Abstract
What do epilepsy, migraine headache, deafness, episodic ataxia, periodic paralysis, malignant hyperthermia, and generalized myotonia have in common? These human neurological disorders can be caused by mutations in genes for ion channels. Many of the channel diseases are “paroxysmal disorders” whose principal symptoms occur intermittently in individuals who otherwise may be healthy and active. Some of the ion channels that cause human neurological disease are old acquaintances previously cloned and extensively studied by channel specialists. In other cases, however, disease-gene hunts have led the way to the identification of new channel genes. Progress in the study of ion channels has made it possible to analyze the effects of human neurological disease-causing channel mutations at the level of the single channel, the subcellular domain, the neuronal network, and the behaving organism.
Some of the most thrilling moments in science occur when workers from distinct disciplines converge on questions of newly recognized mutual interest. Recent publications build on one such convergence, between clinical neurology, human genetics, and ion channel studies (1–11). Clinicians and geneticists seeking the causes of neurological disorders have mapped chromosomal loci for hereditary diseases and have found at these loci both previously unknown channel genes and pathogenic mutations in known channel genes. This work has proceeded rapidly. The first ion channel disease mutations, associated with hyperkalemic periodic paralysis, were identified in 1991; now the list includes hundreds of disease alleles responsible for more than 20 nerve and muscle disorders (Table 1). In the meantime, advances in many disciplines, including electrophysiology, cell biology, genomics, neuroanatomy, and structural biology have deepened our understanding of how ion channels function at the molecular and cellular level. It is now possible to begin to understand how channel gene mutations cause particular types of neurological symptoms. Here, we provide an update on some of the progress in ion channel studies. Then, because disorders causing abnormalities in skeletal muscle contraction remain the best understood of the human ion channel diseases, we briefly will review what has been learned about the biology and treatment of these disorders. This survey allows us to introduce concepts that are important in considering some very recent discoveries linking channel mutations with disorders of the central nervous system (1–11).
Table 1.
Nerve and muscle diseases caused by mutations in ion channel genes
| Disorder | Channel gene | Ref. |
|---|---|---|
| Neurons | ||
| Episodic ataxia with myokymia (EA-1) | KCNA1 | (12) |
| Episodic ataxia (EA-2) | CACNL1A4 | (13) |
| Hemiplegic migraine | CACNL1A4 | (13) |
| Spinocerebellar ataxis (SCA6) | CACNL1A4 | (14, 15) |
| Benign neonatal familial convulsions (20) | KCNQ2 | (2, 3) |
| Benign neonatal familial convulsions (8) | KCNQ3 | (1) |
| Generalized epilepsy with febrile seizures plus | SCN1B | (5) |
| A.D. nocturnal frontal lobe epilepsy | CHRNA4 | (16) |
| Startle disease (hyperekplexia) | GLRA1 | (17) |
| Jervell and Lange-Neilson syndrome | KCNE1, KCNQ1 | (18–20) |
| Autosomal dominant deafness (DFNA 2) | KCNQ4 | (8) |
| Skeletal muscle | ||
| Hyperkalemic periodic paralysis | SCN4A | (21, 22) |
| Paramyotonia congenita | SCN4A | (23) |
| Potassium-activated myotonia | SCN4A | (24) |
| Hypokalemic periodic paralysis | CACNL1A3 | (25) |
| A.R. (Becker’s) myotonia | CLCN1 | (26) |
| A.D. (Thomsen’s) myotonia | CNCN1 | (27) |
| Malignant hyperthermia | RYR1 | (28) |
| Central core disease | RYR1 | (29) |
| Congenital myesthenia syndrome | CHRNA1 | (30) |
| Cardiac muscle | ||
| Long QT syndrome 1 | KCNQ1 | (31) |
| Long QT syndrome 2 | HERG | (32) |
| Long QT syndrome 3 | SCN5A | (33) |
| Long QT syndrome 5 | KCNE1 | (19) |
| Idiopathic ventricular fibrillation | SCN5A | (34) |
| Jervell and Lange-Neilson syndrome | KCNE1, KCNQ1 | (18–20) |
Ion Channels and Cellular Excitability
All cells maintain concentration gradients for inorganic ions between their interior and exterior. Typically, potassium ion is at higher concentration in cytoplasm than in extracellular fluid; sodium, chloride, and calcium ions exhibit the opposite distribution. These gradients make possible a system for electrical signaling based on the activity of ion channel proteins embedded in the cell membrane (35). Ion channels form pores that allow ions to move rapidly through cell membranes down their electrochemical gradients. Channels transport ions at rates of 1,000,000 to 100,000,000 ions per sec. This flow of ions creates electrical current on the order of 10−12 to 10−10 amperes per channel. Such currents are large enough to produce rapid changes in the membrane potential, the electrical potential difference between the cell interior and exterior. Because calcium and sodium ions are at higher concentration extracellularly than intracellularly, openings of calcium and sodium channels cause these cations to enter the cell and depolarize the membrane potential. For analogous reasons, when potassium leaves or chloride enters the cell through open channels, the cell interior becomes more negative, or hyperpolarized.
