Abstract
When the major ampulate spidroins (MaSp1) are called upon to form spider dragline silk, one of nature’s most amazing materials, a small drop in pH must occur. Using a state-of-the-art simulation technique, constant pH molecular dynamics, we discovered a few residues that respond to the pH signal in the dimerization of the N-terminal domain (NTD) of MaSp1 which is an integral step in the fiber assembly. At neutral pH the deprotonation of Glu79 and Glu119 leads to water penetration and structural changes at the monomer-monomer binding interface. At strongly acidic pH, the protonation of Asp39 and Asp40 weakens the electrostatic attraction between the monomers. Thus, we propose a “trap-and-trigger” mechanism whereby the intermolecular salt-bridges at physiologically relevant pH conditions always act as a stabilizing “trap” favoring dimerization. As pH is lowered to about 6, Glu79 and Glu119 become protonated, triggering the dimerization and subsequent silk formation. We speculate that this type of mechanism is operative in many other pH-sensitive biological processes.
Keywords: electrostatics, pKa, dimer, stability, pH, molecular dynamics, replica exchange
Spider dragline silk is one of nature’s most spectacular materials. The toughness of spider silk surpasses synthetic rubber and its strength is comparable to high-tensile steel1. These exceptional properties combined with biocompatibility make silk and silk biomimetics attractive materials for future bio- and material engineering applications. Silk based products have been envisioned as drug delivery systems2 and scaffolds for tissue engineering3,4, adhesives, and microfluidic devices5. Dragline silk is made of proteins known as the major ampullate spidroins 1 and 2 (MaSp1 and MaSp2), which are stored in the major ampullate gland of spiders as a microemulsion6,7. The MaSp’s are comprised of a repetitive domain containing polyalanine, glutamine- and glycine-rich motifs as well as the non-repetitive C-terminal and N-terminal domains6,7. During silk spinning the spidroins pass from the gland through the exit duct while being subjected to several chemical and physical forces leading to nano-composite fibers6. A major factor contributing to the transition from soluble proteins to silk fibers is acidification8. While solution pH drops from 7.2 in the gland to 6.3 within the first millimeter of the 20 mm exit duct9, the pH at the distal end of the duct may be substantially lower8,10. Acidification has also been found to increase the rate of self-assembly of silk proteins from Euprosthenops australis spiders11 and Bombyx mori silkworms12, suggesting that the acid-bath treatment is a general method employed by nature for producing delicate, yet durable fibers.
Recently, several experiments have demonstrated that the NT domain of MaSp1 (NTD) is the pH sensing portion, which dimerizes allowing spidroins to self-assemble by a relay-like mechanism into silk fiber13,7,11,14. However, a detailed mechanism of the pH-dependent assembly process remains unclear. Askarieh et al. solved a crystal structure at pH 7 which showed that the NTD from Euprosthenops australis exists as a stable homodimer11. However, experiments based on electrospray ionization mass spectrometry with the same protein14 and NMR with the protein from Latrodectus15 indicated that NTD is mainly monomeric at neutral pH. The dimeric form is more stable at acidic pH and low concentrations of salt, consistent with the pulldown experiments using NTD’s from both Latrodectus and Nephila7. Moreover, the interpretation of a pH-dependent red shift in the tryptophan fluorescence differs as to whether it is the result of the conformational change in the dimer11 or monomer7.
The aforementioned inconsistency has prompted us to investigate the pH effect on the dimerization of NTD using a state-of-the-art simulation technique, continuous constant pH molecular dynamics (CpHMD)16,17 with conformational sampling in explicit solvent and pH-based replica exchange (pH-REX)18, which allows us to determine the pKa values of all ionizable sidechains and probe the pH-dependent conformational dynamics in atomistic details. Our data indicates that the dimer becomes destabilized as the pH is increased above 6, as a result of the ionization of key residues Glu79 and Glu119 which leads to water penetration into the monomer-monomer interface.
Considering the experimental findings11,14,7,15, we hypothesized that the origin of the pH-dependent dimerization may be explained by the intermolecular interactions between monomers, and that ionization of a few residues could shift the monomer-dimer equilibrium. To test this hypothesis, we carried out pH titration simulations using the pH-REX/CpHMD method in explicit solvent for the dimer and monomer forms of NTD starting from the crystal structure of the dimer (PDB ID: 3LR2) and two monomer units, respectively. Molecular dynamics at pH conditions of 0 to 10 was performed for 5.5 ns while simultaneously titrating all acidic and histidine residues and periodically attempting exchanges between pH replicas. The cumulative simulation time was 77 ns and 93.5 ns for the dimer and monomers, respectively. These simulations allowed us to determine pKa values and obtain details of the conformational dynamics at each pH condition.
