Synaptic activity–dependent remodeling of the glutamate receptor scaffold complex generates a negative feedback loop that limits further NMDA receptor activation.
Abstract
Scaffolding proteins interact with membrane receptors to control signaling pathways and cellular functions. However, the dynamics and specific roles of interactions between different components of scaffold complexes are poorly understood because of the dearth of methods available to monitor binding interactions. Using a unique combination of single-cell bioluminescence resonance energy transfer imaging in living neurons and electrophysiological recordings, in this paper, we depict the role of glutamate receptor scaffold complex remodeling in space and time to control synaptic transmission. Despite a broad colocalization of the proteins in neurons, we show that spine-confined assembly/disassembly of this scaffold complex, physiologically triggered by sustained activation of synaptic NMDA (N-methyl-d-aspartate) receptors, induces physical association between ionotropic (NMDA) and metabotropic (mGlu5a) synaptic glutamate receptors. This physical interaction results in an mGlu5a receptor–mediated inhibition of NMDA currents, providing an activity-dependent negative feedback loop on NMDA receptor activity. Such protein scaffold remodeling represents a form of homeostatic control of synaptic excitability.
Introduction
Protein–protein interactions play key roles in cellular processes. Identifying such multiprotein complexes has been a major issue in the discovery of key biological pathways. Indeed, in their natural environment, membrane receptors are associated with scaffolding proteins that link them to their intracellular signal transduction pathways and cytoskeleton. Such receptor-associated scaffolds are relatively stable structures, but exchange of individual adaptor proteins can occur at a fast time scale and in a highly regulated manner, which provides fine tuning, speed, and specificity of the receptor signaling (Zeke et al., 2009). Therefore, understanding how proteins are activated as free molecules or part of complexes is an essential biological concern. Currently, the molecular detail of the dynamics of these interactions and the roles that they play in various cellular functions are poorly defined because of the dearth of methods for acutely and specifically controlling the binding interactions.
For instance, at brain synapses, scaffolding proteins function not only as anchors but also as signaling proteins for neurotransmitter receptors. As synapses are dynamic structures, it is a fundamental issue to study the dynamics of such synaptic receptor scaffolds and their role in neurotransmission. Synaptic transmission involves neurotransmitter receptors and ion channels. Their targeting, functioning, and dynamic exchanges at synapses depend on their interaction with synaptic scaffolding proteins (Renner et al., 2008). Glutamatergic neurotransmission in the mammalian brain is mainly mediated by the ligand-gated AMPA (2-amino-3-(5-methyl-3-oxo-1,2-oxazol-4-yl)propanoic acid) and kainate receptor channels and modulated by NMDA (N-methyl-d-aspartate) receptor channels and G protein–coupled/metabotropic receptors (mGlu). Interestingly, group I mGlu receptor (mGlu1 and mGlu5) and NMDA receptors are physically linked together in the postsynaptic density by a Homer protein–containing complex (Brakeman et al., 1997; Scannevin and Huganir, 2000). Functional interaction between the NMDA and group I mGlu receptors has been extensively studied (Gerber et al., 2007; Homayoun and Moghaddam, 2010), but whether dynamic exchange of Homer proteins controls functional cross talk between those receptors has received little attention (Bertaso et al., 2010).
The Homer family of postsynaptic proteins displays an EVH1 (Ena/Vasp homology) N-terminal domain, which recognizes a proline-rich sequence (PPxxF) of protein partners. The long forms of Homer proteins (Homer1b, Homer1c, Homer2, and Homer3) display a central hinge region followed by a C-terminal coiled-coil domain that is organized in two separate regions, CC1 and CC2 (Hayashi et al., 2006). These proteins are constitutively expressed in neurons and other cell types. They form a tetrameric hub structure composed of two antiparallel dimers at the postsynaptic density (Hayashi et al., 2006). This structure confers slow turnover rates and great efficiency in coordinating dendritic spine functions. Shank proteins are major components of the postsynaptic density that are assembled in high-order complexes through self-association of their sterile α-motif C-terminal domain. Together with Homer multimers, they are major constituents of a platform that cross-links group I mGlu receptors to the guanylate kinase–associated protein (GKAP)–PSD95-NMDA receptor assembly in dendritic spines (Baron et al., 2006). The short form of Homer proteins, Homer1a, also displays an EVH1 domain but lacks of a coiled-coil domain. Therefore, this protein can virtually bind to the same targets as the long forms of Homer proteins but cannot multimerize. This confers to Homer1a dominant-negative properties and the ability to disrupt Homer-associated multiprotein complexes. In contrast to the long forms of Homer proteins, Homer1a is an immediate early gene that is transiently expressed after elevated neuronal activity or intake of drug of abuse such as cocaine, amphetamine, LSD, phencyclidine, and nicotine (Xiao et al., 2000; Fagni et al., 2002; Szumlinski et al., 2006, 2008). Homer1a plays fundamental physiological functions, such as modulating synaptic plasticity and temporal encoding of spatial memory (Celikel et al., 2007) or enabling homeostatic scaling (Hu et al., 2010).
Because of its dominant-negative properties, Homer1a is a viable mean that may trigger dynamic changes in the association/dissociation of mGlu receptors to the GKAP-PSD95-NMDA receptor assembly. Therefore, it is fundamental to depict the spatiotemporal dynamics of mutually exclusive interactions of long and short forms of Homer proteins with mGlu receptors. Bioluminescence resonance energy transfer (BRET) is a very sensitive technology that became in the past decade a technology of choice to study the dynamic of protein–protein interactions in living cells (see Boute et al. [2002] and Pfleger and Eidne [2006] for reviews about BRET). In this assay, the efficacy of the energy transfer depends on the close proximity (<10 nm) and orientation of the donor and acceptor entities. The mean radius of proteins being 5 nm, the occurrence of resonance energy transfer is interpreted as a strong indication that the proteins attached to the energy donors and acceptors, respectively, are indeed in direct contact. We recently adapted BRET to single-cell imaging analysis to study subcellular localization of protein–protein interactions under microscope in living cells (Coulon et al., 2008; Perroy, 2010). Here, we investigated by single-cell BRET imaging the spatiotemporal remodeling of the interactions between Homer proteins and mGlu5a receptor in cultured hippocampal neurons and its consequence on NMDA receptor function. We found that competition between Homer1a and multimeric Homer proteins on mGlu5a receptor binding is virtually restricted to dendritic spines. Such a scaffold remodeling triggers a direct physical interaction between mGlu5a and NMDA receptors and inhibition of NMDA currents. Our data also evidence that such a regulation occurs after sustained activation of synaptic NMDA receptors to restore synaptic NMDA current. Thus, this remodeling controls synapse excitability. To our knowledge, this is the first study showing temporal dynamic remodeling of a multiprotein scaffold and its function at a subcellular level in living cells.
Results
Two different adaptor proteins, the long and short Homer isoforms, competitively interact with mGlu5a receptor in living cells
The proline-rich Homer ligand domain (PPxxFr) identified on mGlu5a receptor indifferently coimmunoprecipitates with the short and long forms of Homer (Tu et al., 1998), suggesting that competitive interaction may occur between the two Homer isoforms. We investigated the modality of these mutually exclusive interactions in HEK-293 living cells by BRET. We fused the C terminus of mGlu5a receptor to the energy donor Renilla luciferase (Rluc8) and the N terminus of the long Homer3 or short Homer1a protein to the acceptor YFP (Myc-mGlu5a-Rluc8, YFP-Homer, and YFP-Homer1a, respectively). The obtained tagged proteins were functional (Fig. S1, A and B). Under the condition of a constant level of Myc-mGlu5a-Rluc8 expression, BRET signal increased hyperbolically as a function of YFP-Homer1a or YFP-Homer expression level (Fig. 1 A). Saturation of the BRET signal when all the donor was linked to the acceptor indicated a specific interaction between the mGlu5a receptor and Homer proteins. For a constant donor/acceptor ratio, the BRET signal between Myc-mGlu5a-Rluc8 and either YFP-Homer1a or YFP-Homer remained constant whatever the total level of protein expression (Fig. 1 B). By opposition, the point mutant Myc-mGlu5a-P1124K-Rluc8, which could not interact with Homer proteins (Tu et al., 1998), did not display BRET signal neither with YFP-Homer1a nor YFP-Homer (Fig. 1 A). NMDA receptor (NR1A-Rluc + NR2B) and YFP-Homer proteins did not display BRET signal either, confirming the specificity of interaction between Homer proteins and the mGlu5 receptor.
Figure 1.
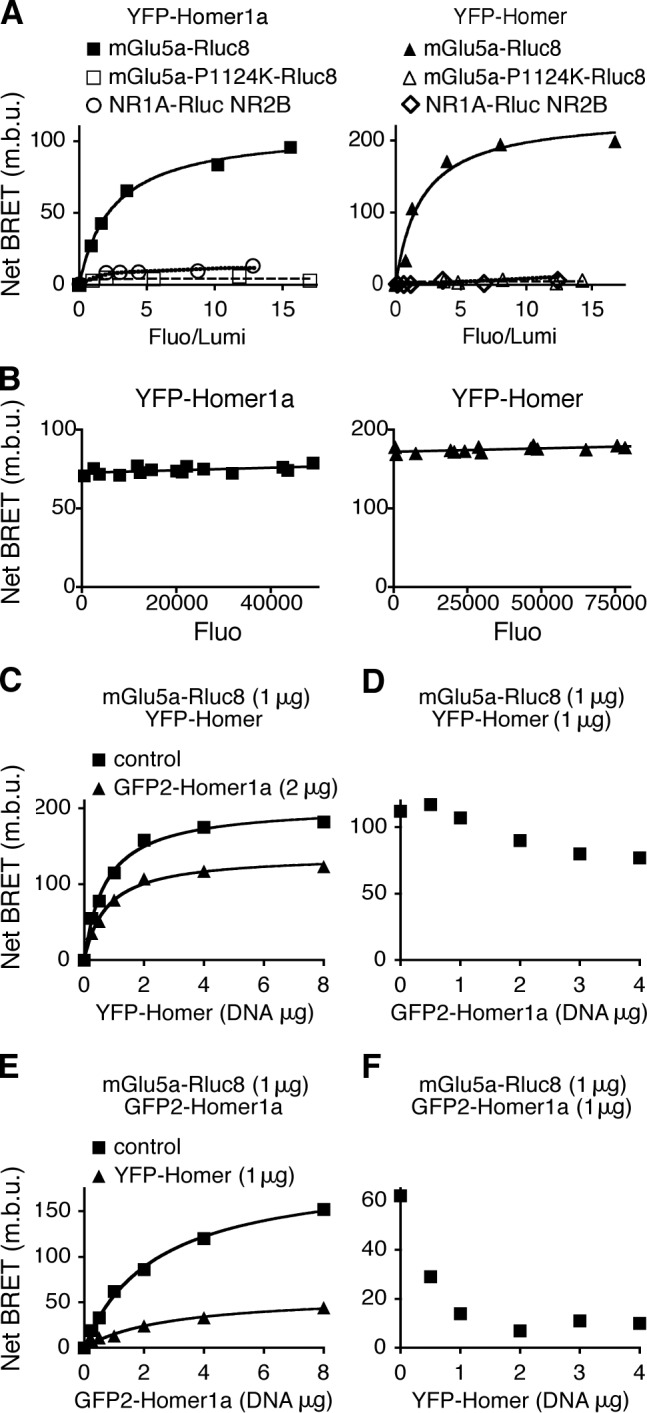
The long and short Homer isoforms specifically and competitively interact with mGlu5a receptor in living cells. (A) Cells were cotransfected with a constant concentration of mGlu5a-Rluc8 or mGlu5a-P1124K-Rluc8 and increasing concentrations of YFP-Homer1a (left) or YFP-Homer3 (referred to as YFP-Homer; right) expression plasmids. To exclude a direct action of Homer proteins on NMDA receptors, the BRET between NR1A-Rluc + NR2B and YFP-Homer1a or YFP-Homer3 was also monitored. Fluo, fluorescence; Lumi, luminescence; m.b.u., milli-BRET unit. (B) The BRET signal is independent of the total level of protein expression, thus reporting a specific interaction, providing that the ratio of acceptor/donor remains constant. Therefore, cells were transfected with a constant ratio (fluorescence/luminescence of 5.1 ± 0.5 [left] and 5.6 ± 0.5 [right]) of YFP-Homer1a/mGlu5a-Rluc8 (left) or YFP-Homer/mGlu5a-Rluc8 (right) and various concentrations of total plasmids. (C–F) Cells were transfected with the following plasmids: mGlu5a-Rluc8, GFP2-Homer1a, and YFP-Homer. Coelenterazine H (BRET1; C and D) or DeepBlueC coelenterazine (BRET2; E and F) was added on two distinct pools of the same population of transfected cells to monitor concomitantly BRET between mGlu5a-Rluc8 and YFP-Homer or GFP2-Homer1a, respectively. Titration curves (C and E) were obtained in the presence of constant concentration of mGlu5a-Rluc8 and increasing concentrations of YFP-Homer, with or without GFP2-Homer1a (C) or increasing concentrations of GFP2-Homer1a with or without YFP-Homer (E). Dose-response competitions (D and F) were measured between constant concentration of mGlu5a-Rluc8 and YFP-Homer for increasing concentration of GFP2-Homer1a (D) or constant concentration of mGlu5a-Rluc8 and GFP2-Homer1a in the presence of increasing concentration of YFP-Homer (F). Displacement of the interaction between mGlu5a receptor and a given isoform by the other isoform of Homer indicates a competitive interaction between the two Homer proteins for the receptor. (A–F) For each condition, data are representative of five independent experiments performed in triplicate.
