EMBO Rep (2012) 13 8, 716–723. doi:; DOI: 10.1038/embor.2012.82
MicroRNAs (miRNAs) are a large family of endogenous non-coding RNAs that, together with the Argonaute family of proteins (AGOs), post-transcriptionally silence the expression of complementary mRNA targets [1,2]. Perfectly complementary targets are cleaved by AGOs [2]. However, to silence partly complementary targets, AGOs must recruit a protein of the GW182 family [2]. GW182 proteins play an essential role, as most animal miRNA targets contain partly complementary miRNA binding sites [2].
…common key finding that translational repression precedes complete deadenylation and degradation of the targeted mRNAs
Virtually all cellular processes investigated thus far involve miRNAs. Despite their widespread role as regulators of gene expression, the mechanisms by which miRNAs repress their targets remain a source of scientific debate [1,2]. The emerging consensus in the field is that miRNAs repress translation and promote deadenylation and subsequent degradation of their target mRNAs. However, the precise contribution and timing of these effects has not been clearly determined.
Three studies, including one published in this issue of EMBO reports by Filipowicz's group, investigate the kinetics of miRNA-mediated translational repression and target deadenylation in Drosophila melanogaster and human cells, and in zebrafish embryos [3,4,5]. Although these studies differ in their details, they all converge on the common key finding that translational repression precedes complete deadenylation and degradation of the targeted mRNAs.
The earliest studies addressing the mechanism of miRNA regulation suggested that miRNAs repress the translation of their targets after initiation, with little or no influence on mRNA levels [1,2]. Subsequent studies reported three further mechanisms by which the translational repression of miRNA targets could be achieved: (i) co-translational protein degradation; (ii) premature ribosome dissociation; and (iii) the inhibition of translation initiation [1,2]. Furthermore, it became clear that miRNAs promote mRNA destabilization [2].
More recently, genome-wide studies demonstrated that degradation of miRNA targets is a widespread effect, which, at steady state, accounts for most of the miRNA-mediated repression in mammalian cell cultures [6,7,8]. These studies did not rule out that target degradation occurred as a consequence of an initial block in translation, but they indicate that if translational repression does occur it probably occurs at initiation, rather than at a subsequent step of translation.
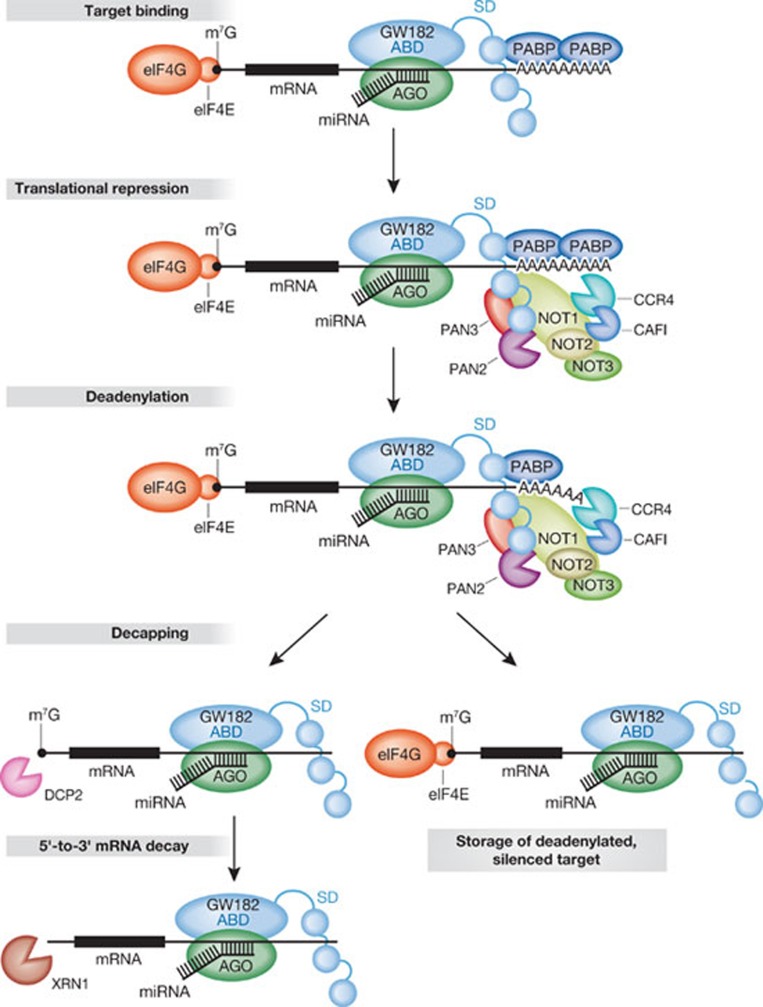
How are miRNA targets degraded? It is known that miRNAs accelerate target destruction by recruiting enzymes of the 5′-to-3′ mRNA decay pathway [2]. In this pathway, mRNAs are first deadenylated, then decapped and, finally, degraded from the 5′ end by the exonuclease XRN1 (Fig 1). mRNA deadenylation is catalysed by the sequential action of two deadenylase complexes: the PAN2–PAN3 and the CCR4–NOT complexes (Fig 1; [2]). Both of these complexes have been shown to interact directly with GW182 proteins [9,10,11].
Figure 1. The mechanism of microRNA-mediated gene silencing in animals.