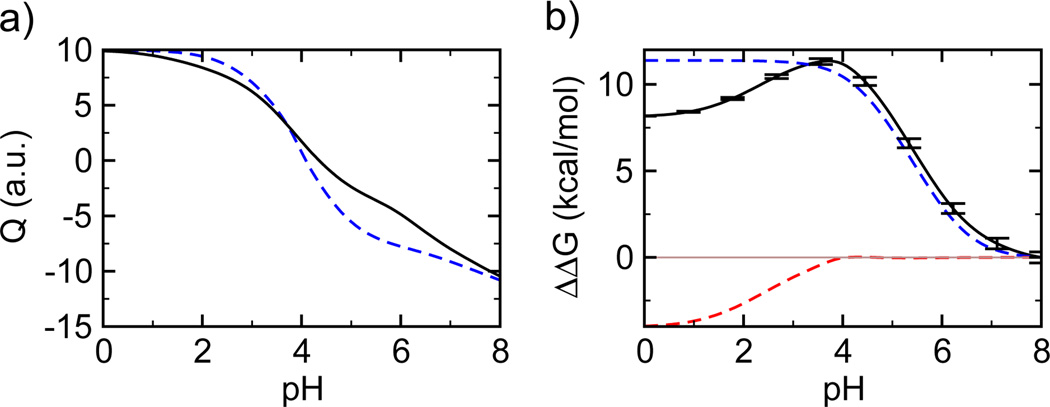
Using the pKa values of all titratable residues in the dimer and unbound monomers (Table 1 and Table S1) we calculated the total charge of the dimer and monomers (Figure 1a). Using the pKa shifts upon dimerization we obtained the pH-dependent changes in the dimer stability (Figure 1b) by analytically integrating the Wyman-Tanford linkage equation19, ∂ΔG/∂pH = ln(10)RTΔQdiss, where ΔG is the free energy of dimer dissociation (or dimer stability) and ΔQdiss is the change in the total charge as dimer dissociates. Our calculated isoelectric point (pI), 4.35 and 4.15 for the dimer and the two monomers, respectively (Figure 1a), are in good agreement with the value of 4.25 for the dimer determined by the measurements of electrophoretic mobility11. According to the calculated stability change (black curve in Figure 1b), the dimer is least stable at pH 8. As the solution pH is decreased to 6, the dimer is stabilized by about 3.5 kcal/mol. The stabilization continues as pH is lowered to near 4 where dimerization is favored by 11 kcal/mol as compared to pH 8. As the solution pH is reduced further the dimer is again destabilized.
Table 1.
Calculated pKa valuesa in the monomer and dimer states
| Residue | pKamono | pKadimer | ΔpKab |
|---|---|---|---|
| Monomer (Subunit) A | |||
| Asp39 | 3.05 ± 0.05 | 1.31 ± 0.06 | −1.74 ± 0.07 |
| Asp40 | 4.11 ± 0.05 | 4.62 ± 0.04 | 0.51 ± 0.06 |
| Glu79 | 4.42 ± 0.04 | 6.26 ± 0.05 | 1.84 ± 0.07 |
| Glu119 | 4.23 ± 0.05 | 6.12 ± 0.04 | 1.89 ± 0.05 |
| Monomer (Subunit) B | |||
| Asp39 | 2.80 ± 0.04 | 2.03 ± 0.06 | −0.77 ± 0.07 |
| Asp40 | 4.19 ± 0.05 | 3.13 ± 0.10 | −1.06 ± 0.11 |
| Glu79 | 4.43 ± 0.05 | 6.73 ± 0.06 | 2.30 ± 0.08 |
| Glu119 | 4.32 ± 0.03 | 6.71 ± 0.05 | 2.39 ± 0.06 |
Only pKa’s with ΔpKa values greater than 0.5 are listed. Error bars are the standard deviations from 100 bootstrap trial fittings. A complete list of calculated pKa’s is given in Table S1.
ΔpKa = pKadimer - pKamono.
Figure 1.
pH-dependent change in the total charge and stability of the dimer NTD. a) Total charge of the dimer (solid) and the two monomers (dashed) calculated at different pH conditions. b) Stability change (free energy of dimer dissociation), ΔΔG) relative to the pH 8 condition calculated using all pKa values (black), Glu79 and Glu119 (blue), and Asp39 and Asp40 (red). A horizontal line is drawn at ΔΔG of zero to guide the eye.
