Abstract
A subset of amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD) patients present pathological redistribution and aggregation of the nuclear protein fused in sarcoma (FUS) in the cytoplasm. Although FUS associates with the spliceosomal complex, no endogenous neuronal splicing targets have been identified. Here we identify Tau mRNA as a physiological splicing target of FUS. In mouse brain, FUS directly binds to Tau pre-mRNA, and knockdown of FUS in hippocampal neurons leads to preferential inclusion of Tau exons 3 and 10. FUS knockdown causes significant growth cone enlargement and disorganization reminiscent of Tau loss of function. These findings suggest that disturbed cytoskeletal function and enhanced expression of the neurodegeneration-associated Tau exon 10 might contribute to FTLD/ALS with FUS inclusions.
Keywords: neurodegeneration, FUS, RNA-binding protein, splicing, Tau (MAPT)
Introduction
Frontotemporal lobar degeneration (FTLD) and amyotrophic lateral sclerosis (ALS) have recently been recognized as opposite ends of a disease spectrum with overlapping clinical symptoms, genetics and pathology [1, 2]. These devastating neurodegenerative diseases are now subdivided by specific marker proteins identified in abnormal aggregates, including Fused in sarcoma (FUS) [2–4]. The RNA/DNA-binding protein FUS normally resides predominantly in the nucleus, but is found in cytosolic inclusions of both FTLD and ALS patients. FUS binds to numerous mRNAs in HEK293 cells, however with little correlation to transcript abundance [5]. Although FUS associates with the spliceosomal complex [6], no endogenous neuronal splicing target is known so far and the role of FUS-mediated splicing in FTLD/ALS remains unclear. Two observations suggest that the loss of nuclear FUS activity contributes to FTLD/ALS pathophysiology: First, mutations disrupting the C-terminal nuclear targeting sequence of FUS cause early-onset ALS [3, 4, 7]. Second, even in the absence of FUS mutations, a partial nuclear clearing of FUS is apparent in neurons with FUS aggregates [1].
Here we identify the microtubule-associated protein Tau (MAPT) as a physiological splicing target of FUS in neurons and found morphological alterations in axons and growth cones that are reminiscent of Tau-knockout mice [8]. The MAPT gene consists of 16 exons and gives rise to six main transcripts in the central nervous system (supplementary Fig S1A online) [9, 10]. The shortest Tau isoform, termed 0N3R, lacks exons 2, 3 and 10. Inclusion of exon 2 or exons 2 and 3 leads to insertion of one or two 29-amino acid regions (1N, 2N). Inclusion of exon 10 gives rise to a fourth microtubule-binding region (4R). The composition of the N-terminal projection domain determines microtubule spacing and might affect cellular signalling [9, 11]. Somatodendritic Tau aggregates with characteristic isoform composition are found in Alzheimer’s disease and a subset of FTLD collectively termed FTLD–Tau, including Pick’s disease and corticobasal degeneration [2, 12]. Mutations that lead to the preferential inclusion of exon 10 cause frontotemporal dementia and parkinsonism [13], showing the importance of exon 10 inclusion for FTLD–Tau disease pathology. Our data show that FUS knockdown enhances expression of Tau exon 10. Tau aggregation has so far not been described in patients with FUS pathology. However, there is evidence that non-fibrillar Tau can cause neurodegeneration and cognitive symptoms [14, 15]. For example, the H1 haplotype at the MAPT locus moderately enhances exon 10 inclusion and is a strong genetic risk factor for Parkinson’s disease, which also lacks overt Tau pathology [16, 17].
