Abstract
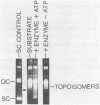
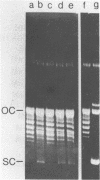
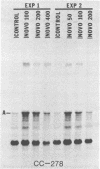
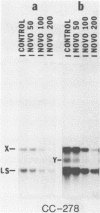
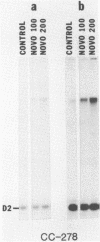
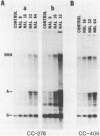
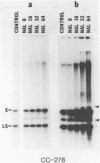
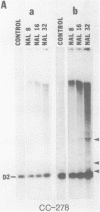
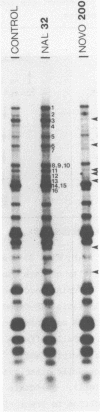
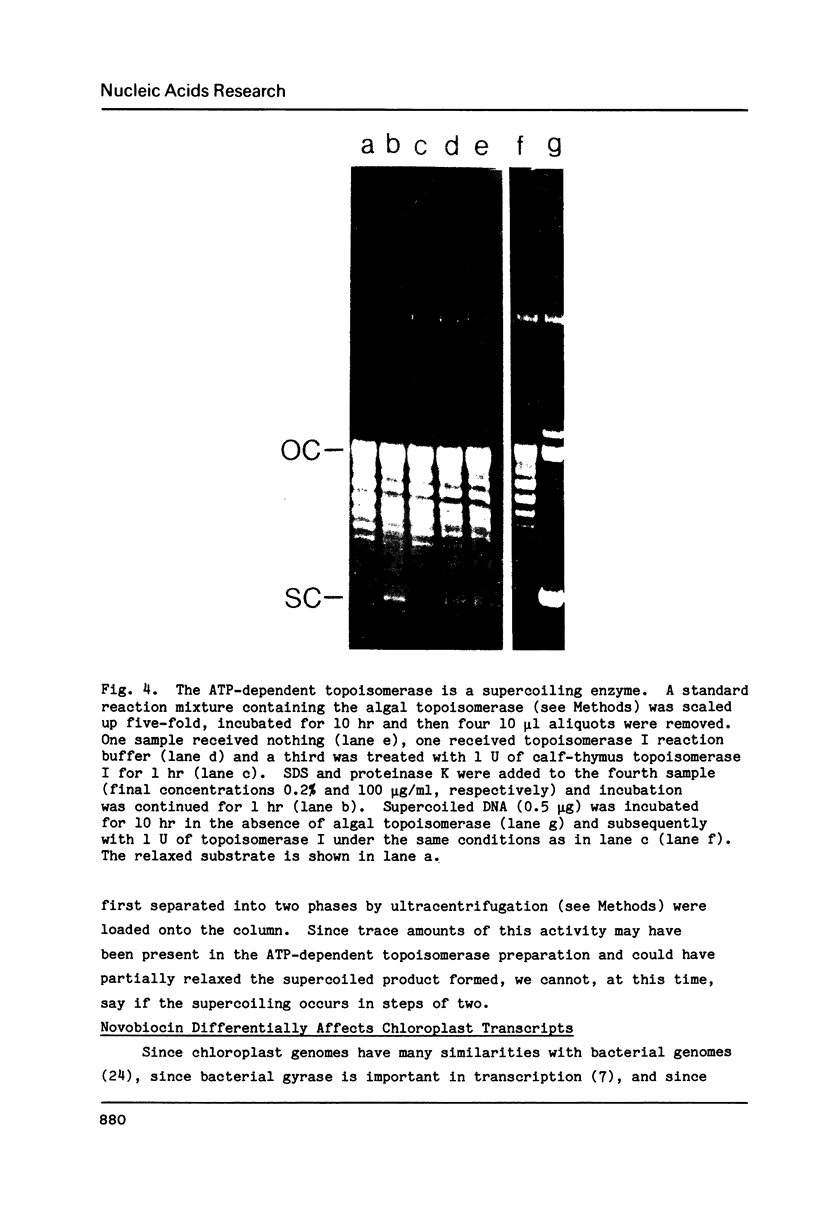
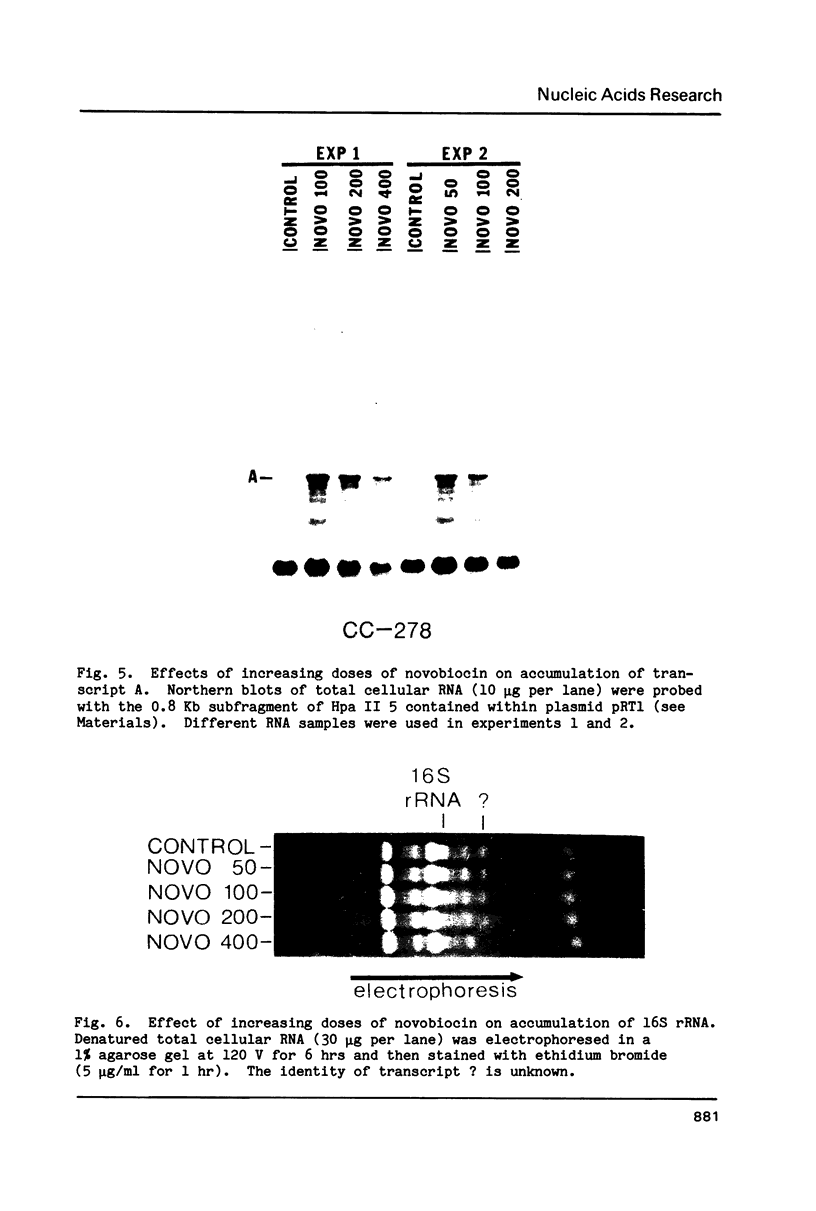
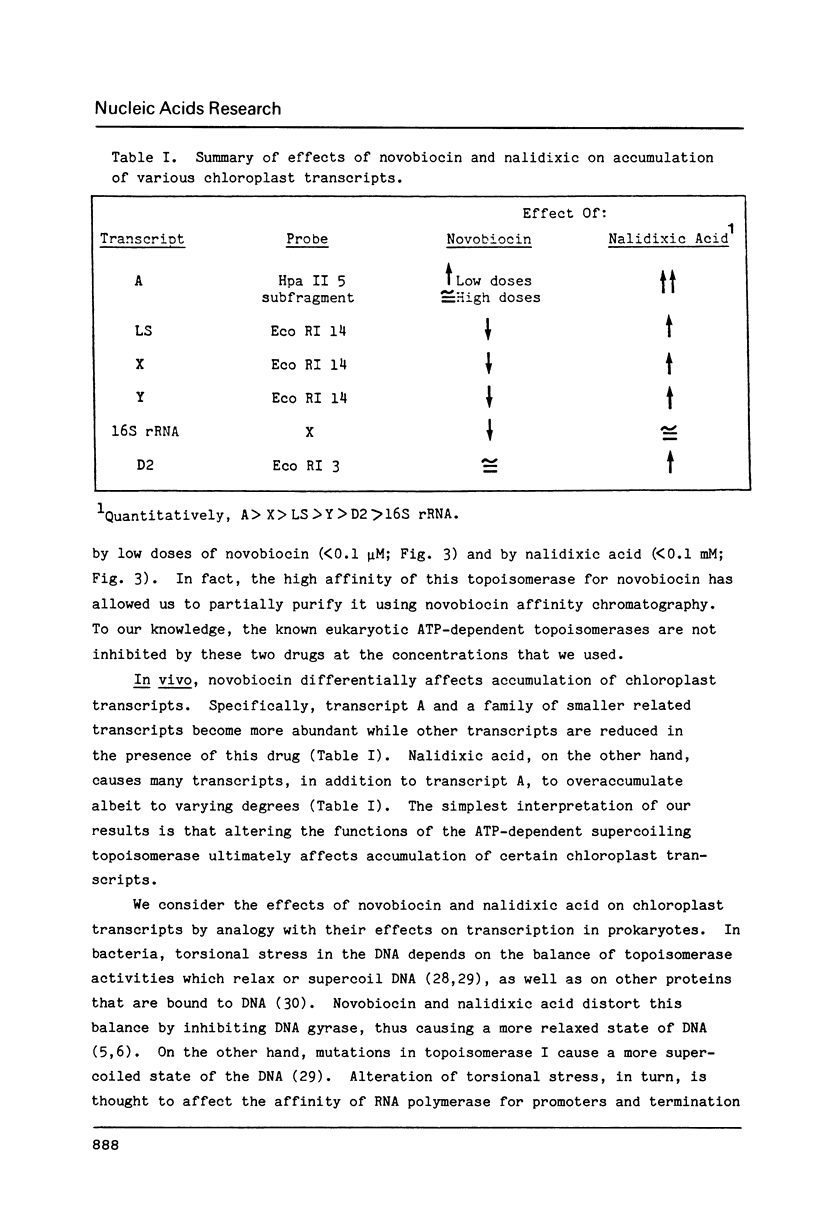
We have found that Chlamydomonas reinhardtii cells contain an ATP-dependent topoisomerase activity that supercoils circular DNA in vitro. Subsequent addition of a type I topoisomerase eliminates the supercoils. Like bacterial gyrase, this activity is inhibited by low concentrations of novobiocin (less than 0.1 microM) and by nalidixic acid (less than 0.1 microM). We have examined the effects of these topoisomerase inhibitors on accumulation of various chloroplast transcripts in vivo. Novobiocin differentially affected such transcripts; some transcripts became more abundant while many others were reduced in the presence of this drug. Nalidixic acid on the other hand caused many transcripts to become more abundant albeit to varying degrees. Inhibitors of this algal topoisomerase specifically stimulate a family of related transcripts which we have previously shown to be under light-dark control. We discuss how the inhibitors of this topoisomerase might exert their in vivo effects.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Colwill R. W., Sheinin R. ts A1S9 locus in mouse L cells may encode a novobiocin binding protein that is required for DNA topoisomerase II activity. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4644–4648. doi: 10.1073/pnas.80.15.4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzarelli N. R. DNA gyrase and the supercoiling of DNA. Science. 1980 Feb 29;207(4434):953–960. doi: 10.1126/science.6243420. [DOI] [PubMed] [Google Scholar]
- DiNardo S., Voelkel K. A., Sternglanz R., Reynolds A. E., Wright A. Escherichia coli DNA topoisomerase I mutants have compensatory mutations at or near DNA gyrase genes. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):779–784. doi: 10.1101/sqb.1983.047.01.089. [DOI] [PubMed] [Google Scholar]
- Drlica K., Snyder M. Superhelical Escherichia coli DNA: relaxation by coumermycin. J Mol Biol. 1978 Apr 5;120(2):145–154. doi: 10.1016/0022-2836(78)90061-x. [DOI] [PubMed] [Google Scholar]
- Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin S., Heizmann P., Howell S. H. Identification and cloning of the chloroplast gene coding for the large subunit of ribulose-1,5-bisphosphate carboxylase from Chlamydomonas reinhardi. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3193–3197. doi: 10.1073/pnas.74.8.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glikin G. C., Ruberti I., Worcel A. Chromatin assembly in Xenopus oocytes: in vitro studies. Cell. 1984 May;37(1):33–41. doi: 10.1016/0092-8674(84)90298-8. [DOI] [PubMed] [Google Scholar]
- Grant D. M., Gillham N. W., Boynton J. E. Inheritance of chloroplast DNA in Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1980 Oct;77(10):6067–6071. doi: 10.1073/pnas.77.10.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi A., Asai K. Reverse gyrase--a topoisomerase which introduces positive superhelical turns into DNA. Nature. 1984 Jun 21;309(5970):677–681. doi: 10.1038/309677a0. [DOI] [PubMed] [Google Scholar]