The stability change of the dimer can be decomposed into contributions from each titratable residue assuming no coupling between their protonation equilibria. The contributions from residues Glu79 and Glu119 dominate the stability change in the pH range of 4 to 8 (blue curve in Figure 1b) and they are also the major source for the change in the total charges upon dimerization in the same pH range (Figure 1a). This is because the pKa’s of Glu79 and Glu119 have the largest positive shifts upon dimerization. The pKa’s in the monomers are similar to the model value, around 4.4, but in the dimer they are shifted to above 6. Ionization of these residues significantly destabilizes the dimer. On the contrary, the pKa values of Asp39 and Asp40 have the largest negative shifts upon dimerization. Therefore, they are together responsible for the stability change in the pH range 0–4 (red curve in Figure 1b). They are also major contributors to the change in the total charge upon dimerization.
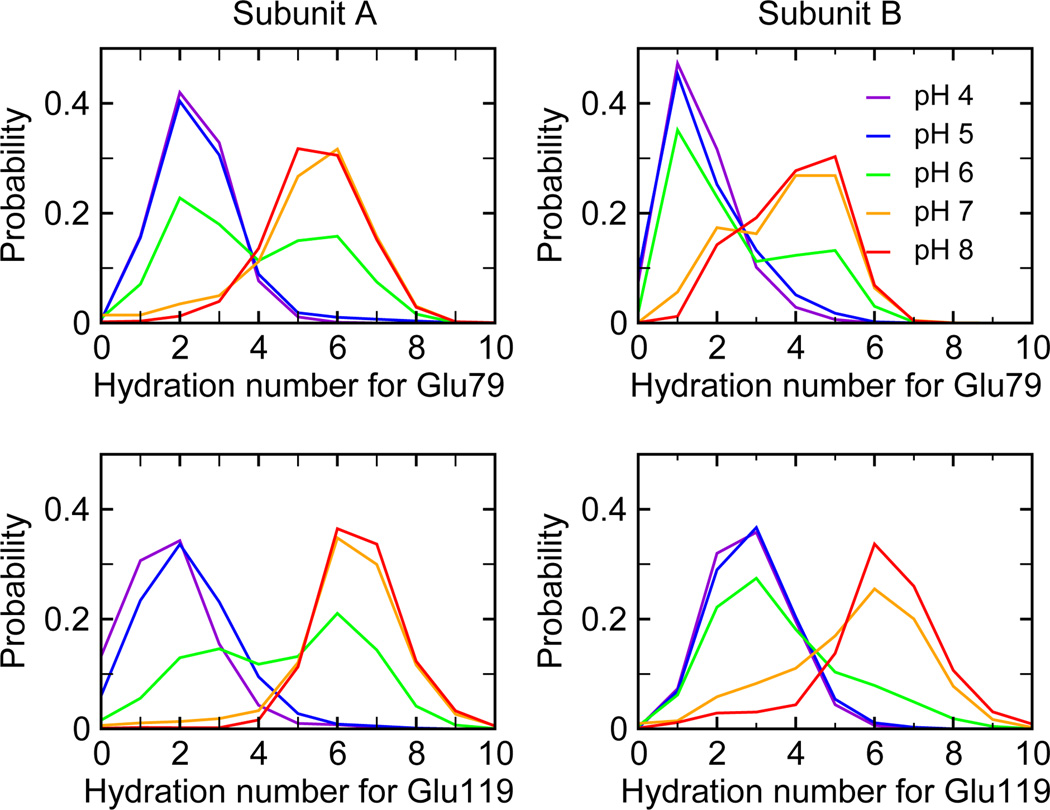
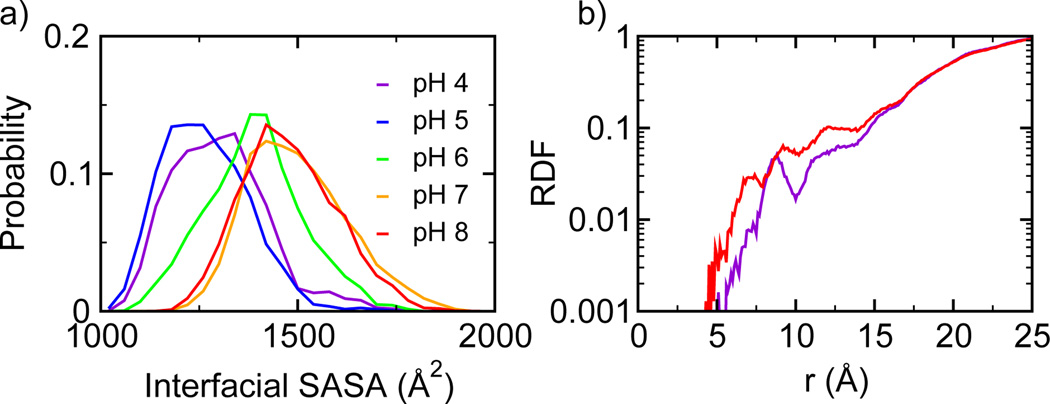
We next examine in detail how ionization of Glu79 and Glu119 leads to the destabilization of the dimer at elevated pH. According to our calculation, as solution pH is increased from pH 4 to pH 8, the ionization of Glu79 and Glu119 destabilizes the dimer by more than 10 kcal/mol. When solution pH is 4 or 5, which is below the pKa values of Glu79 and Glu119 (between 6.1 and 6.7), these residues are only weakly solvated. The distribution of the hydration number is centered around 2 (Figure 2). However, as pH is increased, the hydration number increases due to ionization of the sidechains of Glu79 and Glu119. The distribution becomes bimodal at pH 6, while at pH 7 and 8, the maximum probability is shifted to around 6 or 7. The hydration of Glu79 and Glu119 at the pH above 6 is associated with the entrance of water molecules into the dimerization interface. The latter can be quantified by the change in the interfacial solvent accessible surface area (SASA) as a function of pH (Figure 3a). At pH 4 and 5, the interfacial SASA fluctuates around 1200–1300 Å2 but the distribution shifts to larger values as pH is increased to 6. When pH is further increased to 7 and 8, the maximum probability for SASA is around 1500 Å2. Thus, an increase of pH from 4 to 8 results in an increased solvent-exposure of the dimer interface by at least 200 Å2. To further characterize the pH-induced water penetration, we calculated the radial distribution function (RDF) for water relative to the dimer center (Figure 3b). At pH 8, the RDF shows an increased density of water near the dimer center as compared to pH 4. The notable accumulation of water at around 10 Å and 12 Å corresponds to the positions of Glu79 and Glu119, respectively.
