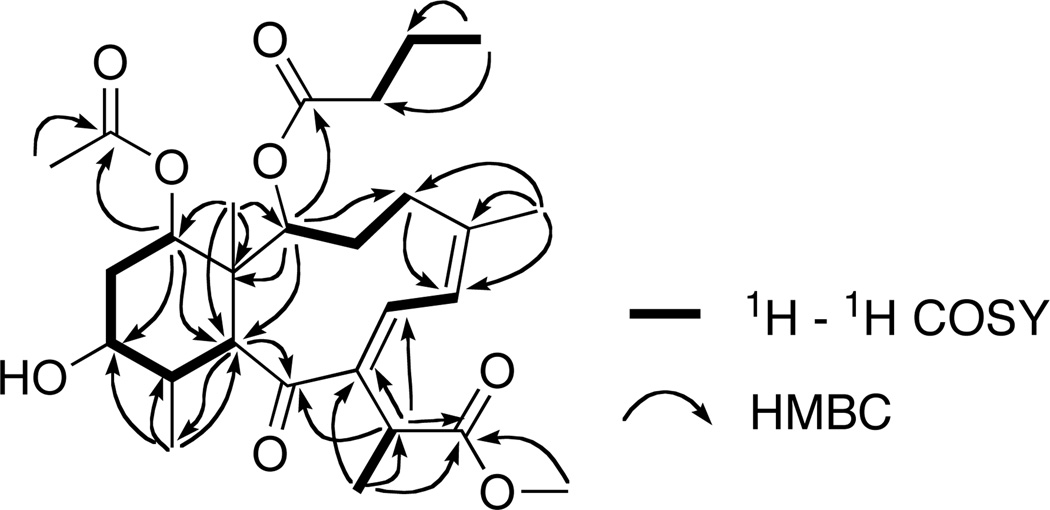Abstract
Three new briarane diterpenoids, briareolate esters L – N (1 – 3) have been isolated from a gorgonian Briareum asbestinum. Briareolate esters L (1) and M (2) are the first natural products possessing a 10-membered macrocyclic ring with a (E,Z)-dieneone and exhibit growth inhibition activity against both human embryonic stem cells (BG02) and a pancreatic cancer cell line (BxPC-3). Briareolate ester L (1), was found to contain a “spring-loaded” (E,Z)-dieneone Michael acceptor group that can form a reversible covalent bond to model sulfur-based nucleophiles.
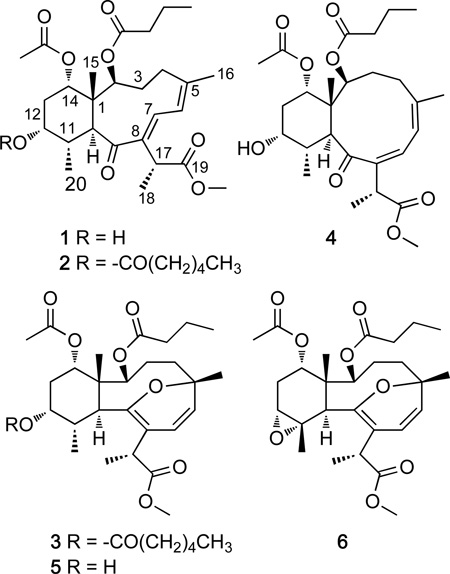
The marine gorgonian of the genus Briareum is an abundant source of highly oxygenated diterpenoids belonging to the eunicellin, asbestinane, cembrane, and briarane classes and exhibit numerous biological activities (e.g., cytotoxicity, anti-microbial, anti-inflammatory, antiviral, immunomodulatory, antifouling, and ichthotoxicity).1 The briareolate esters are a small group of unusual briarane diterpenoids isolated from Briareum asbestinum off the coast of Tobago that contain a C-19 methyl ester instead of the γ-lactone ring.2,3,4 In the course of screening pre-fractionated and semi-purified extracts of marine invertebrates to discover compounds that impact human embryonic stem cell (hESC) growth using a 96-well plate real-time cell electronic sensing (RT-CES) system, activity was found for the extract of the gorgonian Briareum asbestinum. Further purification of the active fractions led to the isolation of three new briarane diterpenoids, briareolate ester L – N (1 – 3), together with three known briareolate esters B (5), C (6) and G (4). In this paper we report the isolation, biological activity and possible mechanism of bioactivity.