Results and discussion
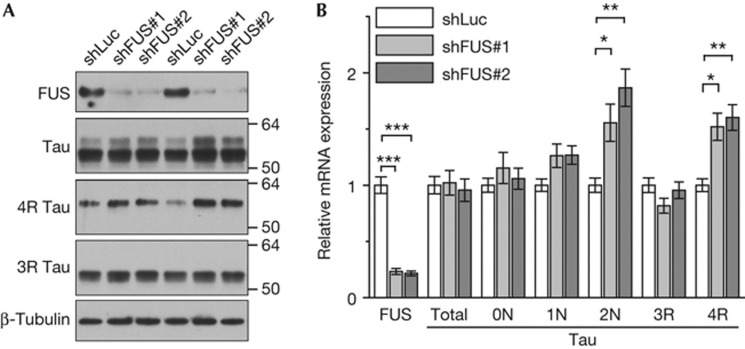
FUS knockdown affects levels of 4R Tau
Tau has been linked to many neurodegenerative disorders including FTLD–Tau. Therefore, we analysed Tau expression upon FUS knockdown in rat primary hippocampal neurons using lentiviral expression of short hairpin RNAs (shRNAs). We transduced neurons at day 2 in vitro and collected proteins 7 days later (DIV2+7). Two FUS-specific shRNAs (shFUS#1 and shFUS#2) specifically suppressed FUS protein expression compared with a control shRNA targeting the luciferase transcript (shLuc; Fig 1A). An XTT-based cell viability assay confirmed that FUS knockdown showed no overt toxicity in this context (supplementary Fig S2A online). Although the main Tau isoform (lower band) was unchanged, a larger isoform (upper band) appeared upregulated in FUS-knockdown neurons (Fig 1A). A Tau exon 10-specific antibody showed robustly increased levels of 4R Tau (comigrating with the upper Tau band), suggesting an effect of FUS on Tau splicing. In contrast, 3R Tau expression was not significantly affected by FUS knockdown. The amount and running behaviour of several other key synaptic proteins were unaffected by FUS knockdown, including synaptic scaffold proteins (PSD95), glutamate-receptor subunits (NR1, NR2A and GluR2), synaptic vesicle proteins (synaptophysin) and cytoskeletal proteins (β-actin and β-tubulin, Fig 1A; supplementary Fig S2B online). Together, these data strongly indicate that FUS knockdown affects splicing of Tau exon 10.
Figure 1.
FUS knockdown enhances expression of Tau exons 3 and 10. Hippocampal neurons (DIV2+7) were infected with lentivirus to knockdown FUS (shFUS#1 and #2) or non-targeting control (shLuc). (A) Immunoblots with the indicated antibodies. Note that the two main Tau isoforms comigrate with 4 R and 3 R Tau. Two replicates are shown. (B) Total RNA was analysed by quantitative PCR for FUS, total Tau and Tau isoforms. FUS and total Tau expression was normalized to the housekeeping gene YWHAZ. Tau isoform expression was normalized to total Tau levels. n=4, mean±s.e.m., one-way analysis of variance with Dunnett’s post-test: *P<0.05, **P<0.01, ***P<0.001. FUS, Fused in sarcoma; shFUS, short hairpin FUS; shLuc, short hairpin RNA targeting the luciferase transcript; YWHAZ, tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta/14-3-3-zeta.
FUS knockdown affects Tau mRNA splicing
To further explore how FUS knockdown modifies Tau splicing in neurons, we designed isoform-specific primers for the alternatively spliced exons 2, 3 and 10 (supplementary Fig S1B online). Reverse transcription and quantitative real-time PCR (qPCR) allowed for quantitative comparison of exon insertion between FUS knockdown and control shRNA-infected neurons. About 80% reduction of FUS levels did not significantly affect total Tau mRNA expression (Fig 1B), but caused significant upregulation of the 2N and 4R Tau isoforms.
As FUS and TDP-43 aggregation cause similar clinical symptoms in FTLD and ALS patients, it is tempting to hypothesize that FUS and TDP-43 regulate overlapping mRNA targets [1]. We therefore used lentiviral shRNA to downregulate TDP-43 in hippocampal neurons and analysed the expression of Tau isoforms by qPCR. Consistent with previous results [18], splicing of exons 2, 3 and 10 was not significantly affected by TDP-43 knockdown (supplementary Fig S3 online). Thus, aberrant Tau splicing might contribute to FTLD/ALS pathophysiology with FUS inclusions, but not with TDP-43 inclusions.