Figure 2.
pH-dependent hydration of Glu79 and Glu119 in the dimer. Probability distributions of the hydration numbers for Glu79 and Glu119 in both monomer units at different pH conditions. Hydration number is defined as the number of water molecules within 3 Å of the sidechain.
Figure 3.
pH-dependent solvent exposure of the dimer interface. a) Probability distribution of the interfacial solvent-accessible surface area (SASA) at different pH conditions. b) Radial distribution function (RDF) of water to the dimer center (defined as the center of the Cα atoms in Phe73 and Ala74) at pH 4 (purple) and 8 (red).
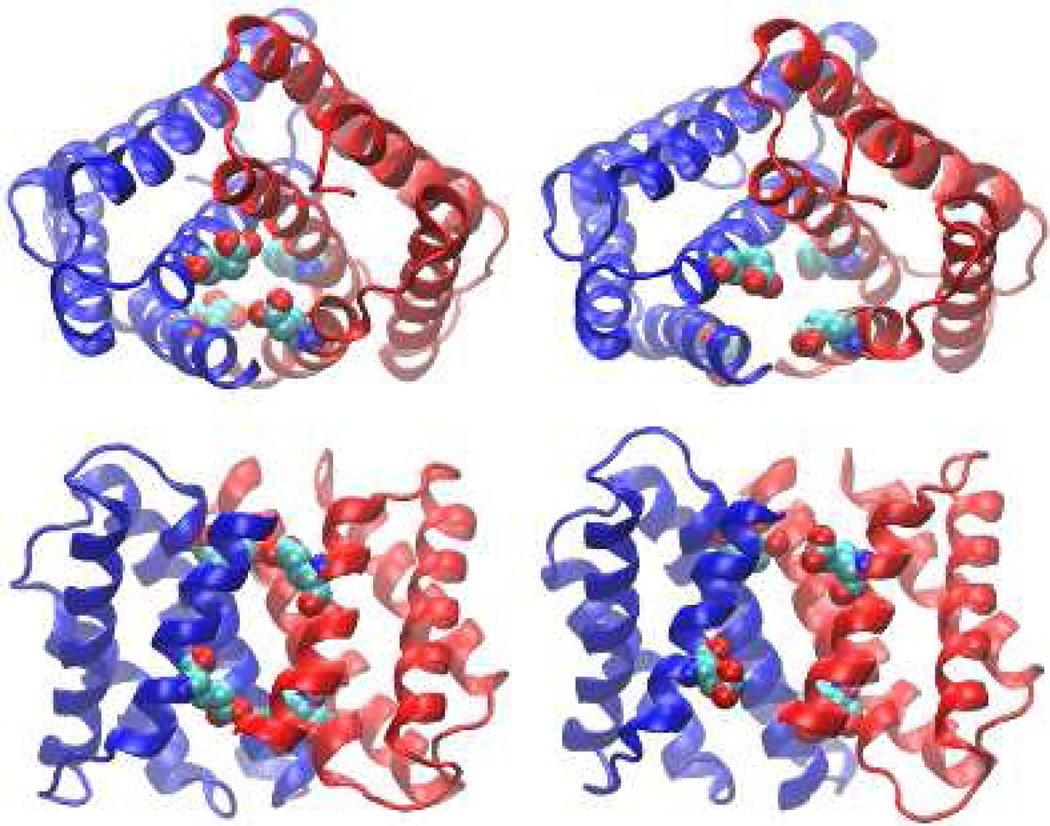
As water molecules enter and the monomer-monomer interface opens up, conformational rearrangement occurs. Although our simulation can not describe the full extent of the conformational change to due to the limited sampling time, it offer a glimpse at the initial events. At pH 6 and above, the contacts between residues from the opposite monomer subunits (A and B) are weakened. Most notably, the contact probabilities for Glu79(A)–Met71(B) and Met71(A)–Glu79(B), as well as for Glu119(A)–Met126 (B) and Met126(A)–Glu119(B) are significantly reduced (see Figure 2 in the Supplemental Information). The general trend of the weakened inter-molecular interactions at elevated pH conditions is another indication of the destabilization of the dimer. Moreover, we observe that upon ionization Glu119 rotates out of the binding interface into solution. The most probable distance between Glu119 and the dimer center is around 12 Å at pH 4, but at pH 8 the distribution becomes bimodal with a population centered at just greater than 15 Å (Figure S3). For Glu79 there is a very slight shift outward, which is a result of the expansion of the dimer structure (Figure 4). For Glu119, along with the overall movement of the dimer, there is a distinct change in the χ1 angle at basic pH and a rotation of Helix 5 towards solution which allows Glu119 to become more solvated (see Figure S4). At pH 4, the χ1 angle of Glu119 samples ±180° and there is a minor population at 70°, but at pH 8 χ1 is predominately 70° and the position at ±180° is not sampled. The related conformational rearrangement can be readily seen by comparing the snapshots taken from the simulation at pH 4 and pH 8 (Figure 4).
Figure 4.