The GW182 proteins consist of an amino-terminal AGO-binding domain (ABD) and a silencing domain (SD) [2]. The silencing domain interacts with the cytoplasmic PABP and with the deadenylase complexes through multiple sequence motifs, which are represented as circles [2,9,10,11]. Two deadenylase complexes interact with GW182 proteins: the dimeric PAN2–PAN3 complex and the CCR4–NOT complex [2]. The mRNA is shown in a linear conformation with the translation factors eIF4E and eIF4G bound to the cap structure (m7G) and PABP bound to the poly(A) tail. In animal cell cultures, deadenylated mRNAs are decapped by the decapping enzyme DCP2 and further co-factors (not shown). Please refer to the text for more information. AGO, Argonaute; PABP, poly(A) binding protein.
Despite the progress made in understanding how miRNAs trigger target degradation, the question of whether translation is inhibited before deadenylation and decay remained unresolved. To dissect the temporal effects of miRNAs on mRNA translation, deadenylation and decay Filipowicz's and Green's groups used inducible luciferase reporters [3,4]. At early time points, both studies reported the repression of protein production before deadenylation. At later time points, both groups observed mRNA degradation that correlates with full silencing [3,4]. In both studies, poly(A) tail length measurements indicate no significant shortening at early time points [3,4].
Giraldez's group investigated the order of events in early zebrafish embryos [5], in which the most abundant miRNA is miR-430 [12]. miR-430 expression increases substantially after fertilization, and this correlates with the degradation of a large number of maternal mRNAs containing miR-430 binding sites [12]. Giraldez and colleagues found that at the onset of miR-430 expression (4 h after fertilization), miR-430 targets are translationally repressed before deadenylation is completed. Six hours after fertilization, however, most targets are degraded, as previously reported by these authors [12]. In fact, targets that were translationally repressed at early time points (4 h) subsequently underwent decay, indicating that most targets experience both regulatory effects. Giraldez and colleagues therefore conclude that translational repression occurs before complete deadenylation.
One tantalizing possibility is that the recruitment of deadenylase complexes […] triggers both translational repression and deadenylation
In combination with the previously published data, the three new studies discussed above allow for the formulation of the following stepwise model of miRNA-mediated regulation in animals (Fig 1). First, a miRNA bound to an AGO protein recognizes an mRNA target and recruits a protein of the GW182 family. The AGO–GW182 complex represses translation; this repression probably occurs at initiation through an unknown mechanism. The repressed mRNA is then deadenylated by deadenylase complexes, which are recruited through interactions with GW182 proteins [9,10,11]. Depending on the cell type and the specific target, deadenylated mRNAs might be stored in their translationally repressed state [2]. In animal cell cultures, deadenylated mRNAs are generally decapped and rapidly degraded by the major 5′-to-3′ exonuclease XRN1 [2].
As suggested by Filipowicz and colleagues, the stepwise model of silencing detailed above allows for regulation and reversibility at multiple steps of the process. For instance, the transitions between translational repression and deadenylation, or deadenylation and decapping, could be regulated in a target- and cell-specific manner, leading to different outcomes for miRNA-mediated regulation.
This model raises several important questions that represent challenges to be addressed by future work. The most urgent of these questions is the issue of how translational repression is achieved. It is also unclear whether translational repression and deadenylation are interconnected or independent mechanisms—with different rates—used by miRNAs to silence mRNA targets. Indeed, although the observation that translational repression precedes deadenylation is interpreted by Giraldez and colleagues as evidence for two parallel repressive mechanisms, the two processes could still be mechanistically linked and could therefore represent two consecutive outcomes of a single molecular mechanism that both interferes with translation and triggers deadenylation.
What might be the initial trigger for such a mechanism? One tantalizing possibility is that the recruitment of deadenylase complexes to the 3′ UTR of miRNA targets triggers both translational repression and deadenylation. This possibility is supported by the observation that NOT1 depletion suppresses the silencing of reporters lacking a poly(A) tail [9,10]. Furthermore, tethering subunits of the CCR4–NOT complex to mRNA reporters lacking a poly(A) tail induces translational repression in the absence of deadenylation [10]. These data suggest that deadenylase complexes could not only promote deadenylation but also contribute to translational repression. If this scenario is correct, it would be important to determine the mechanism by which the deadenylase complexes repress translation and the ways in which these complexes interact with the translation and silencing machineries.
Footnotes
The author declares that she has no conflict of interest.
References
- Djuranovic S et al. (2011) Science 331: 550–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E (2011) Nat Rev Genet 12: 99–110 [DOI] [PubMed] [Google Scholar]
- Béthune J et al. (2012) EMBO Rep (in the press) [Google Scholar]
- Djuranovic S et al. (2012) Science 336: 237–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA et al. (2012) Science 336: 233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M et al. (2008) Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Baek D et al. (2008) Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H et al. (2010) Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun JE et al. (2011) Mol Cell 44: 120–133 [DOI] [PubMed] [Google Scholar]
- Chekulaeva M et al. (2011) Nat Struct Mol Biol 18: 1218–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR et al. (2011) Nat Struct Mol Biol 18: 1211–1217 [DOI] [PubMed] [Google Scholar]
- Giraldez AJ et al. (2006) Science 312: 75–79 [DOI] [PubMed] [Google Scholar]