Briareolate ester L (1) was isolated as a colorless oil. The molecular formula of briareolate ester L (1), C27H40O8, that was determined from the HRESIMS of the [M + Na]+ ion at m/z 515.2596, required 8 degrees of unsaturation. An initial analysis of the 13C NMR data revealed four carbonyl carbons (δC 201.4, 176.5, 175.5, 173.2), and two C-C double bonds (δC 148.1, 147.0, 141.7, 118.5) (see Table S1, Supporting Information). These data accounted for six of the eight double bond equivalents, and indicated that 1 was bicyclic. The presence of a ketone conjugated with two double bonds (α,β,γ,δ-unsaturated ketone) was indicated from a carbonyl carbon with a chemical shift of δC 201.4 (C-9), and the C–C double bond carbons at δC 148.1, 147.0, 141.7 and 118.5. The observation of UV absorption maxima at λmax = 284 and 232 nm and a C=O stretching band at 1650 cm−1 in the IR spectrum were consistent with this assignment.
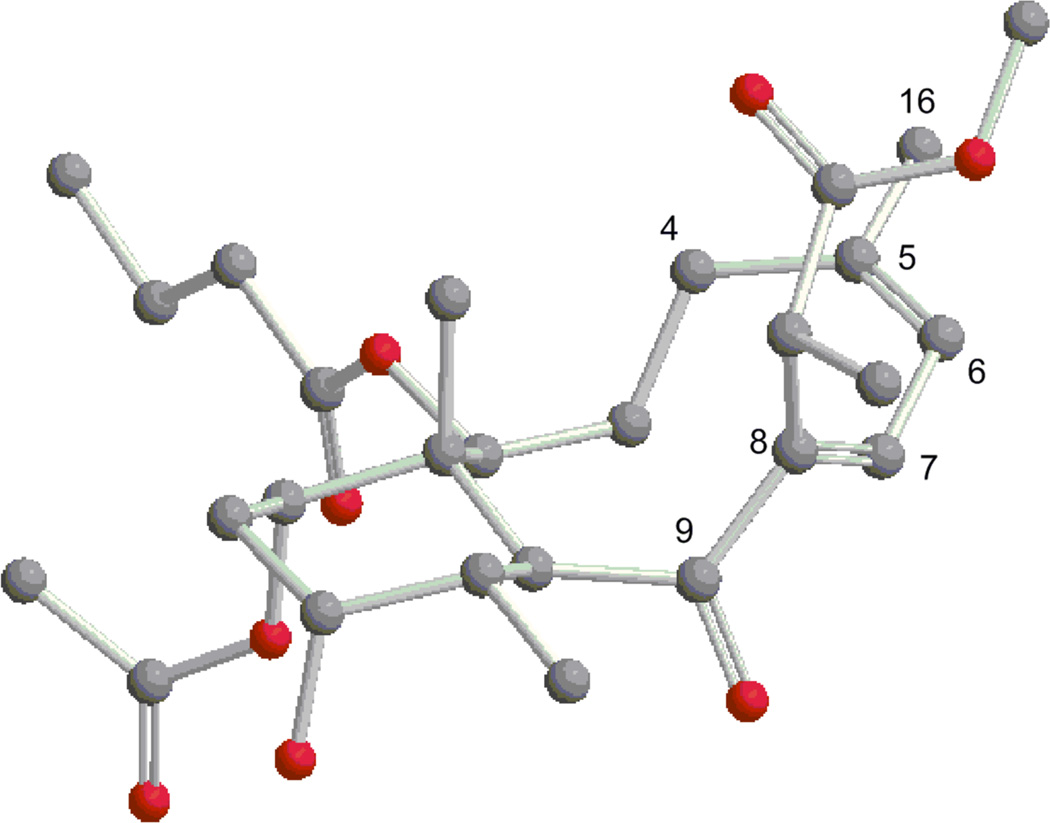
The structure of 1 was determined by a detailed analysis of the NMR data (Figure 1). The HSQC experiment allowed the assignment of all the protons to the corresponding carbon atoms. The 1H–1H COSY and HMBC experiments allowed the gross structure of 1 to be determined and revealed that 1 had the same planar structure as the C-19 methyl ester briarane diterpenoid briareolate ester G (4). However, the signals of 1, at δH 6.21 (br s) and δH 7.68 (br s) appeared more downfield when compared with the analogous signals of briareolate ester G 4 [H-6 (δH 6.07) and H-7 (δH 6.62)]4 suggesting a change in the geometry of the double bonds.