pH-dependent conformational rearrangement of the dimer. Top (upper) and side (lower) views of the snapshots taken from the simulation at pH 4 (left) and pH 8 (right). Glu79 and Glu119 are explicitly shown.
Our simulated titration data reveals two key residues responsible for the pH-dependent dimerization of NTD-MaSp1. At slightly acidic pH Glu79 and Glu119 are protonated, but at pH above 7 these residues become ionized. Burial of these charged residues in the hydrophobic environment of the dimer interface is unfavorable which results in a destabilization of the dimer by about 3.5 kcal/mol when pH is raised from 6 to 8 (Figure 1). Thus, according to our data, mutation of Glu79 to a neutral residue Gln should favor the dimerization at neutral or elevated pH, consistent with the electrospray ionization mass spectrometry data which showed that mutant D79N is able to dimerize at pH 6.8 and 7 in contrast to the wild type14.
Our dynamics data shows that ionization of the interfacial residues Glu79 and Glu119 at or above pH 7 causes the dimer interface to open up, which allows water molecules to enter, thereby weakening the intermolecular interactions that are responsible for holding the two monomer units together (Figure 2, 3 and 4). The pH-induced conformational change in the dimer is consistent with the deuterium exchange data which showed decreased deuteration level at pH 6 relative to pH 714. The same set of experiments also showed that mutation D40N, E84Q or D40N/E84Q inhibits the dimer formation at low pH. However, these data do not necessarily imply that ionization of Asp40 or Glu84 promotes the dimer stability at low pH. This is because Asp39, Asp40 and Glu84 are clustered together in the crystal structure, and mutation of one sidechain likely perturbs the electrostatic interactions of the other two. To rationalize these experimental data, additional simulation based on the mutant structures would be necessary, which is beyond the scope of the current paper.
Our simulation also reveals that the dimer interface is further stabilized by electrostatic interactions. Asp39 forms salt-bridges with Arg60 or Lys65 of the opposite subunit (see Figure S5). These favorable interactions are reflected in the negative pKa shift of Asp39 upon dimerization. Asp40 also interacts with Lys65 of the other subunit, although the extent of the pKa shift is less than that of Asp39. As pH is decreased below 4, Asp39 and Asp40 become protonated, the fraction of the tightly bound interactions with Arg60 and Lys65 is drastically reduced. Thus, ionization of Asp39 and Asp40 is responsible for the reduced dimer stability at pH below 4 (blue curve in Figure 1b).