FUS reintroduction rescues altered Tau splicing
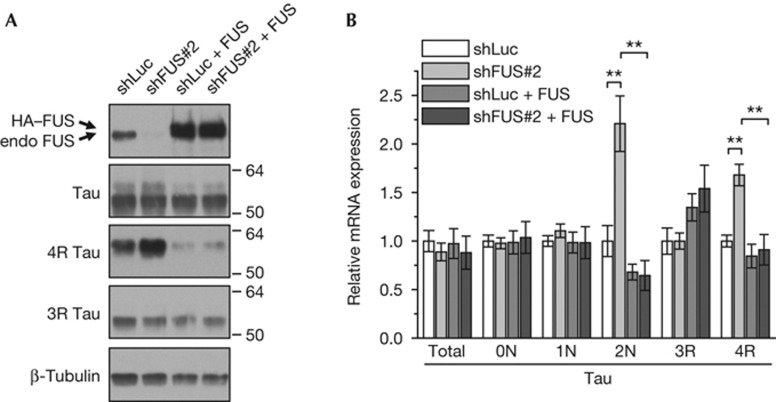
To confirm that altered Tau splicing upon shFUS treatment is mediated by loss of FUS and not by off-target effects, we performed rescue experiments by reintroduction of FUS in shRNA-treated cells. We doubly infected neurons with lentiviral vectors expressing shRNA (shLuc or shFUS#2) and haemagglutinin (HA)-tagged wild-type human FUS resistant to shFUS#2. Transduction with HA–FUS strongly increased FUS-expression compared with the endogenous levels of shLuc control infected cells (Fig 2A). Importantly, HA–FUS co-transduction not only prevented the shFUS#2-mediated upregulation of 4R Tau, but even suppressed its expression below shLuc control levels (Fig 2A). Similarly, HA–FUS coexpression completely abolished the induction of 2N and 4R isoforms by shFUS on mRNA level (Fig 2B). The full rescue of the shFUS#2-mediated splicing effects by overexpression of FUS strongly argues for a FUS-specific effect.
Figure 2.
FUS overexpression fully rescues the Tau splicing phenotype. Hippocampal neurons (DIV2+7) were coinfected with lentivirus expressing the shLuc or shFUS#2 and HA-tagged shRNA-resistant FUS. (A) Immunoblots with the indicated antibodies. (B) Total RNA was analysed by quantitative PCR for total Tau and isoforms. Total Tau expression was normalized to YWHAZ. Tau isoform expression was normalized to total Tau levels. n=3, mean±s.e.m, analysis of variance with Bonferroni correction: **P<0.01. Note that samples from shLuc- and shFUS-transduced neurons were statistically indistinguishable on coexpression of shFUS-resistant FUS. FUS, Fused in sarcoma; HA, haemagglutinin; shFUS, short hairpin FUS; shLuc, short hairpin RNA targeting the luciferase transcript.
FUS is associated with Tau transcripts in the brain
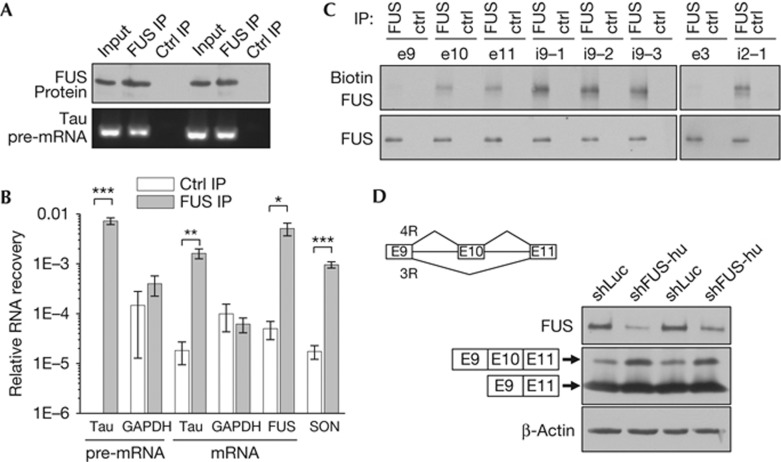
To test whether FUS is associated with Tau RNA, we performed immunoprecipitations from mouse brain (P15) extracts using a FUS antibody and a GST antibody as a negative control, followed by isolation of the bound RNA. By reverse transcription and qPCR we amplified Tau pre-mRNA using intronic primers and compared the recovery of Tau pre-mRNA from the immunoprecipitates. While the control immunoprecipitates contained no detectable Tau pre-mRNA (Fig 3A,B), FUS-immunoprecipitates recovered about 1% of the input material, which strongly suggests a specific interaction. Using intron-spanning primers, we also detected spliced Tau mRNA specifically enriched (but to a lesser extent than pre-mRNA) in FUS-immunoprecipitates. The enrichment of Tau mRNA was comparable to two positive controls recently identified by crosslinking and immunoprecipitation (CLIP) analysis of FUS-bound RNAs in HEK293 cell, its own mRNA and SON (Fig 3B; [5]). In contrast, glyceraldehyde-3-phosphate dehydrogenase pre-mRNA and mRNA were not enriched in FUS-immunoprecipitates. Taken together, these data show that FUS is associated with Tau pre-mRNA and mRNA in mouse brain.