To monitor competition between Homer1a and the long form of Homer proteins for the binding to the mGlu5a receptor in the same cell, we took advantage of the distinct spectral properties of two luciferase substrates, coelenterazine H and DeepBlueC coelenterazine (see Materials and methods). The coexpression of Homer1a slightly reduced the binding of cotransfected Homer3 to the receptor (Fig. 1, C and D). Reciprocally, Homer3 expression impaired the interaction between Homer1a and mGlu5a receptor but with significantly higher efficacy (Figs. 1 [E and F] and S1 [C–F]). Thus, concomitant detection of Homer1a and Homer3 interactions with mGlu5a receptor in cells coexpressing the three partners revealed that the competitive binding between the two Homer proteins on mGlu5a receptor occurred in living cells. The shift in interaction between mGlu5a and Homer induced by the presence of a concurrent Homer isoform emphasized the apparent higher affinity of the receptor for the multimeric Homer than for the monomeric Homer1a isoform. Both long multimeric isoforms of Homer tested (Homer3 and Homer1c) displayed similar affinity for the mGlu5a receptor (in all the following experiments, the multimeric Homer3 was used and referred to as Homer; Fig. S1 G).
In living neurons, the competition between long and short Homer isoforms on mGlu5a is confined to dendritic spines
We further studied mGlu5a receptor–Homer interactions in cultured hippocampal neurons by BRET imaging. The highest BRET signals between mGlu5a-Rluc8 and the multimeric form of Homer, YFP-Homer, were found in the neurites, as compared with the cell body. Despite a similar mean BRET intensity, quantification of the high SD indeed indicates a clusterization in neurites (Fig. 2 A). We then compared BRET signals in the dendritic spine and shaft. Spines were chosen according to their morphological criteria using the YFP-Homer fluorescence: a protrusion composed of a large spine head (diameter ranging from 0.5 to 1.0 µm) connected to the dendrite via a membranous neck. BRET images revealed the highest intensity of BRET signals between mGlu5a-Rluc8 and YFP-Homer confined to spines (image 535/480 in Fig. 2 [B–D]). Interestingly, this occurred despite colocalization and equal abundance of the two proteins in both the dendritic shaft and spine (image Em480 for mGlu5a-Rluc8 and image GFP for YFP-Homer in Fig. 2 B). In contrast to the interaction between multimeric Homer and mGluR5, the interaction between the mGlu5a-Rluc8 and the monomeric form, YFP-Homer1a, was equally distributed in the cell body and neurites (Fig. 2 A). Importantly, the long-term expression of transfected Homer1a induced dramatic decrease in spine number and size, which was consistent with the previously described inhibitory effect of the protein on synaptic transmission (Sala et al., 2003). This effect on dendritic spines hampered reliable experiments of precise dendritic localization and physical interaction between mGlu5a-Rluc8 and YFP-Homer1a.
Figure 2.
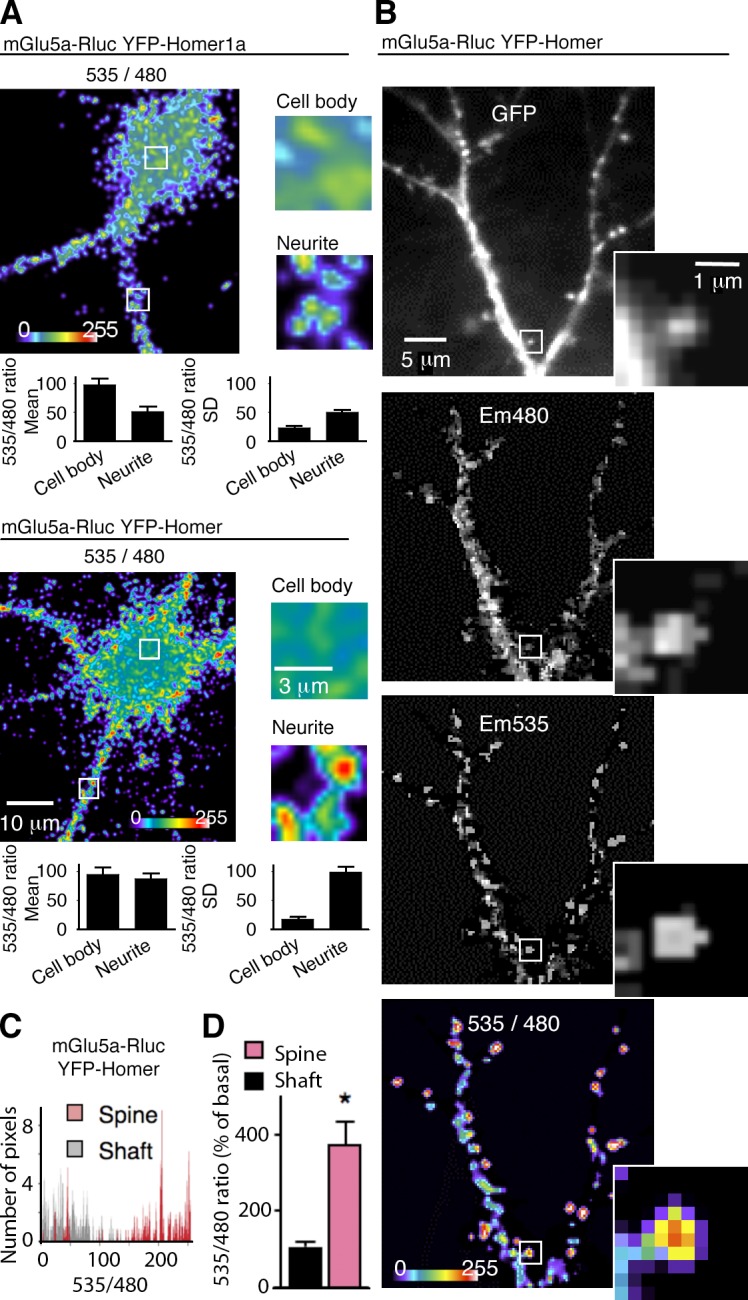
mGlu5a interaction with multimeric Homer in neurons is confined to dendritic spines. (A–D) In this and the following figures, experiments were performed in cultured hippocampal neurons, unless specified otherwise. Neurons were transfected with mGlu5a-Rluc8 and YFP-Homer (except for the top image in A, in which neurons expressed mGlu5a-Rluc8 and YFP-Homer1a). (A) BRET was imaged on neurons and zoomed in a 5 × 5–µm area on the neurite and cell body. Insets show magnified images of the boxed areas on the left. Histograms show the quantification of the mean BRET intensity and the SD in each area. Note that the highest BRET signals between mGlu5a and Homer were found in the neurites, as compared with the cell body, and that the high SD indicates a clusterization in dendrites (bottom). By opposition, the BRET signal between mGlu5a and Homer1a appeared homogenous in neurons (top). (B) The pictures show expression of YFP-Homer (GFP), mGlu5a-Rluc8 (Em480), YFP-Homer excited by energy transfer (Em535), and BRET signals generated by the two tagged proteins (535/480) within a 2.5 × 2.5–µm area zoomed on the framed spine. Note that although mGlu5a and Homer proteins were both present in the dendritic shaft and spine, the highest intensity BRET signals are located in the spines, indicating a preferential interaction of the two proteins in this cell compartment. (C) The histogram represents BRET pixel distribution as a function of BRET intensity in spines and adjacent dendritic shaft regions (n = 10) of a representative neuron. (D) BRET intensity was measured in the dendritic spine and shaft (mean ± SEM of 10 neurons; 7–10 regions per neuron; *, P = 0.05).
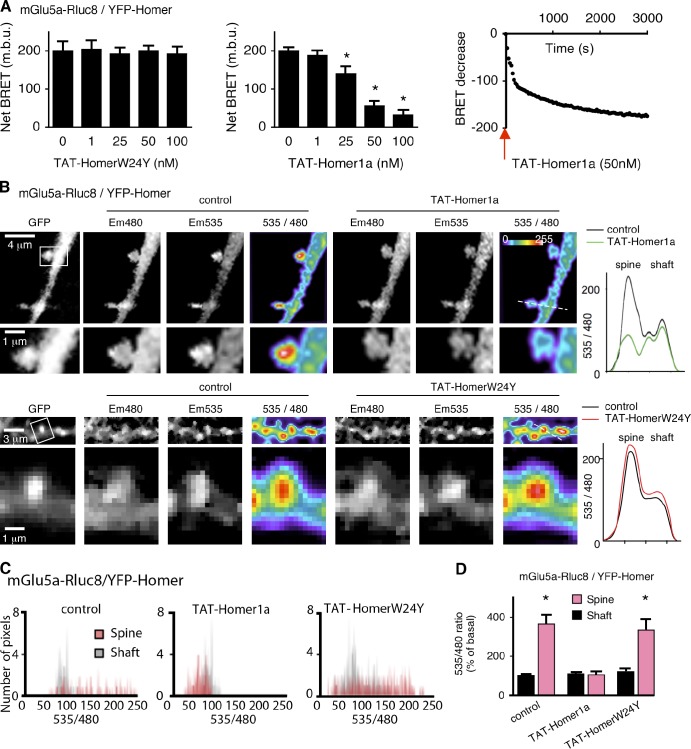
To bypass this long-term effect of Homer1a, the protein was conjugated to the cell membrane transduction domain of the HIV-1 TAT protein (TAT-Homer1a). TAT-conjugated proteins can cross the plasma membrane, thus allowing their acute cell internalization (Dietz and Bähr, 2005). We verified that this also applied to TAT-Homer1a (Fig. S3). A 10-min perfusion of TAT-Homer1a, but not a TAT-HomerW24Y protein (a point mutation that selectively abolished the interaction of Homer1a with mGlu5a; Fig. S1 H; Beneken et al., 2000), decreased the BRET signal between mGlu5a-Rluc8 and YFP-Homer in a dose- and time-dependent manner (Fig. 3 A). Consistently, BRET imaging revealed that the spine-confined basal interaction between YFP-Homer and mGlu5a-Rluc8 was disrupted by acute perfusion of TAT-Homer1a but not a TAT-HomerW24Y protein (Fig. 3 B). In basal condition, BRET pixel distribution in dendritic shaft versus spines, expressed as a function of BRET intensity, further showed the existence of high-intensity BRET signals between the receptor and Homer in spine solely, whereas TAT-Homer1a shifted the pixel distribution in the spine to lower BRET values undistinguishable from the basal signal in the shaft (in the presence of Homer1a, BRET ratio in the spine was shifted from 364 + 57% to 101 + 19% of mean BRET in the shaft; n = 8; Fig. 3, C and D). These experiments put emphasis on the efficiency of Homer1a to disrupt the association of mGlu5a receptor with multimeric forms of Homer specifically in the spine.