In summary, we have identified a pH-modulated electrostatic system that controls the dimerization of NTD-MaSp1 (see Figure 5). In the pH range 4–6, the dimer is stabilized by the salt bridges (Asp39-Arg60, Asp39-Lys65, Asp40-Lys65) formed at the opposite poles of the subunits. Glu79 and Glu119 are protonated and buried in the interface. At pH above 6, these two residues become deprotonated and introduce a large excess negative potential resulting in a large desolvation penalty for the formation of the homodimer. Thus, we propose that a “trap-and-trigger” mechanism controls dimerization where the opposite poles at physiologically relevant pH conditions always act as a stabilizing “trap” favoring dimerization. However, the acidic residues at the hydrophobic interface must be protonated, and protonation of these residues is the “trigger” that causes the NTD to dimerize and act as a pH-sensitive relay during silk formation.
Figure 5.
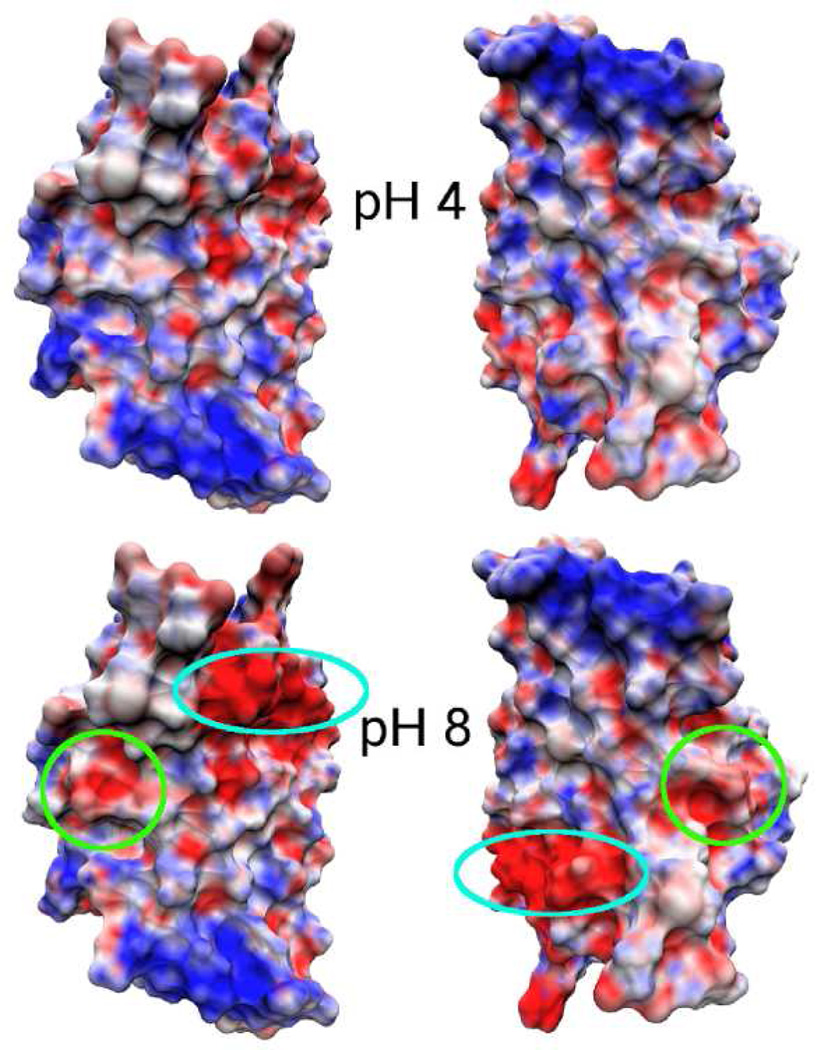
pH-dependent electrostatic potential of the dimer subunits. Electrostatic surface potential maps are calculated using the average charges derived from the unprotonated fractions of titratable residues at each pH (details see Supporting Information) The subunits are separated and oriented to show the dimer interface. The locations of Glu79 (turquoise) and Glu119 (green) are circled.