Figure 3.
FUS is associated with Tau pre-mRNA in brain and affects splicing of a minigene reporter construct. (A) Anti-FUS immunoblot of mouse brain (age 15 days) extracts, FUS- and control-immunoprecipitates. FUS-associated RNA was reverse transcribed and analysed by quantitative PCR. pre-mRNA-specific primers bind in introns 9 and 11 adjacent to Tau exon 10 (compare supplementary Fig S1C online). The end product after quantitative PCR analysis is shown. (B) RNA recovery from FUS-immunoprecipitates compared with control-immunoprecipitates. For both groups, individual immunoprecipitations from six mouse brains were analysed by quantitative PCR. SON primers spanning a verified FUS-binding motif within a large exon amplify both pre-mRNA and mRNA and served as positive control together with FUS mRNA [5]. n=6, mean±s.e.m, Student’s t-test: *P<0.05, **P<0.01, ***P<0.001. (C) Ultraviolet crosslinking of biotinylated RNA probes with FUS in mouse brain extracts (age 15 days). Detection of biotinylated FUS using strepatavidin and anti-FUS immunoblot of FUS- and control-immunoprecipitates. (D) HEK293 cells co-transfected with Tau exons 9 to 11 minigene construct (depicted in inset) and the indicated shRNAs. HA-tagged alternatively spliced protein product, FUS and β-actin were detected by immunoblotting. Ctrl, control; FUS, Fused in sarcoma; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IP, immunoprecipitation; shFUS, short hairpin FUS; shLuc, short hairpin RNA targeting the luciferase transcript.
FUS directly binds to Tau transcripts at multiple sites
To analyse whether the interaction of FUS with Tau RNA is direct, we incubated mouse brain extracts with biotinylated RNA probes followed by ultraviolet crosslinking, RNase A digest and FUS immunoprecipitation [19]. RNA probes covering the FUS-regulated exon 10 and the neighbouring exon 11 resulted in significant biotinylation of FUS compared with an exon 9 spanning probe, indicating a direct and specific interaction of FUS with these RNA fragments (Fig 3C; supplementary Fig S5B online). CLIP analysis in HEK293 cells had revealed preferential interaction of FUS with AU-rich intronic RNA that was corroborated by in vitro binding assays with artificial AUU-repeats [5]. Interestingly, mouse Tau pre-mRNA contains three clusters with multiple AUU motifs in intron 9 (9–17 AUU within ∼300 nucleotides) and RNA probes spanning these regions showed strong interaction with FUS in the crosslinking assay (compare i9-1, -2 and -3 in Fig 3C). Probe i9-2 is located closest to exon 10 and shows the strongest conservation between rodents and humans (supplementary Fig S5A,C online). Although FUS-knockdown led to preferential inclusion of Tau exon 3, we could not detect significant interaction of an exon 3 spanning RNA probe with FUS. However, the region within intron 2 with the highest conservation between mouse and human Tau (supplementary Fig S5A,D online) was strongly crosslinked to FUS, although it contained only four AUU motifs. Thus, FUS directly binds to Tau RNA at several sites near the FUS-regulated exons 3 and 10.
FUS knockdown promotes exon 10 inclusion in minigene
To further analyse the effects of FUS on pathologically relevant Tau splicing in a heterologous system, we used a minigene construct covering mouse exons 9 to 11 that contains the full intron sequences and expresses short HA-tagged Tau protein fragments, corresponding to the 3R/4R Tau domains. HEK293 cells co-transfected with the 3R/4R minigene and a control shRNA (shLuc) predominantly express fragments corresponding to 3R Tau and some 4R Tau. However, FUS knockdown (using shFUS-hu) led to a shift towards the longer fragment corresponding to 4R Tau, suggesting that endogenous FUS promotes skipping of exon 10 in the heterologous system (Fig 3D). These data corroborate the effects of FUS knockdown on Tau 4R expression in hippocampal neurons (Fig 1).