- Kreuzer K. N., Alberts B. M. Site-specific recognition of bacteriophage T4 DNA by T4 type II DNA topoisomerase and Escherichia coli DNA gyrase. J Biol Chem. 1984 Apr 25;259(8):5339–5346. [PubMed] [Google Scholar]
- Lyman H., Jupp A. S., Larrinua I. Action of Nalidixic Acid on Chloroplast Replication in Euglena gracilis. Plant Physiol. 1975 Feb;55(2):390–392. doi: 10.1104/pp.55.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnoë P., Rochaix J. D., Chua N. H., Spahr P. F. Characterization of the gene and messenger RNA of the large subunit of ribulose 1,5-diphosphate carboxylase in Chlamydomonas reinhardii. J Mol Biol. 1979 Sep 25;133(3):417–434. doi: 10.1016/0022-2836(79)90401-7. [DOI] [PubMed] [Google Scholar]
- Manes S. H., Pruss G. J., Drlica K. Inhibition of RNA synthesis by oxolinic acid is unrelated to average DNA supercoiling. J Bacteriol. 1983 Jul;155(1):420–423. doi: 10.1128/jb.155.1.420-423.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R., Gellert M. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell. 1983 Aug;34(1):105–113. doi: 10.1016/0092-8674(83)90140-x. [DOI] [PubMed] [Google Scholar]
- Orr E., Staudenbauer W. L. Bacillus subtilis DNA gyrase: purification of subunits and reconstitution of supercoiling activity. J Bacteriol. 1982 Jul;151(1):524–527. doi: 10.1128/jb.151.1.524-527.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix J. D. Restriction endonuclease map of the chloroplast DNA of Chlamydomonas reinhardii. J Mol Biol. 1978 Dec 25;126(4):597–617. doi: 10.1016/0022-2836(78)90011-6. [DOI] [PubMed] [Google Scholar]