Figure 3.
Homer1a disrupts the interaction between Homer and mGlu5a receptor in dendritic spines. (A) HEK293 cells were transfected with mGlu5a-Rluc8 and YFP-Homer. BRET signals were monitored 10 min after addition of the indicated concentrations of either TAT-HomerW24Y (left) or TAT-Homer1a (middle) and during 50 min in the presence of 50 nM TAT-Homer1a (right). The graphs show loss of interaction between mGlu5a receptor and Homer in the presence of TAT-Homer1a but not TAT-HomerW24Y (left, middle). This occurred in <5 min after the beginning of application to the TAT-Homer1a protein (right). The red arrow represents the application of the TAT-Homer1a protein. Each bar of histogram represents the mean ± SEM of four experiments performed in triplicate. The asterisk represents significant difference from the BRET in absence of TAT. m.b.u., milli-BRET unit. (B–D) BRET images (B) and measurements (C and D) were obtained in neurons transfected with mGlu5a-Rluc8 and YFP-Homer before (control) and after exposure to TAT-Homer1a or TAT-HomerW24Y (50 nM; 10 min). (B) The pictures show expression of YFP-Homer (GFP), mGlu5a-Rluc8 (Em480), YFP-Homer excited by energy transfer (Em535), and BRET signals generated by the two tagged proteins (535/480). The right graphs indicate the profiles of the 535/480 ratio values along the dotted lines crossing the spine and shaft shown in the image before and after TAT-Homer1a or TAT-HomerW24Y perfusion. Bottom images show magnifications of the boxed areas. (C) BRET pixel distribution in spines and adjacent dendritic shaft regions (n = 10) of a representative neuron before (control) and after perfusion of TAT-Homer1a or TAT-HomerW24Y. (D) Mean BRET intensity in the dendritic spine and shaft before (control) and during exposure to TAT-Homer1a or TAT-HomerW24Y. Each bar of the histogram represents the mean ± SEM obtained from eight neurons and 7–10 regions/neuron. Note that in the presence of Homer1a, BRET signal between mGlu5a-Rluc8 and YFP-Homer decreased in spines but not the shaft. *, P = 0.05.
Dynamics of mGlu5 receptor–Homer complex control physical and functional cross talk between mGlu5a and NMDA receptors in hippocampal neurons
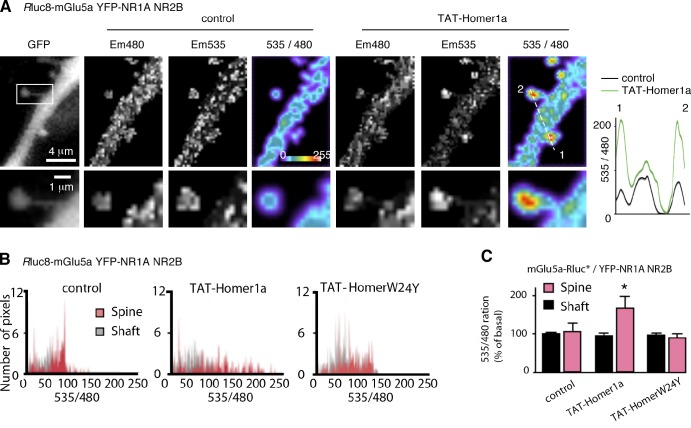
In a heterologous expression system, NMDA and mGlu5a receptors can directly interact and display constitutive mutual inhibition (Perroy et al., 2008). In neurons, however, the constraints that result from the link between the C terminus of these receptors with the Homer-Shank-GKAP-PSD95 scaffold might preclude the direct mGlu5a–NMDA receptor association. Consistent with this hypothesis, although Rluc8-mGlu5a (Em480 image in Fig. 4 A) and YFP-NR1A + NR2B subunits (GFP image in Fig. 4 A) clearly colocalized in the dendritic shaft and spine, BRET signals between these tagged proteins remained close to noise in both cell compartments (control condition 535/480 image in Fig. 4 A). This suggested that the NMDA and mGlu5a receptors did not interact in control hippocampal neurons, although both receptors were present in the same dendritic spine. We reasoned that disassembly of the synaptic multimeric Homer–mGlu5a receptor complex by Homer1a would allow physical and functional interactions between NMDA and mGlu5a receptors. Consistent with this hypothesis, after a 10-min incubation with TAT-Homer1a (50 nM), BRET spots of high intensity appeared in spines (TAT-Homer1a condition; 535/480 image in Fig. 4 A), thus highlighting the occurrence of confined mGlu5a and NMDA receptor putative physical interactions. In basal condition, BRET pixel distribution did not differ in the dendritic shaft versus spines. On the other hand, two distinct distributions clearly appeared after TAT-Homer1a incubation (but not TAT-HomerW24Y), with the highest- and lowest-pixel intensities in spines and the dendritic shaft, respectively (Fig. 4 B). Statistical analyses further confirmed such a Homer1a-induced spine-restricted interaction between Rluc8-mGlu5a and YFP-NR1a + NR2B, as the basal BRET ratio in spine over shaft was shifted from 1.05 ± 0.24 to 1.77 ± 0.12 (n = 8) in the presence of Homer1a (Fig. 4 C). These data show that although mGlu5a and NMDA receptors colocalized in neurons, the receptors were in sufficient proximity to directly interact with each other, only in the presence of Homer1a and specifically in spines.
Figure 4.
Homer1a–mGlu5a interaction enables physical association of mGlu5a with NMDA receptors in spine. (A–C) BRET images (A) and analyses (B and C) obtained in neurons transfected with Rluc8-mGlu5a and YFP-NR1A + NR2B expression plasmids. (A) BRET images acquired before (control) and after incubation with TAT-Homer1a (50 nM; 10 min). From left to right, images revealed expression of YFP-NR1A (GFP) and Rluc8-mGlu5a (Em480), YFP-NR1A excited by the energy transfer (Em535), and BRET between Rluc8-mGlu5a and YFP-NR1A (535/480). The boxed area in the GFP picture was enlarged below. The graph on the right represents the profile of the 535/480 ratio values along the dotted line before (control) and after TAT-Homer1a exposure. (B) Pixel distribution in spines and adjacent dendritic shaft regions (n = 10) before and after application of TAT-Homer1a or TAT-HomerW24Y. (C) Mean BRET intensity in the dendritic spine and shaft before (control) and during exposure to TAT-Homer1a or TAT-HomerW24Y. Each bar of the histogram represents the mean ± SEM obtained from eight neurons and 7–10 regions/neuron. *, P = 0.05.
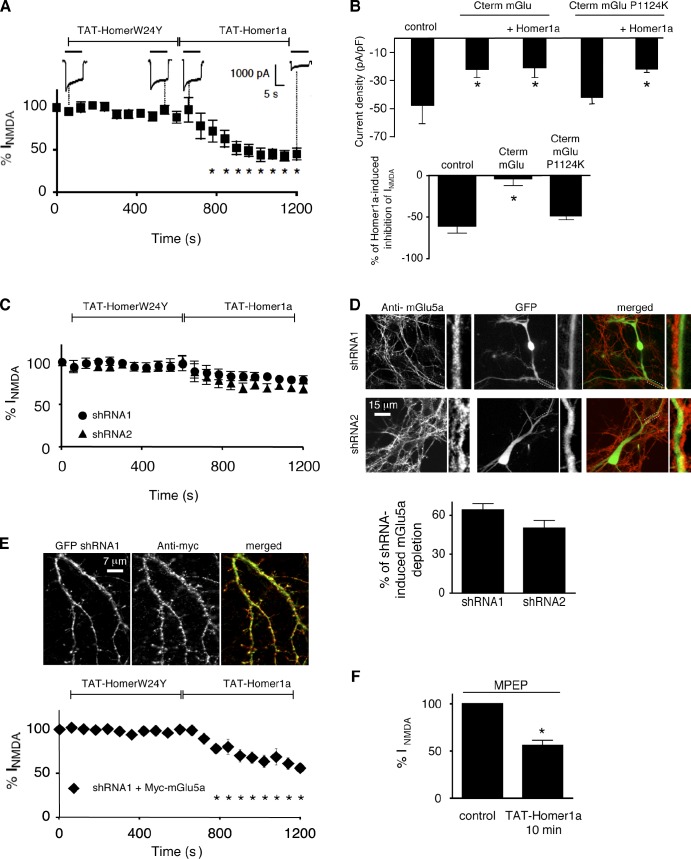
We further investigated the functional consequences of such a protein scaffold remodeling and consequent physical interaction between receptors on NMDA currents. Interestingly, in nontransfected hippocampal neurons, whole-cell NMDA currents were strongly decreased after TAT-Homer1a exposure (57.7 ± 7.1% decrease; n = 8; Fig. 5 A). Based on previous observations showing that, in HEK cells (i.e., in the absence of scaffolding protein expression), NMDA and mGlu5 receptor directly interact, resulting in inhibition of NMDA current (Perroy et al., 2008), we hypothesized that the present Homer1a-induced inhibition of NMDA current could result from disruption of endogenous mGlu5a receptor–multimeric Homer complexes by Homer1a allowing physical interaction of mGlu5a with NMDA receptors. To test this hypothesis, we used the Homer1a point mutant, HomerW24Y, which cannot interact with mGlu5a (Fig. S1 H) and therefore could not disrupt the interaction between mGlu5a and the multimeric Homer (Fig. 3). This Homer1a mutant had no effect on whole-cell NMDA currents (Fig. 5 A), showing that to inhibit NMDA currents, Homer1a needs to interact with mGlu5a receptor. This mGlu5a–Homer1a interaction would relax mGlu5a from the physical constraint of the scaffold. We also used an alternative manner to disrupt the scaffold, achieved by coexpression of the C terminus of the mGlu5a receptor, which quenched mGlu5a receptor partners (Mao et al., 2005). This led to similar NMDA current inhibition (55.3 ± 5.3% decrease; n = 8; Fig. 5 B) and prevented additional effects of Homer1a on NMDA current (Fig. 5 B). By opposition, the mGlu5a–C terminus point mutant (P1124K), which cannot interact with Homer proteins, had no effect on the NMDA currents and did not impair their inhibition by Homer1a (Fig. 5 B). Disengagement of the mGlu5a receptor would favor its direct interaction with NMDA receptor and functional blockade of NMDA receptors. Accordingly, depletion of mGlu5a receptor expression by specific short hairpin RNAs (shRNAs) significantly reduced the TAT-Homer1a–induced inhibition of NMDA currents (Fig. 5 C). Importantly, the potency of Homer1a to induce inhibition of NMDA currents was correlated to the amount of mGlu5a receptor expression (Fig. 5, C and D). To rule out unspecific targets of the mGlu5 shRNAs, we verified that the loss of Homer1a-induced inhibition of NMDA currents could be rescued by overexpression of the mGlu5a receptor (Fig. 5 E). Finally, blocking the activity of mGlu5 receptor with a specific antagonist, MPEP (2-methyl-6-(phenylethynyl)-pyridine; 10 µM), was not sufficient to abolish the Homer1a-dependent suppression of NMDA currents (Fig. 5 F), suggesting that the removal of mGlu5a protein was required. Collectively, these results showed that inhibition of NMDA currents by Homer1a requires the interaction of Homer1a with mGlu5. Homer1a-induced dissociation of the mGlu5a receptor from multimeric Homer complex allowed, in turn, association between mGlu5a and NMDA receptors and inhibition of the NMDA current.
