immunoprecipitation. Cell lysates generated in TNE buffer (10 mM Tris (pH 7.8), 1% Nonidet P-40, 0.15 M NaCl, 1 mM EDTA) were immunoprecipitated with anti-Flag or anti-myc agarose bead conjugates (Sigma), fractionated on SDS–polyacrylamide gel electrophoresis gels, and processed for immunoblotting (supplementary Methods online).
Stable cell lines. TNKS1 shRNA cell lines (11, 12, and 13) and the GFP shRNA line were generated by lentiviral infection of HTC75 cells as previously described [27]. F7 is a HTC75 cell line containing an inducible allele of FlagTNKS1. F7 cells were grown in parallel under uninduced (with doxycyclin) and induced (without doxycyclin) conditions for 16 h (supplementary Methods online).
Indirect immunofluorescence. For analysis of elongated centrioles cells were cold-treated for 1 h at 4 °C, pre-extracted with Triton X-100 (0.5%) in PBS for 1 min, and then fixed in methanol at −20 °C for 10 min. For all other immunofluorescence analysis, cells were fixed in methanol at −20 °C for 10 min (supplementary Methods online).
Supplementary Material
Acknowledgments
We thank K. Bisht for comments on the manuscript and helpful discussion. We are grateful to B. Dynlacht for anti-cp110 antibody and helpful discussion, J. Salisbury for anti-centrin antibody, and J. Visvader for the FlagCPAP plasmid. This work was supported by National Institutes of Health Grant R01 CA095099.
Authors contributions: M.K.K., C.D., and S.S. designed experiments; M.K.K. and C.D. performed experiments; and M.K.K. and S.S. wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Azimzadeh J, Bornens M (2007) Structure and duplication of the centrosome. J Cell Sci 120: 2139–2142 [DOI] [PubMed] [Google Scholar]
- Nigg EA, Stearns T (2011) The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol 13: 1154–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P, Gonczy P (2008) Mechanisms of procentriole formation. Trends Cell Biol 18: 389–396 [DOI] [PubMed] [Google Scholar]
- Leidel S, Gonczy P (2003) SAS-4 is essential for centrosome duplication in C. elegans and is recruited to daughter centrioles once per cell cycle. Dev Cell 4: 431–439 [DOI] [PubMed] [Google Scholar]
- Pelletier L, O'Toole E, Schwager A, Hyman AA, Muller-Reichert T (2006) Centriole assembly in Caenorhabditis elegans. Nature 444: 619–623 [DOI] [PubMed] [Google Scholar]
- Hsu WB, Hung LY, Tang CJ, Su CL, Chang Y, Tang TK (2008) Functional characterization of the microtubule-binding and -destabilizing domains of CPAP and d-SAS-4. Exp Cell Res 314: 2591–2602 [DOI] [PubMed] [Google Scholar]
- Hung LY, Tang CJ, Tang TK (2000) Protein 4.1 R-135 interacts with a novel centrosomal protein (CPAP) which is associated with the gamma-tubulin complex. Mol Cell Biol 20: 7813–7825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA (2007) Plk4-induced centriole biogenesis in human cells. Dev Cell 13: 190–202 [DOI] [PubMed] [Google Scholar]
- Kohlmaier G, Loncarek J, Meng X, McEwen BF, Mogensen MM, Spektor A, Dynlacht BD, Khodjakov A, Gonczy P (2009) Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr Biol 19: 1012–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TI, Kleylein-Sohn J, Westendorf J, Le Clech M, Lavoie SB, Stierhof YD, Nigg EA (2009) Control of centriole length by CPAP and CP110. Curr Biol 19: 1005–1011 [DOI] [PubMed] [Google Scholar]
- Tang CJ, Fu RH, Wu KS, Hsu WB, Tang TK (2009) CPAP is a cell-cycle regulated protein that controls centriole length. Nat Cell Biol 11: 825–831 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA (2011) Centrosomes and cilia in human disease. Trends Genet 27: 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J et al. (2005) A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet 37: 353–355 [DOI] [PubMed] [Google Scholar]
- Thornton GK, Woods CG (2009) Primary microcephaly: do all roads lead to Rome? Trends Genet 25: 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao SJ, Smith S (2008) Tankyrase function at telomeres, spindle poles, and beyond. Biochimie 90: 83–92 [DOI] [PubMed] [Google Scholar]
- Smith S, Giriat I, Schmitt A, de Lange T (1998) Tankyrase, a poly(ADP-ribose) polymerase at human telomeres (see comments). Science 282: 1484–1487 [DOI] [PubMed] [Google Scholar]
- Chang W, Dynek JN, Smith S (2003) TRF1 is degraded by ubiquitin-mediated proteolysis after release from telomeres. Genes Dev 17: 1328–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SM et al. (2009) Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461: 614–620 [DOI] [PubMed] [Google Scholar]
- Guettler S, LaRose J, Petsalaki E, Gish G, Scotter A, Pawson T, Rottapel R, Sicheri F (2011) Structural basis and sequence rules for substrate recognition by Tankyrase explain the basis for cherubism disease. Cell 147: 1340–1354 [DOI] [PubMed] [Google Scholar]
- Sbodio JI, Chi NW (2002) Identification of a tankyrase-binding motif shared by IRAP, TAB182, and human TRF1 but not mouse TRF1. NuMA contains this RXXPDG motif and is a novel tankyrase partner. J Biol Chem 277: 31887–31892 [DOI] [PubMed] [Google Scholar]
- Cook BD, Dynek JN, Chang W, Shostak G, Smith S (2002) Role for the related poly(ADP-Ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol Cell Biol 22: 332–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang WY, Spektor A, Vijayakumar S, Bista BR, Li J, Sanchez I, Duensing S, Dynlacht BD (2009) Cep76, a centrosomal protein that specifically restrains centriole reduplication. Dev Cell 16: 649–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarguaglini G, Duncan PI, Stierhof YD, Holmstrom T, Duensing S, Nigg EA (2005) The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol Biol Cell 16: 1095–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai M, Uchida M, Hanai S, Uematsu N, Uchida K, Miwa M (2000) Poly(ADP-ribose) polymerase localizes to the centrosomes and chromosomes. Biochem Biophys Res Commun 278: 385–389 [DOI] [PubMed] [Google Scholar]
- Augustin A et al. (2003) PARP-3 localizes preferentially to the daughter centriole and interferes with the G1/S cell cycle progression. J Cell Sci 116: 1551–1562 [DOI] [PubMed] [Google Scholar]
- Kanai M, Tong WM, Sugihara E, Wang ZQ, Fukasawa K, Miwa M (2003) Involvement of poly(ADP-Ribose) polymerase 1 and poly(ADP-Ribosyl)ation in regulation of centrosome function. Mol Cell Biol 23: 2451–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao SJ, Smith S (2009) Sister telomeres rendered dysfunctional by persistent cohesion are fused by NHEJ. J Cell Biol 184: 515–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.