Methods
Structure Preparation
The initial structure of the dimeric NTD-MaSp1 was prepared by removing all heteroatoms and adopting the first of the two sidechain orientations based on the crystal structure (PDB ID: 3LR2). Missing residues were built with the program Modeller20. Hydrogens were added with the HBUILD facility in the CHARMM program21 and the structure was placed in a truncated octahedral water box with dimensions 18 Å greater than the largest dimension of the protein. Sodium chloride was added such that the solvent ionic strength was 100 mM. Water molecules within 2.6 Å of any heavy atom were deleted and the system was energy minimized in several stages with progressively smaller harmonic restraint applied to heavy atoms of the protein.
Simulation Details
The structures of the dimer and corresponding subunits as prepared above were used to initiate the pH-REX titration simulations of the dimer and monomers, respectively. All simulations were conducted using the PHMD module16,17,18 in the CHARMM program (version c35b3)21. The most recent extension18 of the PHMD module which includes conformational sampling in explicit solvent and the pH-based replica-exchange (pH-REX) protocol to accelerate barrier crossing and convergence was applied. The all-atom force fields, CHARMM22/CMAP22 and TIP3P23, were used to represent the protein and water atoms, respectively. The generalized-Born (GB) implicit model, GBSW24, with a Debye-Hückel term for taking into account salt screening (ionic strength was set to 100 mM), was applied to calculate solvent-modulated electrostatic energies for the propagation of titration coordinates18. The GB calculation and update of titration coordinates were executed every 10 dynamic steps. In a pH-REX simulation, independent replicas were subject to molecular dynamics runs at ambient temperature and pressure but different pH conditions. An exchange between adjacent pH conditions was attempted every 500 dynamic steps. A total of 14 and 17 replicas was used for the monomer and dimer simulations, respectively. Simulations were carried out for 5500 exchange attempts (5.5 ns) per replica, resulting in the cumulative simulation time of 77 ns for the monomers and 93.5 ns for the dimer. The coordinates and titration states were recorded after each exchange attempt. In the data analysis, the first 500 exchange cycles (0.5 ns per replica) were discarded. Additional details and convergence tests are given in Supporting Information.
Calculations of the pH-dependent Dimer Stability
pKa values were calculated by fitting the unprotonated fractions (S) at each pH to the Hill equation. Analytical integration of the Wyman-Tanford linkage equation leads to the following expression for the pH-dependent free energy of dimer dissociation in analogy to the pH-dependent protein stability25,
where the summation runs over all residues. are the Hill coefficients and pKa values for the ith residue in the dimeric and monomeric forms, respectively. In this work we set pHref = 8.0. Details of the error estimates are given in Supporting Information.
Supplementary Material
ACKNOWLEDGMENT
The authors acknowledge University of Oklahoma and National Institutes of Health (R01 GM098818 to JKS) for financial support.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Additional simulation details and analysis results. This material is available free of charge via http://public.acs.org.
References
- 1.Gosline JM, Guerette PA, Ortlepp CS, Savage KN. The Mechanical Design of Spider Silks: From Fibroin Sequence to Mechanical Function. J. Exp. Biol. 1999;202:3295–3303. doi: 10.1242/jeb.202.23.3295. [DOI] [PubMed] [Google Scholar]
- 2.Spiess K, Lammel A, Scheibel T. Recombinant Spider Silk Proteins for Applications in Biomaterials. Macromol. Biosci. 2010;10:998–1007. doi: 10.1002/mabi.201000071. [DOI] [PubMed] [Google Scholar]
- 3.Altman G, Diaz F, Jakuba C, Calabro T, Horan R, Chen J, Lu H, Richmond J, Kaplan D. Silk-Based Biomaterials. Biomaterials. 2003;24:401–416. doi: 10.1016/s0142-9612(02)00353-8. [DOI] [PubMed] [Google Scholar]
- 4.Zhou S, Peng H, Yu X, Zheng X, Cui W, Zhang Z, Li X, Wang J, Weng J, Jia W, Li F. Preparation and Characterization of a Novel Electrospun Spider Silk Fibroin/Poly(D,L-Lactide) Composite Fiber. J. Phys. Chem. B. 2008;112:11209–11216. doi: 10.1021/jp800913k. [DOI] [PubMed] [Google Scholar]