Figure 1.
Selected 2D NMR correlations for 1.
From the 2D ROESY data, it was found that H-7 exhibited NOE correlations with H-2 and H-10 and placed H-7 on the inside of the 10-membered ring and allowed the geometry of the C-7–C-8 double bond to be assigned as E. In addition, NOE correlations observed from H-6 to H-17, and H3-16 established the geometry of the C-5–C-6 double bond as Z. The relative configuration of C-1, C-2, C-10, C-11, C-12, and C-14 was determined from coupling patterns in the 1H NMR spectrum and NOE correlations observed in a ROESY spectrum (see Figure S1, Supporting Information). The configuration at C-17 could not be definitely determined due to limited NOE data, however a correlation from H-6 to H-17 is consistent with the assignment of the Me-18 being -orientated as observed in all previously reported briareolate esters.2,3,4
The absolute configuration of 1 was assigned by analogy to the known co-isolated compounds briareolate esters G (4), and B (5), whose structure were assigned on the basis X-ray crystallography of compounds in the series, that were found to have identical NMR spectral data and comparable optical rotation values ([α]25D -112, lit[α]D -122 for 4; and [α]25D +147 lit[α]D +150 for 5).3,4 Thus the absolute configuration of briareolate ester L (1) is therefore defined as 1S,2S,5Z,7E,10S,11S,12R,14S,17R.
Briareolate ester M (2) was isolated as a colorless oil. The HRESIMS of briareolate ester M (2) showed an [M + Na]+ ion at m/z 613.3350, corresponding to the molecular formula C33H50O9, 98 mass units higher than that of 1. The 1H NMR spectrum of 2 was similar to that of briareolate ester L (1), except that H-12 [δH 4.82, br q (3.0)] was shifted downfield by 1.05 ppm (Table 1) as compared with that of 1. In the 13C NMR spectrum, the resonance of C-12 (δC 73.0) was shifted downfield by 2.5 ppm and those of C-11 (δC 33.2) and C-13 (δC 29.1) were shifted upfield by 2.7 and 3.6 ppm, respectively, in comparison with those of 1. This suggested that the 12-hydroxy group of 1, was replaced by a hexanoate group at C-12 in 2. The presence of the hexanoate group was confirmed by the NMR data [δH 0.89 (3H, t, J = 7.0 Hz), ca. 1.33 (4H, overlapped), 1.62 (2H, overlapped), 2.28 (2H, m), δC 15.8 (q), 22.4, 24.9, 31.3, and 34.7 (each t), 173.1 (CO)].
Table 1.
1H NMR Data (500 MHz) for Briareolate Esters 1–3
| position | 1 a (J in Hz) | 2 b (J in Hz) | 3 b (J in Hz) |
|---|---|---|---|
| 2 | 5.50, br d (8.0)c | 5.51, br d (7.5) | 4.98, d (9.5) |
| 3α | 2.48, dd (16.5, 11.0) | 2.48, dd (16.0, 11.0) | 2.13, m |
| 3β | 1.26, m | 1.26, m | |
| 4α | 2.29, m | 2.35, m | 2.22, m |
| 4β | 2.17, m | 2.22, m | 1.38, m |
| 6 | 6.21, br s | 6.13, br s | 5.62, d (9.0) |
| 7 | 7.68, br s | 7.64, br s | 5.96, d (9.5) |
| 10 | 3.80, d (11.5) | 3.70, d (11.0) | 3.68, d (12.0) |
| 11 | 2.28, m | 2.15, m | 2.26, m |
| 12 | 3.77, br q (3.0) | 4.82, br q (3.0) | 5.13, m |
| 13α | 2.07, dt (16.0, 3.0) | 2.18, dt (13.0, 3.0) | 2.10, m |
| 13β | 1.89, dt (16.0, 3.0) | 1.87, dt (13.0, 3.0) | 2.10, m |
| 14 | 4.81, br s | 5.08, br s | 4.92, t (3.0) |
| 15 | 0.87, s | 0.87, s | 1.42, m |
| 16 | 1.73, s | 1.75, s | 1.38, m |
| 17 | 3.52, q (6.0) | 3.51, q (6.5) | 3.76, q (7.0) |
| 18 | 1.24, d (7.0) | 1.28, d (7.0) | 1.36, d (7.0) |
| 20 | 0.89, d (7.0) | 0.78, d (7.0) | 0.89, d (7.0) |
| OCH3 | 3.60, s | 3.69, s | 3.87, s |
| C-2 ester | 2.20, m | 2.22, m | 2.20, m |
| 1.60, sextet (8.0) | 1.64, m | 1.61, m | |
| 0.93, t (7.0) | 0.93, t (7.0) | 0.96, t (7.0) | |
| C-12 ester | 2.28, m | 2.32, m | |
| 1.62, m | 1.69, m | ||
| 1.33, m | 1.34, m | ||
| 1.33, m | 1.34, m | ||
| 0.89, t (7.0) | 0.93, t (7.0) | ||
| C-14 ester | 2.00, s | 2.04, s | 1.98, s |
Measured in CD3OD,
Measured in CDCl3.