FUS knockdown disturbs cytoskeletal organization
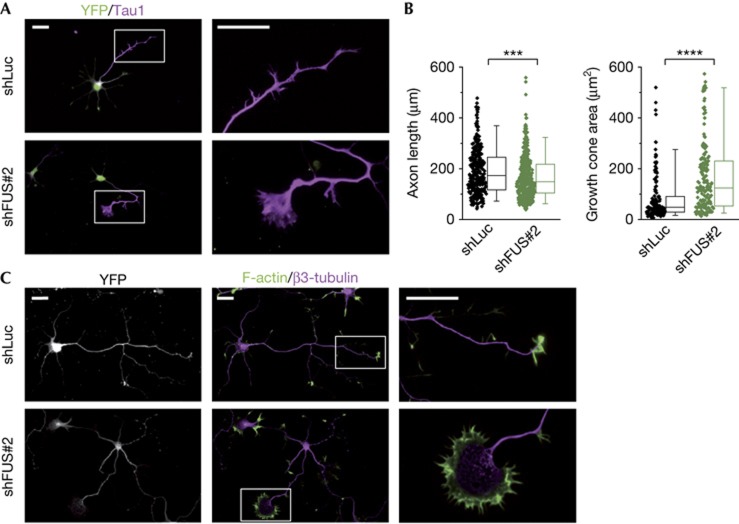
To address whether FUS knockdown has functional consequences on neuronal development, we transfected hippocampal neurons before plating with shFUS#2 or an unspecific control (shLuc). FUS knockdown was apparent by immunostaining at DIV4 (supplementary Fig S4A online). Toxicity assays performed showed no difference between groups even after prolonged knockdown (supplementary Fig S4C online). After 4 days in culture, both neurons transfected with shLuc or shFUS sent out a similar number of neurites (Fig 4A; supplementary Fig S4B online) and had developed an axon (89% and 88%, respectively) that was identified by immunostaining with the axonal marker Tau1 [20]. Furthermore, neurons lacking FUS showed normal axonal branching (supplementary Fig S4B online). However, axons in FUS-knockdown neurons were slightly, but significantly shorter than in control cells (Fig 4B, left panel) and interestingly, the shFUS-transfected neurons typically developed severely enlarged growth cones. We found that the growth cone area of the shFUS-transfected neurons was in average twice as large as in the control cells (Fig 4B, right panel). When analysing tubulin and actin localization in axonal growth cones (Fig 4C), we observed that in shFUS-transfected neurons microtubules spread further into the enlarged growth cone area and appeared less bundled, which resembles findings in Tau- and MAP1B-knockout neurons [8]. Together, these data indicate that FUS has an important role in cytoskeletal organization, particularly in the organization of microtubule network at axonal tips.
Figure 4.
FUS knockdown affects axon and growth cone morphology. Hippocampal neurons co-transfected with the shLuc or shFUS#2 and pEYFP-C1 before plating. (A) Immunostaining at day 4 with anti-YFP as transfection control and anti-Tau1 as axonal marker. Right panels show a high-magnification view of growth cones. For morphometric analysis processes with proximal-to-distal Tau1 gradient were defined as axons. (B) Quantification of axonal length (left, n=325 for shLuc, n=388 for shFUS#2) and growth cone area (right, n=147 for shLuc, n=164 for shFUS#2) measured blinded to the experimental condition. Box-plots represent lower quartile, median and upper quartile. Whiskers represent the 5th and 95th percentile. Mann–Whitney test: ***P<0.001, ****P<0.0001. (C) Staining for F-actin (using phalloidin) and β3-tubulin. Right panels show a high-magnification view of axonal growth cones. Scale bars, 25 μm. FUS, Fused in sarcoma; shFUS, short hairpin FUS; shLuc, short hairpin RNA targeting the luciferase transcript; YFP, yellow fluorescent protein.