Figure 5.
Homer1a–mGlu5a interaction enables inhibition of NMDA currents. (A) NMDA-induced currents recorded in nontransfected neurons before (100% NMDA current at time 0 s) and during sequential perfusion of TAT-HomerW24Y and TAT-Homer1a proteins. Horizontal bars above each trace represent application of 100 µM NMDA. Each plot represents the mean ± SEM obtained from eight neurons (asterisks represent significant difference from NMDA current before TAT perfusion). (B) NMDA current density (top) and percentage of TAT-Homer1a–induced inhibition of NMDA current (bottom) measured in neurons transfected or not with mGlu-Cterm (to quench Homer1a) or mGlu-Cterm-P1124K (which cannot interact with Homer proteins). Each bar of the histogram represents the mean ± SEM obtained from eight neurons. *, P = 0.05. (C and D) Neurons transfected with shRNA1 or shRNA2 and GFP (transfection reporter) were used for either NMDA-induced current recording (C; same legend as in A) or immunostaining with an anti-mGlu5 antibody (D; boxed areas are magnified to the right of each image). (D, bottom) Quantification within a dendritic area of 5 µm × 15 µm. (E) Neurons transfected with shRNA1, GFP (transfection reporter), and Myc-mGlu5a were immunolabeled with an anti-Myc antibody in nonpermeabilized conditions (top) or used to record NMDA-induced currents (bottom; same legend as in A). (F) Percentage of NMDA current in the presence of the mGlu5a antagonist (MPEP; 10 µM) before and after TAT-Homer1a perfusion. *, P = 0.05. (D and F) Each bar of histogram represents the mean ± SEM obtained from eight neurons.
Molecular Homer–mGlu5 receptor complex reorganization occurs after sustained activation of synaptic NMDA receptors
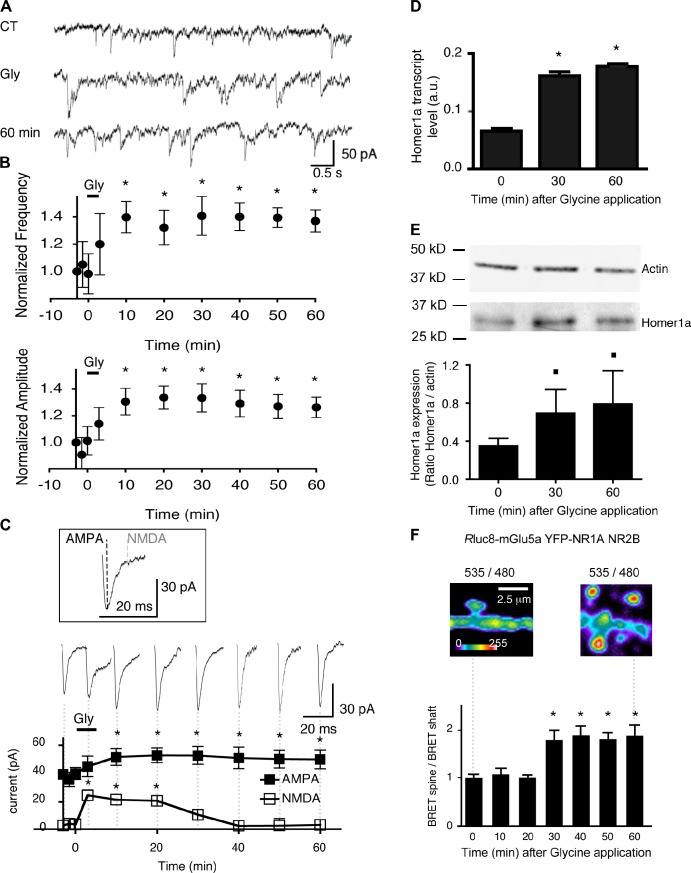
Homer1a protein expression is very low or virtually absent in resting neurons. Elevated neuronal activity induced by psychostimulants (Brakeman et al., 1997) or sustained NMDA receptor stimulation (Sato et al., 2001), which eventually results in long-term potentiation (LTP) of synaptic transmission (Kato et al., 1997), can induce expression of the protein in neurons. Therefore, we examined whether Homer1a-induced mGlu5a–NMDA receptor interaction occurred after NMDA receptor–sustained activity. As previously shown (Lu et al., 2001; Park et al., 2004), prolonged stimulation of NMDA receptors by the coagonist glycine (200 µM; 3 min) successfully induced LTP in primary culture of hippocampal neurons (Figs. 6 [A–C] and S4). Indeed, the glycine treatment significantly increased the amplitude and frequency of miniature excitatory postsynaptic currents (mEPSCs), and this effect remained stable for >60 min after glycine washout (mEPSC frequency and amplitude were increased by 40 ± 10 and 34 ± 7% [mean ± SEM; n = 8], respectively; Fig. 6 B). Consistently, glycine treatment also increased the size of the spine head and the level of AMPA receptors expressed at the cell surface (Fig. S4, B and C). Interestingly, this sustained activation of synaptic NMDA receptor successfully triggered Homer1a transcription and expression (Fig. 6, D and E). We found that sustained activation of synaptic NMDA receptor (a 3-min application of glycine/strychnine followed by washout of the drugs) also triggered BRET signals between NMDA (YFP-NR1a + NR2B) and Rluc8-mGlu5a receptors in the spine. The BRET was detectable 30 min after the glycine application and persisted for at least 60 min (Fig. 6 F). Such a kinetic correlated with the time course of Homer1a induction (Fig. 6 E). To characterize the effect of glycine treatment on synaptic AMPA and NMDA receptors, we analyzed the fast and slow components of mEPSCs (Fig. 6 C), which are carried by AMPA and NMDA currents, respectively (Fig. S4 A). The glycine treatment increased the amplitude of the synaptic AMPA component of mEPSCs for at least 60 min (which was consistent with the aforementioned increase in mEPSCs amplitude). The NMDA component of mEPSCs also strongly increased but only transiently. Indeed, the current progressively recovered to its control value (Fig. 6 C). Thus, after sustained activation of synaptic NMDA receptor, there is a strong temporal correlation between Homer1a induction, mGlu5a–NMDA receptor interaction in the spine head, and inhibition of mEPSC NMDA receptor component (Fig. 6). All of these evidences favor the hypothesis of a role of the Homer1a-dependent mGlu5–NMDA receptor interaction to inhibit synaptic NMDA current.
Figure 6.
Synaptic NMDA receptor activation triggers Homer1a endogenous expression, physical interaction between NMDA and mGlu5a receptors in the spine, and inhibition of postsynaptic NMDA currents. (A) mEPSCs recorded at −60 mV before (C terminal), during (Gly), and 60 min after (60 min) synaptic NMDA receptor stimulation (200 µM glycine for 3 min). (B) Amplitude and frequency of mEPSCs, normalized to values obtained at time 0 s before and 60 min after application of glycine (mean ± SEM; n = 8 neurons). Asterisks represent significant differences from the signal before glycine application. (C) Mean of 50 mEPSCs showing the AMPA and NMDA postsynaptic current components. Dotted lines show the times (at the peak and 7 ms later for the AMPA and NMDA components, respectively) at which measurements were performed on the averaged mEPSCs. The sequential traces and graph below show persistent potentiation of the AMPA component and prominent NMDA current component during and after application of glycine. Asterisks represent significant differences from the signal before glycine application. Error bars are SEM. (D and E) Level of Homer1a mRNA assessed by quantitative PCR (D) and Homer1a protein quantified by Western blot and divided by the expression of actin as a control for protein loading (E) in hippocampal neurons before (time 0), 30, and 60 min after glycine application. The histograms represent the mean ± SEM obtained from three individual experiments. Squares represent significant differences at P < 0.10. *, P = 0.05. a.u., arbitrary unit. (F) Neurons transfected with Rluc8-mGlu5a, YFP-NR1A, and NR2B expression plasmids. BRET images were acquired before (left) and 60 min after (right) glycine application. Note the increase in BRET signal in dendritic spines. The histogram represents the ratio between dendritic spine and shaft BRET values before (time 0), 30, and 60 min after glycine application. The histogram represents the mean ± SEM obtained from eight neurons and four to seven regions per neuron for each time condition. *, P = 0.05.
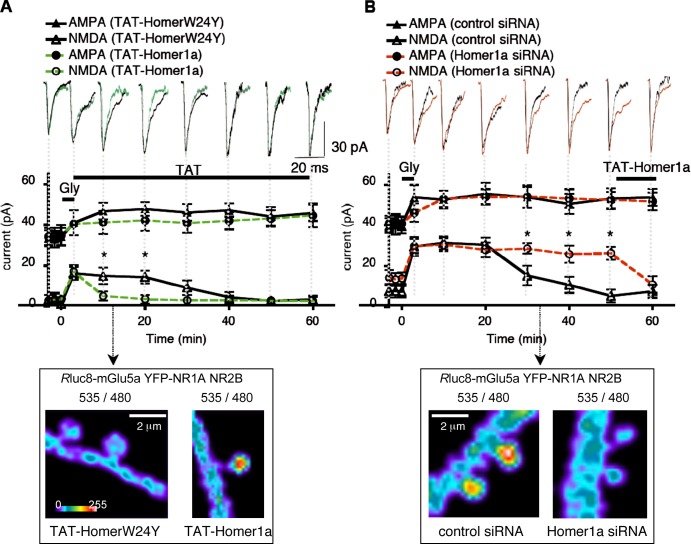
To further examine the direct implication of Homer1a in synaptic mGlu5a–NMDA receptor interaction and inhibition of synaptic NMDA current, we perfused the TAT-Homer1a protein immediately after glycine application. Remarkably, the TAT-Homer1a, but not TAT-HomerW24Y, decreased NMDA synaptic currents 10 min after glycine washout (Fig. 7 A). This delay corresponded to the time required for the TAT-Homer1a protein to enter the cell and to abolish BRET signals between Homer and mGlu5a receptor (Fig. 3 A). By opposition, the knockdown of Homer1a by a mix of two specific siRNAs (see Materials and methods) blocked the induction of Homer1a expression after sustained activation of synaptic NMDA receptors (Fig. S5) and abolished the recovery of synaptic NMDA current (Fig. 7 B). However, this siRNA-induced loss of postsynaptic NMDA current inhibition was rescued by perfusion of TAT-Homer1a at the end of the experiment. These electrophysiological experiments were systematically correlated to BRET imaging between mGlu5a and NMDA receptors. In the presence of TAT-HomerW24Y, which does not interact with mGlu5a receptor, no BRET signal was detected 10 min after glycine washout. By opposition, TAT-Homer1a treatment induced BRET signals between mGlu5a and NMDA receptors in the spine (Fig. 7 A). On the other hand, knockdown of endogenous Homer1a by siRNA abolished BRET signals between the two receptors 30 min after glycine treatment (inset in Fig. 7 B). These results strongly suggest that induction of Homer1a by sustained activation of synaptic NMDA receptors can trigger physical and functional interaction between mGlu5a and NMDA receptors, thus leading to down-regulation and recovery of synaptic NMDA currents while leaving synaptic AMPA current potentiation unaffected.
Figure 7.