The relative configuration of briareolate ester M (2) was assumed to be the same as that of 1 due to the similarity of proton–proton coupling constants and 1H and 13C chemical shifts. Thus, briareolate ester M was assigned as 2.
Briareolate ester N (3) was isolated as a colorless oil. The molecular formula of briareolate ester N (3), C33H50O9, that was determined from the HRESIMS of the [M + Na]+ ion at m/z 613.3350 and required 9 degrees of unsaturation. Initial analysis of the NMR suggested that briareolate ester N (3) had the same carbon skeleton as briareolate ester B (5), except that the hydroxy group at C-12 was replaced by a hexanoate group. The 1H and 13C NMR data of 3 were similar to that of 5, except that H-12 (δH 5.13, m) was shifted downfield by 1.27 ppm as compared with that of 5.2 In addition, in the 13C NMR spectrum, resonances of C-12 (δC 72.4) were shifted downfield by 1.8 ppm and those of C-11 (δC 32.3) and C-13 (δC 31.4) were shifted upfield by 1.2 and 2.2 ppm, respectively, in comparison with those of 5. An HMBC correlation observed from H-12 (δH 5.13) to the ester carbonyl carbon at δC 172.9 allowed placement of the hexanoate group at the C-12 position. The similarity of proton–proton coupling constants and 1H and 13C chemical shifts together with a ROESY spectrum of 3 showed the same relative configuration as 5 at all eight chiral centers. Thus, briareolate ester N was assigned as 3.
In addition to isolating three new briareolate esters the previously reported compound briareolate ester G (4) was isolated and identified from identical spectroscopic data (MS, IR, UV, optical rotation, 1H and 13C NMR data).4 In the original publication the geometry of the C-7–C-8 double bond was not determined due to decomposition. In the ROESY experiment of 4, the olefinic proton H-6 exhibited NOE correlations to both H-7 and the olefinic methyl H3-16 and allowed assignment of the geometry of the C-7–C-8 double bond as Z. NOE correlations from H-7 to H-17 placed H-7 on the outside of the 10-membered ring confirmed the Z configuration of the C-5–C-6 double bond. Further NOE correlations confirmed the relative configuration of the compound with that of the previously reported compound 4.
hESCs are a unique cell type isolated from the inner cell mass of pre-implantation blastocysts.5,6 The advent of defined media and controlled differentiation has enabled screening of hESCs to discover molecules that impact growth, differentiation or apoptosis in undifferentiated and differentiating populations. The cell growth inhibitory activities of compounds 1 - 6 were evaluated against hESCs (BG02) and a pancreatic cancer cell line (BxPC-3) using a RT-CES system.7 Briareolate ester L (1) showed the greatest growth inhibition against both the BG02 and BxPC-3 cells with EC50 values of 2.4 and 9.3 µM, respectively (see Figure S2, Supporting Information). Briareolate ester G (4) did not show any cytotoxic effects against the BG02 or the BxPC-3 cells at 20 µM (see Figure S3, Supporting Information). Compound 2 showed reduced cytotoxic activity against the BG02 cells with an EC50 value of 8.0 µM and only some cytostatic effects at 13.0 and 17.0 µM against the BxPC-3 cells. No inhibitory activity was found for compounds 3, 5, and 6 at 40 µM.