Conclusion
In this study, we established Tau as the first physiological splice target of FUS in neurons. This was corroborated on both mRNA and protein level. FUS knockdown promotes inclusion of exons 3 and 10 and thus expression of 2N- and 4R-containing Tau isoforms. Moreover, regulation of exon 10 was confirmed in a minigene reporter assay. We found that Tau pre-mRNA is associated with FUS in the brain and demonstrated direct binding to several intronic and exonic regions of Tau RNA, suggesting a direct role in Tau splicing. Consistent with recent CLIP analysis [5], we found the most robust binding of FUS to intronic regions containing multiple AUU motifs, although this was no absolute prerequisite for binding.
Enhanced expression of 4R Tau is a well-established cause of neurodegeneration both in the presence and absence of overt Tau aggregation [13–16]. Our data indicate that FTLD–Tau and FTLD–FUS share disease mechanisms through increased Tau 4R expression. Increased expression of 4R Tau has been linked to Parkinson’s disease and is a strong predictor of progression to dementia [16, 17, 21]. Interestingly, FUS is downregulated in brains from Parkinson patients [22]. Furthermore, Tau variants are a risk factor for the Guam variant of ALS [23]. The consequences of enhanced 2N expression are poorly understood. Because the interaction of the Tau N-terminal region with Fyn has been linked to excitotoxicity, altered Tau splicing might contribute to the loss of mature dendritic spines observed in FUS-knockout mice potentially causing synaptic dysfunction in FTLD/ALS [9, 24, 25].
We observed shortened axonal length and growth cone enlargement on FUS-knockdown that are reminiscent of Tau-knockout mice [8]. Although we cannot exclude that other FUS-regulated genes contribute to this phenotype, altered Tau isoform expression might disturb cytoskeletal function in the axon and thus affect growth cone organization. It will be crucial to investigate Tau splicing and cytoskeletal aberrations in FTLD/ALS cases with FUS pathology and vice versa look for altered FUS expression in other tauopathies.
Methods
Antibodies. FUS (Bethyl, A300-292A), Tau (Dako, A 0024), 3R Tau (Millipore, 8E6/C11), 4R Tau (Millipore, 1E1/A6), Tau1 (Millipore, PC1C6), β-actin (Sigma, AC-15), β3-Tubulin (Sigma, SDL.3D10 and Covance, Tuj1), synaptophysin (Millipore, SY38), PSD95 (Neuromab, K28/43), NR1 (BD, 54.1), NR2A (Covance, PRB-513P), GluR2 (Neuromab, L21/32), and GFP (rabbit, Fitzgerald Industries International) were used.
DNA constructs and transfection. shRNA targeting FUS (rat #1 5′-ggcctaggcgagaatgtta-3′, rat #2 5′-gtgcaaggcctaggcgaga-3′, human 5′-ggacagcagcaaagctatg-3′), TDP-43 (5′-gtagatgtcttcattcccaaa-3′) and firefly luciferase (5′-cgtacgcggaatacttcga-3′) at the indicated sites were cloned into pSUPER (Oligoengine). For lentiviral knockdown, the shRNA-expression cassette was subcloned into a lentiviral vector coexpressing mCherry from human ubiquitin C promoter [26]. HA-tagged human FUS complementary DNA containing silent mutations in the shRNA-binding sites were expressed under the human synapsin promoter in a lentiviral vector [26].A Tau minigene construct containing exons 9 to 11 was cloned by homologous recombination using the Red/ET system (Genebridges) from a mouse genomic BAC (Imagenes) into an expression vector (derived from pBUD-CE4, Invitrogen) containing an N-terminal myc-tag and a C-terminal HA-tag. For the splicing assay, the minigene construct was co-transfected in HEK293 cells that had been transfected with shRNAs two days earlier using Lipofectamine 2000 (Invitrogen) to allow sufficient FUS knockdown.To generate RNA probes for the crosslinking experiments, 250–400 bp fragments of Tau pre-mRNA were cloned from mouse genomic DNA into pGEM3 (Promega) or pCR-blunt2 TOPO (Invitrogen) and transcribed into RNA using the MAXIscript T7 kit (Invitrogen) in the presence of biotin-16-UTP (Biotin RNA labelling mix, Roche) according to the manufacturer’s instruction.All constructs were verified by DNA sequencing. All oligos are listed in supplementary Table S1 online.