After activation of synaptic NMDA receptors, Homer1a is necessary and sufficient to induce mGlu5a–NMDA interaction and inhibition of NMDA receptor. (A and B) Mean of 50 mEPSCs recorded at different times before, during, and after the application of glycine (top). The graphs represent the mean (±SEM; n = 8) of AMPA and NMDA components of mEPSCs (middle). BRET images were obtained at the indicated times after washout of glycine (bottom). Calibration bars apply to A and B. (A) TAT-HomerW24Y (black traces) or TAT-Homer1a (green traces) was perfused immediately after glycine application. Note that TAT-Homer1a accelerated the inhibition of the NMDA component of mEPSCs after washout of glycine, as compared with the inactive TAT-HomerW24Y. Asterisks indicate that NMDA current is significantly different in TAT-Homer1a and TAT-HomerW24Y conditions at the same time after glycine application. BRET images show that interaction between mGlu5a and NMDA receptor in the presence of TAT-Homer1A (but not TAT-HomerW24Y) occurs in spines at the same time as postsynaptic NMDA current is inhibited. (B) 2 d before electrophysiological recordings, neurons were transfected with control siRNA (black traces) or Homer1a siRNA (red traces). Note that Homer1a siRNA (but not control siRNA) prevented the recovery of NMDA current. Asterisks indicate that NMDA current is significantly different in Homer1a siRNA and control siRNA conditions at the same time after glycine application. TAT-Homer1a perfusion restored the inhibition of postsynaptic NMDA currents. 30 min after washout of glycine, BRET images show that Homer1A siRNA (but not control siRNA) prevented mGlu5a–NMDA receptor interaction in the spine.
Discussion
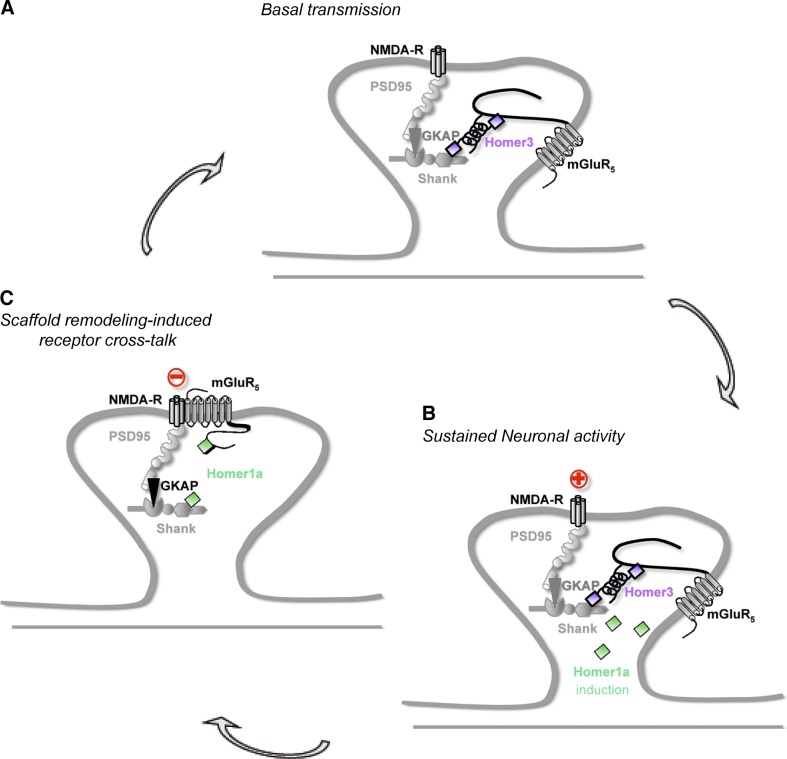
Several studies have suggested that the temporal and spatial dynamics of protein–protein interactions is crucial to specify synaptic membrane receptor and channel signaling (Sala et al., 2001; Fagni et al., 2002; Renner et al., 2008; Zeke et al., 2009; Sainlos et al., 2011). Thanks to recent development in single-cell BRET imaging (Coulon et al., 2008; Perroy, 2010), we have examined this issue by studying the competitive binding of the monomeric Homer1a and multimeric Homer with the neurotransmitter mGlu5a receptor at synapses of living neurons. We found that the competitive interactions between mGlu5a receptor and Homer proteins was restricted to specific subcellular compartments (dendritic spines), precisely where synaptic transmission takes place. Interaction with Homer1a and dissociation with Homer resulted in mGlu5a–NMDA receptor association and NMDA current inhibition. We further showed that a sustained activation of synaptic NMDA receptors triggered the expression of endogenous Homer1a, interaction between synaptic mGlu5a and NMDA receptors, and inhibition of postsynaptic NMDA currents. These results provide, for the first time, a physiological role for dynamic remodeling of a receptor scaffold (the postsynaptic glutamate receptor scaffold) and its cellular function (the control of synaptic excitability; Fig. 8).
Figure 8.
Model of molecular scaffold remodeling and NMDA–mGlu5a receptor cross talk to control synaptic excitability. (A) Under basal condition, multimeric forms of Homer cross-link group I mGlu receptors to the GKAP–PSD95–NMDA receptor complex in the spine and prevent mGlu–NMDA receptor interaction. In this condition, mGlu5a and NMDA receptors do not directly interact. (B) Intense neuronal activity triggers Homer1a expression, which competes with multimeric forms of Homer on the mGlu5a receptor C terminus, resulting in the disruption of the interaction between mGlu5a receptor and GKAP–PSD95–NMDA receptor complex. (C) Homer1a would trigger disengagement of mGlu5a from the multimeric Homer complex, thus allowing association of mGlu5a with NMDA receptor in spines. In this condition, mGlu5a receptors directly inhibit NMDA receptors.
Despite nonpreferential distribution of the transfected Homer protein and mGlu5 receptor in mature hippocampal neurons, we found that the mGlu5a receptor interacts with Homer exclusively in spines. Consistent with this finding, transfection of long or short Homer isoforms results in spine enlargement or drastic reduction of spine density, respectively, in hippocampal neurons (Sala et al., 2001; Szumlinski et al., 2005; Tappe et al., 2006; Jaubert et al., 2007). This drastic loss of spines as a result of the long-term expression of transfected Homer1a prevented the direct analysis of the BRET between Homer1a and mGlu5a receptor in spines. However, it is important to note that the competition between TAT-Homer1a and Homer to interact with the receptor in spines indeed attested the occurrence of Homer1a–mGlu5a interaction in the spine as well (Fig. 3, B–D). The occurrence of Homer1a–mGlu5a interaction in the spine was further supported by the spine-confined interaction between mGlu5a and NMDA receptors after Homer1a expression induced by glycine stimulation (Fig. 7 B). These observations further support the notion that competition between long and short Homer isoforms on mGu5 receptors at synaptic sites may control neurotransmission.
Competition between the monomeric Homer1a and multimeric Homer proteins on mGlu5 receptor depends on the level of Homer1a expression, the relative affinity of the Homer proteins for their target, and the subcellular localization of these partners. The apparent higher affinity of multimeric versus monomeric Homer for the mGlu5a receptor might be explained by the dimeric state of the receptor, which would favor cooperative binding of the multimeric over monomeric Homer isoform (Sainlos et al., 2011). Nevertheless, native Homer1a, even at a relatively low level, can affect group I mGlu receptor signaling in neurons (Kammermeier, 2008). Consistent with this result, the displacement of multimeric Homer interaction with the mGlu5a receptor by coexpressed Homer1a was quite efficient. In light of these results, sufficient Homer1a expression would be required to displace Homer–mGlu5a receptor interaction, and this may restrain the phenomenon to particular conditions, such as induction of synaptic plasticity (Ronesi and Huber, 2008) or after drug-of-abuse intake, epilepsy, or chronic pain (Tappe et al., 2006; Szumlinski et al., 2008).
Our results also show that the delicate balance of Homer protein interactions would control mGlu5a receptor functions. Thus, interaction of Homer1a with mGlu5a receptor triggers a direct inhibition of NMDA channel by mGlu5a receptor. The functional modulation of NMDA receptor channels by group I mGlu receptors is highly debated. Both synergism (Aniksztejn et al., 1992; Harvey and Collingridge, 1993; Doherty et al., 1997; Ugolini et al., 1997; Awad et al., 2000; Attucci et al., 2001) and mutual antagonism (Anwyl, 1999; Bortolotto et al., 1999; Kotecha et al., 2003; Perroy et al., 2008; Bertaso et al., 2010) have been observed in neurons of various brain regions. The mGlu5a receptor is physically linked to the Shank–GKAP–PSD95–NMDA receptor complex by multimeric Homer proteins. Herein, we suggest that competition between monomeric Homer1a and multimeric Homer on the binding domain of mGlu5 receptor would disrupt the interaction between this receptor and the PSD95–GKAP–Shank–Homer scaffold complex, thus allowing the mGlu5 receptor to directly interact with and inhibit the NMDA receptor. The dual role of group I mGlu receptors on NMDA receptors in neurons may rely on the presence or absence of Homer1a and on the integrity of the mGlu5–Homer–Shank–GKAP–PSD95–NMDA receptor complex. By disrupting the scaffolding complex, Homer1a may affect mGlu5a as well as NMDA receptor complex localization. Studies are currently in progress in our laboratory to further address the precise movement of glutamate receptors and their rearrangement in the postsynaptic density.
Homer1a expression is regulated by neuronal activity (Brakeman et al., 1997; Kato et al., 1997). Here, we could induce Homer1a expression by sustained activation of synaptic NMDA receptors, and this was necessary and sufficient to promote physical interaction between mGlu5a and NMDA receptors in dendritic spines as well as to scale down postsynaptic NMDA currents. It has been previously reported that NMDA-dependent LTP is blocked in transgenic mice expressing hippocampal-specific tetracycline-inducible Homer1a (Celikel et al., 2007). This may result from the herein-described Homer1a-induced inhibition of NMDA currents, as in this transgenic mouse, Homer1a was induced before application of the LTP stimulus. This hypothesis was further confirmed by applying TAT-Homer1a before glycine stimulation. In this condition, TAT-Homer1a disrupted the integrity of the scaffold in spines (Fig. 3), enabled NMDA and mGlu5a receptor interaction (Fig. 4), and totally prevented the induction of LTP (Fig. 9). Indeed, in the presence of TAT-Homer1a, neither NMDA nor AMPA current was affected by glycine stimulation (Fig. 9). These effects were consistent with the blockade of NMDA receptors and NMDA-dependent AMPA receptor LTP by Homer1a. Thus, during elevated synaptic activity and synaptic plasticity, induced Homer1a would trigger mGlu5a receptor–mediated down-regulation of synaptic NMDA currents and scale down synaptic excitability. Dysfunction of such a mechanism may engender neuropathologies. It has been shown that Homer1a overexpression reduces susceptibility to amygdala kindling (Potschka et al., 2002) and decreases the magnitude of LTP in the hippocampus, presumably by diminishing postsynaptic responses during tetanization (Celikel et al., 2007). Electroconvulsive therapy, which represents an induced seizure used for treatment of psychosis, promotes Homer1a expression, which reduces seizure susceptibility and drives homeostasis of pyramidal cell excitability (Sakagami et al., 2005). By opposition, in a mouse model of human Fragile X mental retardation syndrome, it has been shown that less mGlu5 receptors are associated with the multimeric Homer isoforms, and this could be the molecular basis of the receptor functional alteration in the pathology (Ronesi and Huber, 2008). Dysregulation of the herein-identified molecular control of synaptic excitability could therefore have dramatic consequences. Such a Homer1a-induced negative feedback control is thus crucial for homeostasis of synaptic excitability and tenacity.
Figure 9.