The screening results raised the question why briareolate G (4), which differs only in the geometry of the double bond between C-7–C-8, was not biologically active. The most significant spectral differences observed between 1 and 4 were the chemical shifts of the protons and carbons of the dienone. In particular the downfield shift of the olefinic proton H-7 at δH 6.62 (δC 138.1) in 4 to δH 7.64 (δC 145.2), the upfield shift of the carbons of the C-5-C-6 double bond [δC 144.8 (C-5), δC 123.5 (C-6) for 4; and δC 140.2 (C-5), δC 116.4 (C-6) for 1], and the upfield shift of the olefinic methyl group H3-16 from δH 2.17 (δC 27.2) in 4 to δH 1.73 (δC 26.5).
The chemical shift differences indicated that the C-5–C-6 double bond in 1 is less conjugated with the C-7–C-8 double bond compared to compound 4 and the (E,Z)-dienone is twisted out of plane due to restriction of the 10-membered macrocycle. The observation of a shorter UV absorption maxima at 284 nm for 1 compared to 288 nm for 4 is consistent with less conjugation. This suggested that 1 could be “spring-loaded” or cocked for nucleophilic attack at the conjugated and electrophilic C-7 position in order to relieve ring strain energy in the macrocycle. To the best of our knowledge, this is the first example of a 10-membered macrocyclic ring containing an (E,Z)-dieneone and could be a possible explanation for the difference in biological activity of compounds 1 and 4.
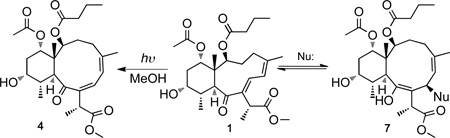
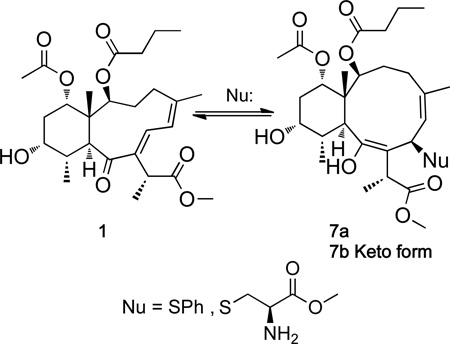
To explore the “spring-loaded” Michael acceptor hypothesis solutions of 1 and 4 were reacted with a series of model nucleophiles (eq 1). Briareolate ester L (1) was found to react with the sulfur-based nucleophile thiophenol at room temperature at 23°C in methanol-d4 to afford the enol 1,4 addition product (7a) quickly (< 5 min), quantitatively (1H NMR analysis; see Figure S4 Supporting Information) and stereoselectively (1D NOESY; see Figure S5, Supporting Information). In addition 1 was found to react with the amino acid derivative cysteine methyl ester in the presence of DMAP over night to afford the keto 1,4 addition product (7b, Nu = L-Cys-OMe; see Figure S6, Supporting Information).
Although the 1,4 addition products were stable enough to obtain full NMR datasets the adducts formed were found to be labile towards purification on HPLC (C18 and PRP-1) and could not be purified chromatographically without inducing reversal (7 → 1). All attempted addition of oxygen- and nitrogen-based nucleophiles (e.g. phenol, aniline, N-Boc L-tyrosine methyl ester, N-Boc L-serine methyl ester, L-histidine methyl ester HCl, and N-benzoyl-L-arginine ethyl ester) with and without DMAP failed to produce any detectable 1,4 addition products in reactions with 1. Briareolate ester G (4) did not react with any of the nucleophiles tested and indicated that the twisted or “spring-loaded” (E,Z)-dienone is required for nucleophilic addition reactions to occur.
To better understand the reactivity of the compounds, the optimized molecular structures and energies of 1 and 4 were calculated using density functional theory in Gaussian 098 employing the B3LYP functional and 6-31G(d) basis set (Figure 2 for 1; and see Figure S7 for 4). The (Z,Z)-dieneone isomer 4 was found to be more stable than the (E,Z)-dieneone isomer 1 with calculated total energies of -1654.75577047 and -1654.69805219 Ha, respectively. This corresponds to an energy difference of 36.22 kcal/mol.