- Siedlecki J., Zimmermann W., Weissbach A. Characterization of a prokaryotic topoisomerase I activity in chloroplast extracts from spinach. Nucleic Acids Res. 1983 Mar 11;11(5):1523–1536. doi: 10.1093/nar/11.5.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden R. R., Pettijohn D. E. Torsional tension in intracellular bacteriophage T4 DNA. Evidence that a linear DNA duplex can be supercoiled in vivo. J Mol Biol. 1982 Dec 15;162(3):659–677. doi: 10.1016/0022-2836(82)90394-1. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Staudenbauer W. L., Orr E. DNA gyrase: affinity chromatography on novobiocin-Sepharose and catalytic properties. Nucleic Acids Res. 1981 Aug 11;9(15):3589–3603. doi: 10.1093/nar/9.15.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewey K. M., Chen G. L., Nelson E. M., Liu L. F. Intercalative antitumor drugs interfere with the breakage-reunion reaction of mammalian DNA topoisomerase II. J Biol Chem. 1984 Jul 25;259(14):9182–9187. [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R. J., Mosig G. Light and Genetic Determinants in the Control of Specific Chloroplast Transcripts in Chlamydomonas reinhardtii. Plant Physiol. 1984 Sep;76(1):1–6. doi: 10.1104/pp.76.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984 Mar;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D. C. Structure and evolution of organelle genomes. Microbiol Rev. 1982 Jun;46(2):208–240. doi: 10.1128/mr.46.2.208-240.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]