- 5.Omenetto F, Kaplan D. New Opportunities for an Ancient Material. Science. 2010;329:528–531. doi: 10.1126/science.1188936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heim M, Keerl D, Scheibel T. Spider Silk: From Soluble Protein to Extraordinary Fiber. Angew. Chem. Int. Ed. 2009;48:3584–3596. doi: 10.1002/anie.200803341. [DOI] [PubMed] [Google Scholar]
- 7.Gaines WA, Sehorn MG, Marcotte WR., Jr Spidroin N-terminal Domain Promotes a pH-Dependent Association of Silk Proteins During Self-Assembly. J. Biol. Chem. 2010;285:40745–40753. doi: 10.1074/jbc.M110.163121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vollrath F, Knight DP, Hu XW. Silk Production in a Spider Involves Acid Bath Treatment. Proc. R. Soc. Lond. B. 1998;265:817–820. [Google Scholar]
- 9.Dicko C, Vollrath F, Kenney JM. Spider Silk Protein Refolding is Controlled by Changing pH. Biomacromolecules. 2004;5:704–710. doi: 10.1021/bm034307c. [DOI] [PubMed] [Google Scholar]
- 10.Dicko C, Kenney J, Knight D, Vollrath F. Transition to a β-Sheet-Rich Structure in Spidroin in Vitro: The Effects of pH and Cations. Biochemistry. 2004;43:14080–14087. doi: 10.1021/bi0483413. [DOI] [PubMed] [Google Scholar]
- 11.Askerieh G, Hedhammar M, Nordling K, Saenz A, Casals C, Rising A, Johansson J, Knight SD. Self-Assembly of Spider Silk Proteins is Controlled by a pH-Sensitive Relay. Nature. 2010;465:236–238. doi: 10.1038/nature08962. [DOI] [PubMed] [Google Scholar]
- 12.Matsumoto A, Chen J, Collette AL, Kim U, Altman GH, Cebe P, Kaplan DL. Mechanisms of Silk Fibroin Sol-Gel Transitions. J. Phys. Chem. B. 2006;110:21630–21638. doi: 10.1021/jp056350v. [DOI] [PubMed] [Google Scholar]
- 13.Hedhammar M, Rising A, Grip S, Martinez AS, Nordling K, Casals C, Stark M, Johansson J. Structural Properties of Recombinant Nonrepetitive and Repetitive Parts of Major Ampullate Spidroin from Euprosthenops australis: Implications for Fiber Formation. Biochemisty. 2008;47:3407–3417. doi: 10.1021/bi702432y. [DOI] [PubMed] [Google Scholar]
- 14.Landreh M, Askarieh G, Nordling K, Hedhammar M, Rising A, Casals C, Astorga-Wells J, Alvelius G, Knight SD, Johansson J, et al. A pH-Dependent Dimer Lock in Spider Silk Protein. J. Mol. Biol. 2010;404:328–336. doi: 10.1016/j.jmb.2010.09.054. [DOI] [PubMed] [Google Scholar]
- 15.Hagn F, Thamm C, Scheibel T, Kessler H. pH-Dependent Dimerization and Salt-Dependent Stabilization of the N-Terminal Domain of Spider Dragline Silk-Implications for Fiber Formation. Angew. Chem. Int. Ed. 2011;45:3795–3800. doi: 10.1002/anie.201003795. [DOI] [PubMed] [Google Scholar]
- 16.Lee MS, Salsbury FR, Jr, Brooks CL., III Constant-pH Molecular Dynamics using Continuous Titration Coordinates. Proteins. 2004;56:738–752. doi: 10.1002/prot.20128. [DOI] [PubMed] [Google Scholar]
- 17.Khandogin J, Brooks CL., III Constant pH Molecular Dynamics with Proton Tautomerism. Biophys. J. 2005;89:141–157. doi: 10.1529/biophysj.105.061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wallace JA, Shen JK. Continuous Constant pH Molecular Dynamics in Explicit Solvent with pH-Based Replica Exchange. J. Chem. Theory Comput. 2011;7:2617–2629. doi: 10.1021/ct200146j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wyman J., Jr Linked Functions and Reciprocal Effects in Hemoglobin: A Second Look. Adv. Protein Chem. 1964;19:223–286. doi: 10.1016/s0065-3233(08)60190-4. [DOI] [PubMed] [Google Scholar]
- 20.Šali A, Blundell TL. Comparative Protein Modeling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 21.Brooks BR, Brooks CL, III, MacKerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartles C, Boresch S, et al. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacKerell AD, Jr, Feig M, Brooks CL., III Extending the Treatment of Backbone Energetics in Protein Force Fields: Limitations of Gas-Phase Quantum Mechanics in Reproducing Protein Conformational Distributions in Molecular Dynamics Simulations. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 23.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 24.Im W, Lee MS, Brooks CL., III Generalized Born Model with a Simple Smoothing Function. J. Comput. Chem. 2003;24:1691–1702. doi: 10.1002/jcc.10321. [DOI] [PubMed] [Google Scholar]
- 25.Shen JK. Uncovering Specific Electrostatic Interactions in the Denatured States of Proteins. Biophys. J. 2010;99:924–932. doi: 10.1016/j.bpj.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.