Figure 2.
Optimized molecular structure of briareolate L (1).
The energies of the two isomers were also calculated in methanol and water solvents at the same level of theory using the CPCM solvent model. In both cases the (Z,Z)-dieneone 4 was found to be the more stable isomer with energy differences of 29.75 and 29.58 kcal/mol in methanol and water, respectively. Although solutions of 1 in benzene or methanol were found to be thermally stable (> 60°C), direct irradiation (200 W Hg lamp) for five days led to complete photoisomerization (see Figure S8, Supporting Information) to give 4 that had identical NMR and optical rotation data and is in agreement with the calculated stability of the (Z,Z)-dieneone.
In conclusion we have isolated three new C-19 methyl ester briarane diterpenoids. Briareolate ester L (1) and M (2) are the first natural products possessing a 10-membered macrocyclic ring with an (E,Z)-dieneone and exhibit growth inhibition activity against both BG02 and BxPC-3 cell lines. We found that the α,β,γ,δ-unsaturated ketone in conjunction with the double bond configuration is required for biological activity. In addition, we showed that 1 contains a “spring-loaded” Michael acceptor that can form a reversible covalent bond to model sulfur-based nucleophiles and provides a possible mechanism of bioactivity of 1 and 2.
Recently there has been a renewed interest in Michael acceptors in particular compounds that can undergo reversible reactions with thiols such bardoxolone methyl (RTA402).9,10 Bardoxolone methyl is a synthetic homotriterpenoid that induces Nrf2 (NF-E2-related factor 2), a transcription factor of the antioxidant response, and is currently in late-stage clinical development for chronic kidney disease.11 Therefore, the identification of novel structural motifs capable of reversible thiol addition reactions such as in 1 and 2 could have utility in the drug discovery process.
Supplementary Material
Acknowledgment
We thank Prof. A. Terentis, FAU, for performing the molecular modeling studies and Prof S. Lepore, FAU, for insightful contributions. This research was supported by the National Institutes of Health Grants (P41GM079597 and P01GM085354).
Footnotes
Supporting Information Available. Experimental procedures, cytotoxic responses of 1 and 4, key NOE correlations of 1, Michael addition reactions, key NOE correlations of 7a-SPh, optimized molecular structure of 4, photoisomerization reaction, and NMR spectra for 1, 2, 3, 7a-SPh and 7b-L-Cys-OMe are available including 1H, 13C, COSY, HSQC, HMBC, and ROESY. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Berrue F, Kerr RG. Nat. Prod. Rep. 2009;26:681–710. doi: 10.1039/b821918b. [DOI] [PubMed] [Google Scholar]
- 2.Dookran R, Maharaj D, Mootoo BS, Ramsewak R, McLean S, Reynolds WF, Tinto WF. Tetrahedron. 1994;50:1983–1992. [Google Scholar]
- 3.Maharaj D, Mootoo BS, Lough AJ, McLean S, Reynolds WF, Tinto WF. Tetrahedron Lett. 1992;33:7761–7764. [Google Scholar]
- 4.Mootoo BS, Ramsewak R, Sharma R, Tinto WF, Lough AJ, McLean S, Reynolds WF, Yang JP, Yu M. Tetrahedron. 1996;52:9953–9962. [Google Scholar]
- 5.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 6.Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Nat. Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- 7.Solly K, Wang X, Xu X, Strulovici B, Zheng W. Assay Drug Dev. Technol. 2004;2:363–372. doi: 10.1089/adt.2004.2.363. [DOI] [PubMed] [Google Scholar]
- 8.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA, Jr, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ö, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ. Gaussian 09, Revision A.1. Wallingford CT: Gaussian Inc.; 2009. [Google Scholar]
- 9.Avonto C, Taglialatela-Scafati O, Pollastro F, Minassi A, Di Marzo V, De Petrocellis L, Appendino G. Angew. Chem. Int. Ed. Engl. 2011;50:467–471. doi: 10.1002/anie.201005959. [DOI] [PubMed] [Google Scholar]
- 10.Sporn MB, Liby KT, Yore MM, Fu L, Lopchuk JM, Gribble GW. J. Nat. Prod. 2011;74:537–545. doi: 10.1021/np100826q. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.