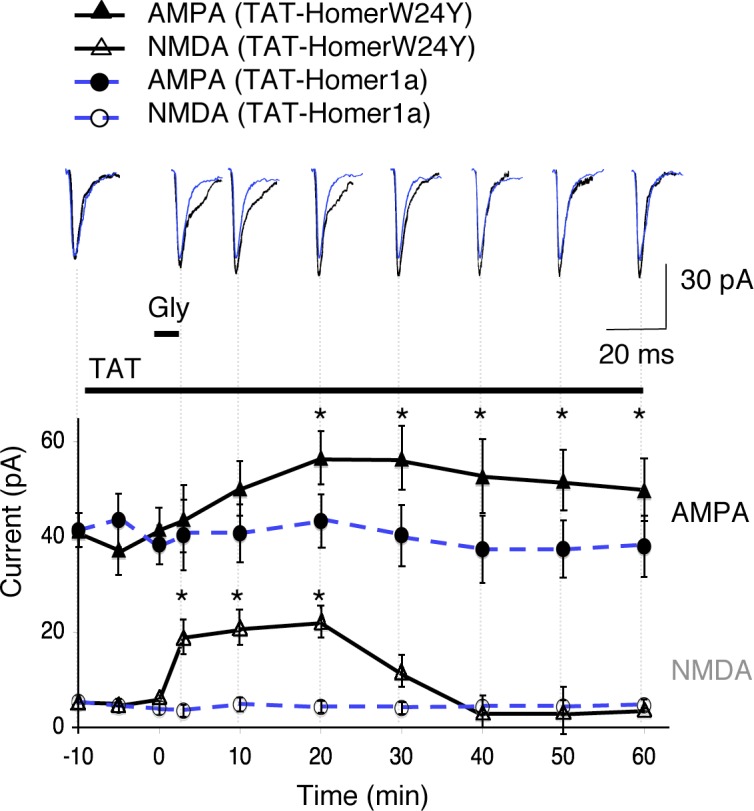
Preapplication of TAT-Homer1a precludes the induction of LTP by activation of synaptic NMDA receptors. Mean of 50 mEPSCs recorded at different times before, during, and after the application of glycine (top). The graph represents the mean (±SEM; n = 8) of AMPA and NMDA components of mEPSCs. TAT-HomerW24Y (black traces) or TAT-Homer1a (blue traces) was perfused at the beginning of the experiment before glycine application. Note that TAT-Homer1a abolished the glycine-induced NMDA current increase as well as AMPA current potentiation, as compared with the inactive TAT-HomerW24Y. Asterisks indicate that NMDA current is significantly different in TAT-Homer1a and TAT-HomerW24Y conditions at the same time after glycine application.
This work highlights the importance of the association/dissociation dynamics of multiprotein complexes in receptor functions and cell physiology. Neurotransmitter and hormone receptors can no longer be seen as cell surface–isolated entities. Regulations of receptor signaling by dynamic changes in receptor-associated complex that we described here can be taken as a template to study other high-order modulatory mechanisms in various systems. Thus, similar remodeling of multiprotein assemblies by environmental factors would define a given cell status, which in turn would influence subsequent cellular responses, depending on the history of the cell.
Materials and methods
Plasmids, shRNA, and siRNA
DNA plasmids containing the ORF for Myc-mGlu5a-Rluc8, Homer1a, Homer3, or the NR2B subunit of NMDA receptors were encoded under control of cytomegalovirus promoter in pcDNA3.1-Myc-mGlu5a-Rluc8, pRK5-Homer1a, pRK5-Homer3, or p3apA-e2-NR2B, respectively, as previously described (Ango et al., 1999; Coulon et al., 2008; Perroy et al., 2008). Homer proteins tagged at their N terminus with either YFP or GFP2 were used for BRET experiments. The coding sequences of Homer1a or Homer3 were added in-frame to the 3′ end coding sequence of YFP or GFP2 in their respective pcDNA3.1 plasmids to obtain pcDNA3.1-YFP-Homer1a, pcDNA3.1-YFP-Homer3, and pcDNA3.1-GFP2-Homer1a. For BRET experiments between receptors, tags were added in the extracellular N terminus part of the receptors to avoid any interference with the protein interactions on their cytosolic tail. The Rluc8 tag (a gift from A. De and A. Loening, Stanford University, Stanford, CA) was added in-frame between the 3′ end of the signal peptide and the 5′ end coding sequence of mGlu5a receptor to obtain pRK5-Rluc8-mGlu5a. The YFP-NR1A construct was engineered by inserting a YFP cDNA fragment in-frame with the NR1-1a subunit after the predicted sequence for signal peptide (MSTMHLLTFALLFSCSFARAA) to obtain the p-YFP-NR1a plasmid. pRK5-cherry-CD4-mGlu-Cterm was generated by PCR amplification of the DNA coding for the C-terminal tail of mGlu5a, which was then subcloned in-frame with the 3′ end of the DNA coding for the ectodomain and transmembrane domain of CD4. The Homer1a coding sequence was fused to the coding sequence of the TAT-permeant peptide to obtain pET-TAT-His-Homer1a. To engineer pcDNA3.1-Myc-mGlu5a-P1124K-Rluc8, pRK5-cherry-CD4-mGlu-Cterm-P1124K, pcDNA3.1-YFP-HomerW24Y, and pET-TAT-His-HomerW24Y, we used primers containing the point mutations coding for P1124K (5′-GAGGAGCTTGTGGCGTTAACTAAACCATCGC-3′) and W24Y (5′-ACAAAGAAGAACTATGTACCCACTAGTAAGCAT-3′) to amplify by PCR the mGlu5a and Homer1a coding sequences, respectively. Two shRNAs subcloned into pLKO.1-CMV-tGFP vector (Sigma-Aldrich) were designed to prevent the expression of mGlu5 receptor specifically: shRNA1, clone ID TRCN0000218761 (5′-GTACCGGACATGCCAGGTGACATTATTACTCGAGTAATAATGTCACCTGGCATGTTTTTTTG-3′), and shRNA2, clone ID TRCN0000219566 (5′-CCGGTCATGGAGCCTCCGGATATAACTCGAGTTATATCCGGAGGCTCCATGATTTTTG-3′). Two siRNAs were designed to prevent the expression of Homer1a specifically: Si1 (5′-GGTGGATAATTGGAAGTCA-3′) and Si2 (5′-CAGCAATCATGATTAAGTA-3′). We used the Eurogentec Universal Negative Control oligonucleotide (reference no. OR-0030-neg05) as a control.
HEK293 cell culture and transfection
HEK293 cell culture and transfection were previously described (Perroy et al., 2003).
Hippocampal primary cell cultures and transfection
Hippocampal cultures were prepared and transfected with Lipofectamine, as previously described (Roussignol et al., 2005). In brief, hippocampal cultures were prepared from embryonic day 17 (E17) to E18 rats and grown in B27-supplemented Neurobasal medium (Life Technologies). We transfected neurons at day in vitro 10 (DIV10) and performed the experiments at DIV14. The amount of cotransfected DNA per 35-mm dishes was as follows: 2 µg pcDNA3.1-Myc-mGlu5a-Rluc8 + 2 µg pcDNA3.1-YFP-Homer1a or pcDNA3.1-YFP-Homer3 in Figs. 2 and 3 and 1 µg pRK5-Rluc8-mGlu5a + 1 µg p-YFP-NR1a + 1 µg p3apA-e2-NR2B in Fig. 4.
BRET measurements in a cell population using a spectrophotometric plate reader
Cell population BRET measurements were previously described (Perroy et al., 2004). In brief, transfected cells were resuspended in PBS with 0.1% glucose and dispersed in 96-well microplates (Corning) at a density of 100,000 cells/well. We initiated BRET by adding 5 µM of the luciferase substrate and measured the ratio of the light emitted by the acceptor to the light emitted by Rluc using the Mithras LB 940 instrument (Berthold Technologies). Values were corrected by subtracting the background ratio detected when the Rluc construct was expressed alone. For concomitant detection of mGlu5a receptor interactions with Homer1a and Homer3/1c, we combined BRET1 and BRET2 generations. Rluc8 catalyzes oxidation of coelenterazine H and DeepBlueC coelenterazine, leading to BRET1 between Rluc8 and YFP and BRET2 between Rluc8 and GFP2, respectively. HEK cells coexpressing Myc-mGlu5a-Rluc8, GFP2-Homer1a, and YFP-Homer were divided into two samples. In one sample, we measured BRET1 signal generated by YFP-Homer and Myc-mGlu5a-Rluc8 interaction, and in the second sample, we monitored BRET2 signals resulting from binding of GFP2-Homer1a with Myc-mGlu5a-Rluc8. These BRET signals were specific to the studied interaction, as no substantial transfer of energy occurred between Rluc8 and GFP2 in the presence of coelenterazine H or between Rluc8 and YFP in the presence of DeepBlueC coelenterazine (Perroy et al., 2004).
BRET imaging
BRET imaging has been previously described (Coulon et al., 2008; Perroy, 2010). In brief, all images were obtained using a bioluminescence-dedicated inverted fluorescence microscope (Axiovert 200M; Carl Zeiss) with a Plan Apochromat 63×/1.40 oil M27 objective at room temperature. Transfected cells were first identified using a monochromatic light and an appropriated filter to excite YFP (exciter HQ480/40 no. 44001 and emitter HQ525/50 no. 42017; Chroma Technology Corp.). The light source was then switched off until the end of the experiment. 20 µM coelenterazine H was applied 5 min before acquisition. Images were collected using a Cascade 512B camera (Photometrics). Sequential acquisitions of 20 s were performed at 5 MHZ (gain of 3,950 and binning of 1, with emission filters D480/60 nm [no. 61274; Chroma Technology Corp.] and HQ535/50 nm [no. 63944; Chroma Technology Corp.]) to select Em480 and Em535 wavelengths, respectively, using MetaMorph software (Molecular Devices). The pixel-by-pixel 535-nm/480-nm ratios were calculated by dividing the absolute light intensities per pixel of images obtained at 535 nm over 480 nm. These numerical ratios (comprised between 0 and 1.5) were translated and visualized with a continuous 256-pseudocolor look-up table, as displayed in the figures. To determine the mean intensity and distribution of the 535-nm/480-nm ratio, we calculated the mean intensity and SD of pixels within a square region of the cell of interest using ImageJ software (National Institutes of Health). To follow TAT-Homer1a–induced BRET changes, 20 µM coelenterazine H was applied for 5 min before the first image acquisition, and 1 µM TAT-Homer1a was added immediately after the first acquisition. A second acquisition was performed 10 min after the TAT-Homer1a application.
Electrophysiology
Hippocampal neurons were recorded in the whole-cell patch-clamp configuration, as previously described (Roussignol et al., 2005). In Fig. 5, whole-cell current density induced by NMDA application was recorded before (control) and after TAT-Homer1a perfusion. NMDA current density was then expressed as the percentage of NMDA current density in control condition, shown as INMDA (%) = (ITAT-Homer1a/Icontrol) × 100.
In Fig. 5 F, hippocampal neurons were incubated with the mGlu5a antagonist (MPEP; 10 µM) before the recording. mEPSCs were recorded on DIV14 hippocampal neurons at room temperature at a holding potential of −60 mV. The recording pipettes had a resistance of around 5 MΩ when filled with the following medium: 140 mM CsCl, 0.5 mM CaCl2, 20 mM EGTA, 10 mM Hepes, and 10 mM d-glucose, pH 7.2, with an osmolarity of 300 mOsm. The high concentration of EGTA avoided slow Ca2+-dependent desensitization of NMDA receptors. Neurons were perfused continuously with the following external medium: 140 mM NaCl, 2 mM CaCl2, 3 mM KCl, 10 mM Hepes, 10 mM d-glucose, 0.01 mM glycine, 0.01 mM bicucullin, and 0.0003 mM tetrodotoxin, pH 7.4, with an osmolarity of 330 mOsm. Currents were recorded through an amplifier (Axopatch 200B), filtered at 1 kHz, digitized at 3 kHz, and analyzed using Axon Instrument pClamp 10.0 software (Molecular Devices). Once a minimal sample of at least 50 mEPSCs had been collected from a neuron, the mean frequency and amplitude of these events were measured on the total duration of the sample. A mean trace was generated from 50 individual mEPSCs to study the fast and slow components (AMPA and NMDA currents, respectively) of the events.