Neuronal culture, transfection and immunostaining. Hippocampal neurons for lentiviral infections were cultured from embryonic day 18 or 19 rats, as described previously, in a feeder-free system using B27 (Invitrogen) [26]. Neurons were lysed in Laemmli buffer and run on 10% denaturing SDS–PAGE gels. Cell viability was measured using the XTT cell proliferation kit (Roche). For the analysis of neuronal morphology (neurite number, axonal length and branching), we cultured rat hippocampal neurons on astrocyte feeder cells using N2 media [27]. The neurons were transfected before plating using an Amaxa 4D-Nucleofector (Lonza) with primary culture kit P3 (program EM110).Hippocampal neurons were fixed with 4% paraformaldehyde, quenched in 50 mM ammonium chloride and permeabilized with 0.1% Triton X-100. After blocking with 2% fetal bovine serum (Invitrogen), 2% bovine serum albumin (Sigma-Aldrich), and 0.2% fish gelatin (Sigma-Aldrich) dissolved in phosphate-buffered saline neurons were incubated with primary antibodies diluted in 10% blocking solution.For the quantification of growth cone area, neurite number and axonal branching, a minimum of 45 cells per condition were analysed in three independent experiments. For axonal length quantifications, a minimum of 99 cells per condition in three independent experiments were measured using AxioVision software (Zeiss).
Reverse transcription and quantitative PCR analysis. Total RNA was extracted using RNeasy Mini Kit (Qiagen) and reverse transcribed with random hexanucleotide primers using TaqMan Reverse Transcription Kit (Applied Biosystems, Invitrogen). The complementary DNA was amplified using qPCR SsoFast Evagreen Supermix (Biorad) and analysed in the CFX384 Real-Time System (Biorad).
FUS immunoprecipitation and RNA isolation. Mouse brains (day 15) were homogenized in 2-ml lysis buffer (20 mM Tris–HCl, pH 7.4, 450 mM NaCl, 0.1 mM EDTA, 1.5 mM MgCl2, 1 mM CaCl2, 0.6% Triton X-100) containing SUPERase RNase inhibitor (Ambion, 1 U/μl) and protease and phosphatase inhibitor cocktails (Sigma). After 70 000 g centrifugation (20 min), supernatants were immunoprecipitated for 1 h with Dynabeads Protein G (Invitrogen) precoupled with FUS or GST antibody (10 μg antibody/75 μl beads). After four washes with lysis buffer, the bound RNA was extracted using RNeasy Mini Kit including treatment with DNase (Qiagen).
Crosslinking of FUS with Tau pre-mRNA. Direct interaction of FUS with RNA was analysed analogous to protocols established for TDP-43 [19]. Mouse brains (day 15) were homogenized as described above (but in the absence of RNase inhibitor). After centrifugation, the supernatant was diluted with 2 volumes of lysis buffer lacking salt (20 mM Tris–HCl, pH 7.4, 0.1 mM EDTA, 1.5 mM MgCl2, 1 mM CaCl2, 0.6% Triton X-100). Endogenously biotinylated proteins in the lysates were depleted using streptavidin agarose (Sigma). Protein extracts (500 μg) were incubated with biotinylated RNA probes for 30 min at room temperature and crosslinked on ice by ultraviolet irradiation (254 nm, 400 mJ/cm2) using a StrataLinker 1800 (Stratagene). After RNase A digestion (15 min, 10 μg/ml, Sigma), the lysates were subjected to immunoprecipitation using anti-FUS or anti-GST antibodies as described above. Crosslinking of biotinylated RNA was detected using Streptavidin-Peroxidase (Sigma).
Supplementary Material
Acknowledgments
We thank S. Kunath and A. Seibel for technical assistance. We are grateful to B. Schmid, M. Müller, S. Lammich, E. Bentmann, D. Dormann, A. Capell and J. Strathmann for helpful discussion and reagents. DE was supported by the Helmholtz Young Investigator program HZ-NG-607.
Author contributions: D.O. performed and analysed most experiments. S.T. analysed the morphological phenotype. K.R. cloned most constructs. B.M.S. performed cytotoxicity assays and cloned constructs. C.H. supervised research. D.E. cloned the minigene construct, designed and supervised the project and wrote the manuscript with input from all authors.
Footnotes
The authors declare that they have no conflict of interest.