Synaptic NMDA receptor stimulation
The selective activation of synaptic NMDA receptors was achieved by briefly (3 min) incubating neurons with the external medium described in the previous section, complemented with saturating levels (200 µM) of the coagonist glycine and 1 µM strychnine (Lu et al., 2001). Neurons were then kept in normal extracellular solution without glycine until the end of the experiment.
mGlu5 receptor immunostaining
DIV14 hippocampal neurons transfected with shRNA to prevent mGlu5 receptor expression and GFP as a transfection reporter were fixed with 4% PFA at 4°C for 20 min, permeabilized with 0.15% Triton X-100 for 5 min, washed, and incubated with a rabbit anti-mGlu5 antibody (EMD Millipore) for 30 min at room temperature. Cells were washed and incubated with Cy3-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, Inc.) for 1 h at room temperature. Cells were then observed under a microscope (Axioimager Z1; Carl Zeiss). Quantification of the immunostaining of endogenous mGlu5 (histogram in Fig. 5 D) was performed on neurons transfected with shRNA1 or shRNA2 and on nontransfected neurons in the same dish. For each condition, we measured the mean fluorescence intensity and substracted the noise measured in an adjacent equivalent area with no cell to obtain netFluo C terminus, net Fluo shRNA1, and netFluo shRNA2. The net fluorescence was expressed as a percentage of the expression of mGlu5a in C-terminal condition, shown as net FluoshRNA (%) = (net FluoshRNA/net FluoC terminal) × 100. The percentage of depletion (D) in mGlu5 receptor expression was calculated as the percentage of DshRNA1 = net FluoC terminal (%) − net FluoshRNA (%) = 100 − (netFluoshRNA1/netFluoC terminal).
Analysis of the expression level of Homer1a
We extracted total RNA from hippocampal neurons with TRIZOL reagent (Invitrogen) according to the manufacturer’s instructions. RT-PCR analysis of total RNA was performed with random hexamer oligonucleotides for reverse transcription. Sequences of the primers used for the determination of Homer1a expression levels are Homer1a-430F (5′-AAAGGCCGAGTAACCTGGCT-3′) and Homer1a-481R (5′-CATTTCGCTCACGTCTTCCAC-3′; available from GenBank under accession no. AF093257). The level of expression of Homer1a was normalized to the geometric mean of the expression levels of three housekeeping genes (GAPD, B2M, and Gus), according to the formula X/geometric mean (R1, R2, R3) = 2(Ct[X]−arithmetic mean [Ct(R1),Ct(R2),Ct(R3)]), where Ct is the threshold cycle, and R1, R2, and R3 are the reference genes (X denotes Homer1a, and R1, R2, and R3 denote GAPD, B2M, and Gus, respectively).
Western blot analyses
Protein samples were resolved on 7.5% PAGE, transferred to nitrocellulose, and subjected to immunoblotting using a rabbit anti-Homer antibody (1:1,000; Synaptic Systems). The nitrocellulose was then incubated with anti–rabbit/HRP conjugate (1:10,000; GE Healthcare) for 30 min and developed using a chemiluminescence kit (RENAISSANCE; PerkinElmer). To quantify the level of Homer protein (Figs. 6 E, S1 C, and S5 C), we measured the mean intensity level in a rectangular area on each band of the Western blot (0, 30, and 60 min after glycine application). We subtracted the noise values obtained in an equal area but in adjacent regions to the bands of interest.
Statistical analyses
Analyses were performed using Prism software (GraphPad Software). Statistical analyses were performed with the nonparametric Kruskal–Wallis test for more than two independent samples or with the Friedman test for paired samples with a p-value risk threshold of 5 or 10%.
Supplemental materials
Quantification of cell surface receptor expression using ELISA.
Cells expressing Myc-mGlu5a-Rluc8 were fixed with 4% PFA in PBS and then blocked with PBS plus 5% FBS. After a 30-min reaction with mouse IgG anti-Myc (1/200; Molecular Probes) primary antibody, goat anti–mouse antibody coupled to HRP (1/500; Jackson ImmunoResearch Laboratories, Inc.) was applied for 30 min. The secondary antibody was detected and instantaneously quantified by chemiluminescence (SuperSignal West Femto; Thermo Fisher Scientific) using a luminescence counter (Wallac VICTOR2; PerkinElmer).
Cell surface AMPA receptor immunostaining.
DIV14 hippocampal neurons transfected with GFP-GluR1 were incubated with rabbit anti-GFP (Invitrogen) for 20 min at 10°C to label surface AMPA receptors. After PBS washing, neurons were fixed with 4% PFA. Cells were washed and incubated with Cy3-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, Inc.) for 1 h at room temperature. Cells were then observed under a microscope (Axioimager Z1) equipped with appropriate epifluorescence and filters (GFP, 475 ± 40 and 530 ± 50 nm for excitation and emission, respectively).
Intracellular Ca2+ measurements.
Transfected HEK293 cells were seeded in polyornithine-coated, black-walled, clear-bottom 96-well plates and cultured for 24 h. Cells were then washed with freshly prepared buffer (20 mM Hepes, 1 mM MgSO4, 3.3 mM Na2CO3, 1.3 mM CaCl2, and 2.5 mM Probenecid in 1× Hank’s balanced salt solution, pH 7.4) supplemented with 0.1% BSA and loaded with 1 µM Ca2+-sensitive fluorescent dye, Fluo-4 AM (Molecular Probes), for 1 h at 37°C. Cells were washed and then incubated with 50 µl buffer. A drug plate was prepared with the mGlu5 receptor agonist dihydroxyphenylglycine (DHPG; 200 µM), and 50 µl of this 2× drug solution was added in each well after 20 s of control recording. Fluorescence signals (excitation of 485 nm and emission of 525 nm) were measured using a fluorescence microplate reader (FlexStation; Molecular Devices) at sampling intervals of 1.5 s for 60 s.
TAT-Homer expression and purification.
BL21 (DE3)–competent cells were transformed with the pET-TAT-His-Homer vector and incubated in lysogeny broth media. Induction of protein synthesis was started by IPTG addition at 500 µM for 3 h. TAT-Homer1a protein was purified under denaturing conditions on Ni–nitrilotriacetic acid (NTA) resin (QIAGEN). Cells were lysed in 8 M urea buffer, pH 8, and centrifuged at 10,000 g for 20 min. Supernatants were loaded on Ni-NTA resin and incubated for 60 min. After the binding, the resin was washed in 8 M urea buffer, pH 6.5. The TAT-Homer1a protein was as follows: MHHHHHHRPGYGRKKRRQRRRGGLDKLNALSFVDPDEQKLISEEDLGGHMGEQPIFSTRAHVFQIDPNTKKNWVPTSKHAVTVSYFYDSTRNVYRIISLDGSKAIINSTITPNMTFTKTSQKFGQWADSRANTVYGLGFSSEHHLSKFAEKFQEFKEAARLAKEKSQEKMELTSTPSQESAGGDLQSPLTPESINGTDDERTPDVTQNSEPRAEPTQNALPFPHRYTFNS.
AIMIK was eluted by addition of 8 M urea buffer, pH 4.5. Just before use, denatured TAT-Homer1a protein was desalted using a PD-10 column (GE Healthcare) and eluted in PBS.
Online supplemental material
Fig. S1 shows that tagged Homer1a and Homer proteins are functional and display different relative apparent affinity for the mGlu5a receptor. Fig. S2 shows that mGlu5a and NMDA receptors’ functions are not altered by addition of the tags. Fig. S3 validates the use of the TAT-Homer1a–permeant protein, its penetration in neurons, and its lack of effect on dendritic spine density. Fig. S4 evidences the glycine-induced increase in spine size as well as the increase in AMPA receptor expression at the cell surface. Fig. S5 demonstrates the specificity of siRNA targeted against Homer1a. Homer1a-siRNA decreased basal Homer1a mRNA and protein expression (but not the long Homer isoform) and blocked glycine-induced Homer1a expression. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201110101/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Abhijit De and Dr. Andreas Loening for providing us the Rluc8 mutant. We thank the Platform of Pharmacology at the Institute of Functional Genomics.
This work was supported by the European Community (Health-F2-2008-222918, REPLACES), Agence Nationale de la Recherche (ANR-08-MNPS-037-01, SYNGEN, and ANR-11-BSV4-018-03, DELTAPLAN), Fondation Jérôme Lejeune, the Fonds Unique Interministériel RHENEPI and DIATRAL, and Fondation Bettencourt Schueller.
Footnotes
Abbreviations used in this paper:
- BRET
- bioluminescence resonance energy transfer
- DHPG
- dihydroxyphenylglycine
- GKAP
- guanylate kinase–associated protein
- LTP
- long-term potentiation
- mEPSC
- miniature excitatory postsynaptic current
- NTA
- nitrilotriacetic acid
- shRNA
- short hairpin RNA
References
- Ango F., Albani-Torregrossa S., Joly C., Robbe D., Michel J.M., Pin J.P., Bockaert J., Fagni L.. 1999. A simple method to transfer plasmid DNA into neuronal primary cultures: Functional expression of the mGlu5 receptor in cerebellar granule cells. Neuropharmacology. 38:793–803. 10.1016/S0028-3908(99)00005-2 [DOI] [PubMed] [Google Scholar]
- Aniksztejn L., Otani S., Ben-Ari Y.. 1992. Quisqualate metabotropic receptors modulate NMDA currents and facilitate induction of long-term potentiation through protein kinase C. Eur. J. Neurosci. 4:500–505. 10.1111/j.1460-9568.1992.tb00900.x [DOI] [PubMed] [Google Scholar]
- Anwyl R. 1999. Metabotropic glutamate receptors: Electrophysiological properties and role in plasticity. Brain Res. Brain Res. Rev. 29:83–120. 10.1016/S0165-0173(98)00050-2 [DOI] [PubMed] [Google Scholar]
- Attucci S., Carlà V., Mannaioni G., Moroni F.. 2001. Activation of type 5 metabotropic glutamate receptors enhances NMDA responses in mice cortical wedges. Br. J. Pharmacol. 132:799–806. 10.1038/sj.bjp.0703904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad H., Hubert G.W., Smith Y., Levey A.I., Conn P.J.. 2000. Activation of metabotropic glutamate receptor 5 has direct excitatory effects and potentiates NMDA receptor currents in neurons of the subthalamic nucleus. J. Neurosci. 20:7871–7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron M.K., Boeckers T.M., Vaida B., Faham S., Gingery M., Sawaya M.R., Salyer D., Gundelfinger E.D., Bowie J.U.. 2006. An architectural framework that may lie at the core of the postsynaptic density. Science. 311:531–535. 10.1126/science.1118995 [DOI] [PubMed] [Google Scholar]
- Beneken J., Tu J.C., Xiao B., Nuriya M., Yuan J.P., Worley P.F., Leahy D.J.. 2000. Structure of the Homer EVH1 domain-peptide complex reveals a new twist in polyproline recognition. Neuron. 26:143–154. 10.1016/S0896-6273(00)81145-9 [DOI] [PubMed] [Google Scholar]
- Bertaso F., Roussignol G., Worley P., Bockaert J., Fagni L., Ango F.. 2010. Homer1a-dependent crosstalk between NMDA and metabotropic glutamate receptors in mouse neurons. PLoS ONE. 5:e9755. 10.1371/journal.pone.0009755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolotto Z.A., Fitzjohn S.M., Collingridge G.L.. 1999. Roles of metabotropic glutamate receptors in LTP and LTD in the hippocampus. Curr. Opin. Neurobiol. 9:299–304. 10.1016/S0959-4388(99)80044-0 [DOI] [PubMed] [Google Scholar]
- Boute N., Jockers R., Issad T.. 2002. The use of resonance energy transfer in high-throughput screening: BRET versus FRET. Trends Pharmacol. Sci. 23:351–354. 10.1016/S0165-6147(02)02062-X [DOI] [PubMed] [Google Scholar]
- Brakeman P.R., Lanahan A.A., O’Brien R., Roche K., Barnes C.A., Huganir R.L., Worley P.F.. 1997. Homer: A protein that selectively binds metabotropic glutamate receptors. Nature. 386:284–288. 10.1038/386284a0 [DOI] [PubMed] [Google Scholar]
- Celikel T., Marx V., Freudenberg F., Zivkovic A., Resnik E., Hasan M.T., Licznerski P., Osten P., Rozov A., Seeburg P.H., Schwarz M.K.. 2007. Select overexpression of homer1a in dorsal hippocampus impairs spatial working memory. Front Neurosci. 1:97–110. 10.3389/neuro.01.1.1.007.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon V., Audet M., Homburger V., Bockaert J., Fagni L., Bouvier M., Perroy J.. 2008. Subcellular imaging of dynamic protein interactions by bioluminescence resonance energy transfer. Biophys. J. 94:1001–1009. 10.1529/biophysj.107.117275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz G.P., Bähr M.. 2005. Peptide-enhanced cellular internalization of proteins in neuroscience. Brain Res. Bull. 68:103–114. 10.1016/j.brainresbull.2005.08.015 [DOI] [PubMed] [Google Scholar]
- Doherty A.J., Palmer M.J., Henley J.M., Collingridge G.L., Jane D.E.. 1997. (RS)-2-chloro-5-hydroxyphenylglycine (CHPG) activates mGlu5, but no mGlu1, receptors expressed in CHO cells and potentiates NMDA responses in the hippocampus. Neuropharmacology. 36:265–267. 10.1016/S0028-3908(97)00001-4 [DOI] [PubMed] [Google Scholar]
- Fagni L., Worley P.F., Ango F.. 2002. Homer as both a scaffold and transduction molecule. Sci. STKE. 2002:re8. 10.1126/stke.2002.137.re8 [DOI] [PubMed] [Google Scholar]
- Gerber U., Gee C.E., Benquet P.. 2007. Metabotropic glutamate receptors: Intracellular signaling pathways. Curr. Opin. Pharmacol. 7:56–61. 10.1016/j.coph.2006.08.008 [DOI] [PubMed] [Google Scholar]
- Harvey J., Collingridge G.L.. 1993. Signal transduction pathways involved in the acute potentiation of NMDA responses by 1S,3R-ACPD in rat hippocampal slices. Br. J. Pharmacol. 109:1085–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M.K., Ames H.M., Hayashi Y.. 2006. Tetrameric hub structure of postsynaptic scaffolding protein homer. J. Neurosci. 26:8492–8501. 10.1523/JNEUROSCI.2731-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homayoun H., Moghaddam B.. 2010. Group 5 metabotropic glutamate receptors: Role in modulating cortical activity and relevance to cognition. Eur. J. Pharmacol. 639:33–39. 10.1016/j.ejphar.2009.12.042 [DOI] [PubMed] [Google Scholar]
- Hu J.H., Park J.M., Park S., Xiao B., Dehoff M.H., Kim S., Hayashi T., Schwarz M.K., Huganir R.L., Seeburg P.H., et al. 2010. Homeostatic scaling requires group I mGluR activation mediated by Homer1a. Neuron. 68:1128–1142. 10.1016/j.neuron.2010.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaubert P.J., Golub M.S., Lo Y.Y., Germann S.L., Dehoff M.H., Worley P.F., Kang S.H., Schwarz M.K., Seeburg P.H., Berman R.F.. 2007. Complex, multimodal behavioral profile of the Homer1 knockout mouse. Genes Brain Behav. 6:141–154. 10.1111/j.1601-183X.2006.00240.x [DOI] [PubMed] [Google Scholar]
- Kammermeier P.J. 2008. Endogenous homer proteins regulate metabotropic glutamate receptor signaling in neurons. J. Neurosci. 28:8560–8567. 10.1523/JNEUROSCI.1830-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A., Ozawa F., Saitoh Y., Hirai K., Inokuchi K.. 1997. vesl, a gene encoding VASP/Ena family related protein, is upregulated during seizure, long-term potentiation and synaptogenesis. FEBS Lett. 412:183–189. 10.1016/S0014-5793(97)00775-8 [DOI] [PubMed] [Google Scholar]
- Kotecha S.A., Jackson M.F., Al-Mahrouki A., Roder J.C., Orser B.A., MacDonald J.F.. 2003. Co-stimulation of mGluR5 and N-methyl-D-aspartate receptors is required for potentiation of excitatory synaptic transmission in hippocampal neurons. J. Biol. Chem. 278:27742–27749. 10.1074/jbc.M301946200 [DOI] [PubMed] [Google Scholar]
- Lu W., Man H., Ju W., Trimble W.S., MacDonald J.F., Wang Y.T.. 2001. Activation of synaptic NMDA receptors induces membrane insertion of new AMPA receptors and LTP in cultured hippocampal neurons. Neuron. 29:243–254. 10.1016/S0896-6273(01)00194-5 [DOI] [PubMed] [Google Scholar]
- Mao L., Yang L., Tang Q., Samdani S., Zhang G., Wang J.Q.. 2005. The scaffold protein Homer1b/c links metabotropic glutamate receptor 5 to extracellular signal-regulated protein kinase cascades in neurons. J. Neurosci. 25:2741–2752. 10.1523/JNEUROSCI.4360-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M., Penick E.C., Edwards J.G., Kauer J.A., Ehlers M.D.. 2004. Recycling endosomes supply AMPA receptors for LTP. Science. 305:1972–1975. 10.1126/science.1102026 [DOI] [PubMed] [Google Scholar]
- Perroy J. 2010. Subcellular dynamic imaging of protein-protein interactions in live cells by bioluminescence resonance energy transfer. Methods Mol. Biol. 591:325–333. 10.1007/978-1-60761-404-3_19 [DOI] [PubMed] [Google Scholar]
- Perroy J., Adam L., Qanbar R., Chénier S., Bouvier M.. 2003. Phosphorylation-independent desensitization of GABA(B) receptor by GRK4. EMBO J. 22:3816–3824. 10.1093/emboj/cdg383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroy J., Pontier S., Charest P.G., Aubry M., Bouvier M.. 2004. Real-time monitoring of ubiquitination in living cells by BRET. Nat. Methods. 1:203–208. 10.1038/nmeth722 [DOI] [PubMed] [Google Scholar]
- Perroy J., Raynaud F., Homburger V., Rousset M.C., Telley L., Bockaert J., Fagni L.. 2008. Direct interaction enables cross-talk between ionotropic and group I metabotropic glutamate receptors. J. Biol. Chem. 283:6799–6805. 10.1074/jbc.M705661200 [DOI] [PubMed] [Google Scholar]
- Pfleger K.D., Eidne K.A.. 2006. Illuminating insights into protein-protein interactions using bioluminescence resonance energy transfer (BRET). Nat. Methods. 3:165–174. 10.1038/nmeth841 [DOI] [PubMed] [Google Scholar]
- Potschka H., Krupp E., Ebert U., Gümbel C., Leichtlein C., Lorch B., Pickert A., Kramps S., Young K., Grüne U., et al. 2002. Kindling-induced overexpression of Homer 1A and its functional implications for epileptogenesis. Eur. J. Neurosci. 16:2157–2165. 10.1046/j.1460-9568.2002.02265.x [DOI] [PubMed] [Google Scholar]
- Renner M., Specht C.G., Triller A.. 2008. Molecular dynamics of postsynaptic receptors and scaffold proteins. Curr. Opin. Neurobiol. 18:532–540. 10.1016/j.conb.2008.09.009 [DOI] [PubMed] [Google Scholar]
- Ronesi J.A., Huber K.M.. 2008. Homer interactions are necessary for metabotropic glutamate receptor-induced long-term depression and translational activation. J. Neurosci. 28:543–547. 10.1523/JNEUROSCI.5019-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussignol G., Ango F., Romorini S., Tu J.C., Sala C., Worley P.F., Bockaert J., Fagni L.. 2005. Shank expression is sufficient to induce functional dendritic spine synapses in aspiny neurons. J. Neurosci. 25:3560–3570. 10.1523/JNEUROSCI.4354-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainlos M., Tigaret C., Poujol C., Olivier N.B., Bard L., Breillat C., Thiolon K., Choquet D., Imperiali B.. 2011. Biomimetic divalent ligands for the acute disruption of synaptic AMPAR stabilization. Nat. Chem. Biol. 7:81–91. 10.1038/nchembio.498 [DOI] [PubMed] [Google Scholar]
- Sakagami Y., Yamamoto K., Sugiura S., Inokuchi K., Hayashi T., Kato N.. 2005. Essential roles of Homer-1a in homeostatic regulation of pyramidal cell excitability: A possible link to clinical benefits of electroconvulsive shock. Eur. J. Neurosci. 21:3229–3239. 10.1111/j.1460-9568.2005.04165.x [DOI] [PubMed] [Google Scholar]
- Sala C., Piëch V., Wilson N.R., Passafaro M., Liu G., Sheng M.. 2001. Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron. 31:115–130. 10.1016/S0896-6273(01)00339-7 [DOI] [PubMed] [Google Scholar]
- Sala C., Futai K., Yamamoto K., Worley P.F., Hayashi Y., Sheng M.. 2003. Inhibition of dendritic spine morphogenesis and synaptic transmission by activity-inducible protein Homer1a. J. Neurosci. 23:6327–6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Suzuki K., Nakanishi S.. 2001. NMDA receptor stimulation and brain-derived neurotrophic factor upregulate homer 1a mRNA via the mitogen-activated protein kinase cascade in cultured cerebellar granule cells. J. Neurosci. 21:3797–3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannevin R.H., Huganir R.L.. 2000. Postsynaptic organization and regulation of excitatory synapses. Nat. Rev. Neurosci. 1:133–141. 10.1038/35039075 [DOI] [PubMed] [Google Scholar]
- Szumlinski K.K., Lominac K.D., Kleschen M.J., Oleson E.B., Dehoff M.H., Schwarz M.K., Seeburg P.H., Worley P.F., Kalivas P.W.. 2005. Behavioral and neurochemical phenotyping of Homer1 mutant mice: Possible relevance to schizophrenia. Genes Brain Behav. 4:273–288. 10.1111/j.1601-183X.2005.00120.x [DOI] [PubMed] [Google Scholar]
- Szumlinski K.K., Kalivas P.W., Worley P.F.. 2006. Homer proteins: Implications for neuropsychiatric disorders. Curr. Opin. Neurobiol. 16:251–257. 10.1016/j.conb.2006.05.002 [DOI] [PubMed] [Google Scholar]
- Szumlinski K.K., Ary A.W., Lominac K.D.. 2008. Homers regulate drug-induced neuroplasticity: Implications for addiction. Biochem. Pharmacol. 75:112–133. 10.1016/j.bcp.2007.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tappe A., Klugmann M., Luo C., Hirlinger D., Agarwal N., Benrath J., Ehrengruber M.U., During M.J., Kuner R.. 2006. Synaptic scaffolding protein Homer1a protects against chronic inflammatory pain. Nat. Med. 12:677–681. 10.1038/nm1406 [DOI] [PubMed] [Google Scholar]
- Tu J.C., Xiao B., Yuan J.P., Lanahan A.A., Leoffert K., Li M., Linden D.J., Worley P.F.. 1998. Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors. Neuron. 21:717–726. 10.1016/S0896-6273(00)80589-9 [DOI] [PubMed] [Google Scholar]
- Ugolini A., Corsi M., Bordi F.. 1997. Potentiation of NMDA and AMPA responses by group I mGluR in spinal cord motoneurons. Neuropharmacology. 36:1047–1055. 10.1016/S0028-3908(97)00103-2 [DOI] [PubMed] [Google Scholar]
- Xiao B., Tu J.C., Worley P.F.. 2000. Homer: A link between neural activity and glutamate receptor function. Curr. Opin. Neurobiol. 10:370–374. 10.1016/S0959-4388(00)00087-8 [DOI] [PubMed] [Google Scholar]
- Zeke A., Lukács M., Lim W.A., Reményi A.. 2009. Scaffolds: Interaction platforms for cellular signalling circuits. Trends Cell Biol. 19:364–374. 10.1016/j.tcb.2009.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.