References
- Mackenzie IR, Rademakers R, Neumann M (2010) TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol 9: 995–1007 [DOI] [PubMed] [Google Scholar]
- Josephs KA, Hodges JR, Snowden JS, Mackenzie IR, Neumann M, Mann DM, Dickson DW (2011) Neuropathological background of phenotypical variability in frontotemporal dementia. Acta Neuropathol 122: 137–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C et al. (2009) Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323: 1208–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ Jr. et al. (2009) Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323: 1205–1208 [DOI] [PubMed] [Google Scholar]
- Hoell JI et al. (2011) RNA targets of wild-type and mutant FET family proteins. Nat Struct Mol Biol 18: 1428–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner M, Lopato S, Gotzmann J, Sauermann G, Barta A (2003) Proto-oncoprotein TLS/FUS is associated to the nuclear matrix and complexed with splicing factors PTB, SRm160, and SR proteins. Exp Cell Res 283: 184–195 [DOI] [PubMed] [Google Scholar]
- Dormann D et al. (2010) ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J 29: 2841–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei Y, Teng J, Harada A, Hirokawa N (2000) Defects in axonal elongation and neuronal migration in mice with disrupted tau and map1b genes. J Cell Biol 150: 989–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris M, Maeda S, Vossel K, Mucke L (2011) The many faces of tau. Neuron 70: 410–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreadis A (2005) Tau gene alternative splicing: expression patterns, regulation and modulation of function in normal brain and neurodegenerative diseases. Biochim Biophys Acta 1739: 91–103 [DOI] [PubMed] [Google Scholar]
- Chen J, Kanai Y, Cowan NJ, Hirokawa N (1992) Projection domains of MAP2 and tau determine spacings between microtubules in dendrites and axons. Nature 360: 674–677 [DOI] [PubMed] [Google Scholar]
- Goedert M, Spillantini MG (2011) Pathogenesis of the tauopathies. J Mol Neurosci 45: 425–431 [DOI] [PubMed] [Google Scholar]
- Hutton M et al. (1998) Association of missense and 5'-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 393: 702–705 [DOI] [PubMed] [Google Scholar]
- Brunden KR, Trojanowski JQ, Lee VM (2008) Evidence that non-fibrillar tau causes pathology linked to neurodegeneration and behavioral impairments. J Alzheimers Dis 14: 393–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Calignon A, Fox LM, Pitstick R, Carlson GA, Bacskai BJ, Spires-Jones TL, Hyman BT (2010) Caspase activation precedes and leads to tangles. Nature 464: 1201–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin JE et al. (2008) Haplotypes and gene expression implicate the MAPT region for Parkinson disease: the GenePD Study. Neurology 71: 28–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey TM, Joachim C, Paracchini S, Esiri MM, Wade-Martins R (2006) Haplotype-specific expression of exon 10 at the human MAPT locus. Hum Mol Genet 15: 3529–3537 [DOI] [PubMed] [Google Scholar]
- Polymenidou M et al. (2011) Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci 14: 459–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiesel FC et al. (2010) Knockdown of transactive response DNA-binding protein (TDP-43) downregulates histone deacetylase 6. EMBO J 29: 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell JW, Banker GA (1996) A spatial gradient of tau protein phosphorylation in nascent axons. J Neurosci 16: 5727–5740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Gray CH et al. (2009) The distinct cognitive syndromes of Parkinson’s disease: 5 year follow-up of the CamPaIGN cohort. Brain 132: 2958–2969 [DOI] [PubMed] [Google Scholar]
- Stamper C et al. (2008) Neuronal gene expression correlates of Parkinson's disease with dementia. Mov Disord 23: 1588–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundar PD et al. (2007) Two sites in the MAPT region confer genetic risk for Guam ALS/PDC and dementia. Hum Mol Genet 16: 295–306 [DOI] [PubMed] [Google Scholar]
- Fujii R, Okabe S, Urushido T, Inoue K, Yoshimura A, Tachibana T, Nishikawa T, Hicks GG, Takumi T (2005) The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr Biol 15: 587–593 [DOI] [PubMed] [Google Scholar]
- Ittner LM et al. (2010) Dendritic function of tau mediates amyloid-beta toxicity in Alzheimer's disease mouse models. Cell 142: 387–397 [DOI] [PubMed] [Google Scholar]
- Edbauer D et al. (2010) Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 65: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S, Banker G (2006) Culturing hippocampal neurons. Nat Protoc 1: 2406–2415 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.