Most ion channels are gated—capable of making transitions between conducting and nonconducting conformations. Channel gating can be induced by extracellular ligands, intracellular second messengers and metabolites, protein–protein interactions, phosphorylation, and other factors. In addition, many ion channels are gated by another regulatory signal—the membrane potential itself. Voltage-gated ion channels respond to and modify the changes in membrane potential produced by the binding of neurotransmitters to ligand-gated ion channels at synapses.
Ion channels colocalized in discrete subcellular compartments function together as signaling elements in excitable cells. These elements amplify weak signals, determine thresholds, propagate signals to other regions of the cell, and generate membrane potential oscillations, among other functions. Physiologists study the channels of tiny neuronal subcellular regions, such as dendrites, axons, and presynaptic terminals, by using refined methods that combine the patch-clamp with sophisticated imaging techniques (36). Such experiments show, for example, that the membranes of dendrites contain unexpectedly high numbers of voltage-gated channels. As a result, signal processing in dendrites now seems to be more dynamic and complex than previously thought (37–39). Potassium channels located in patches along the lengths of central axons may allow differential control of signal propagation into particular axonal branches (40). Presynaptic terminals possess a variety of voltage-gated channels and G-protein linked receptors regulating the last steps leading to neurotransmitter release (41, 42). Anatomists have found that the channels underlying these signaling functions are localized with extraordinary precision. It seems that axons, dendrites, presynaptic terminals, and postsynaptic spines all contain a rather large number of distinct microdomains (e.g., subsynaptic, perisynaptic, nonsynaptic) and that the localization of channels and other signaling proteins respects these borders (43–46). Protein–protein interactions between channels, multivalent scaffolding proteins, and the cytoskeleton appear to control the spatial organization of channels in these microdomains (47).
Molecular cloning has revealed a large number of channel genes. This expansion began early in evolution. The genome of the nematode Caenorhabditis elegans contains about 80 potassium channel genes, 90 ligand-gated channel genes, five voltage-gated calcium channel genes, six cyclic nucleotide-gated channel genes, and six chloride channel genes (48). These statistics do not include the many channel auxiliary subunits, that, though incapable of forming pores themselves, nonetheless make important contributions to channel function (49, 50). Homologues of a large portion of the worm channel genes already have been found in mammals.
Why are there so many different channel genes? We suspect that the large number of channels reflects a diversity of specific signaling needs. This point is illustrated by recent studies of the neurons in the inner ear. Sound vibrations cause specialized sensory neurons in the cochlea (called hair cells) to depolarize repetitively and release their neurotransmitter. Hair cells that respond optimally to different sound frequencies are arranged sequentially along the length of the cochlea, like piano keys. The optimal frequency of each hair cell depends largely on the kinetics of a calcium-activated potassium channel, Slo, that causes hyperpolarization and counters the depolarization caused by the opening of voltage-gated calcium channels (51). Alternative exon usage in Slo genes allows expression of a large number of distinct Slo channels, with differing gating kinetics—these channels are differentially expressed along the length of the cochlea, resulting in cells that oscillate at varying frequencies (52–54). The rules governing signaling between and within neurons in the central nervous system, though poorly understood, are likely to be as complex as within the cochlea, and also may rely on differential expression and localization of channels with distinctive properties. Analysis of mutations is an important approach for identifying these rules.
Ion Channel Disorders of the Motor Nerve and Skeletal Muscle
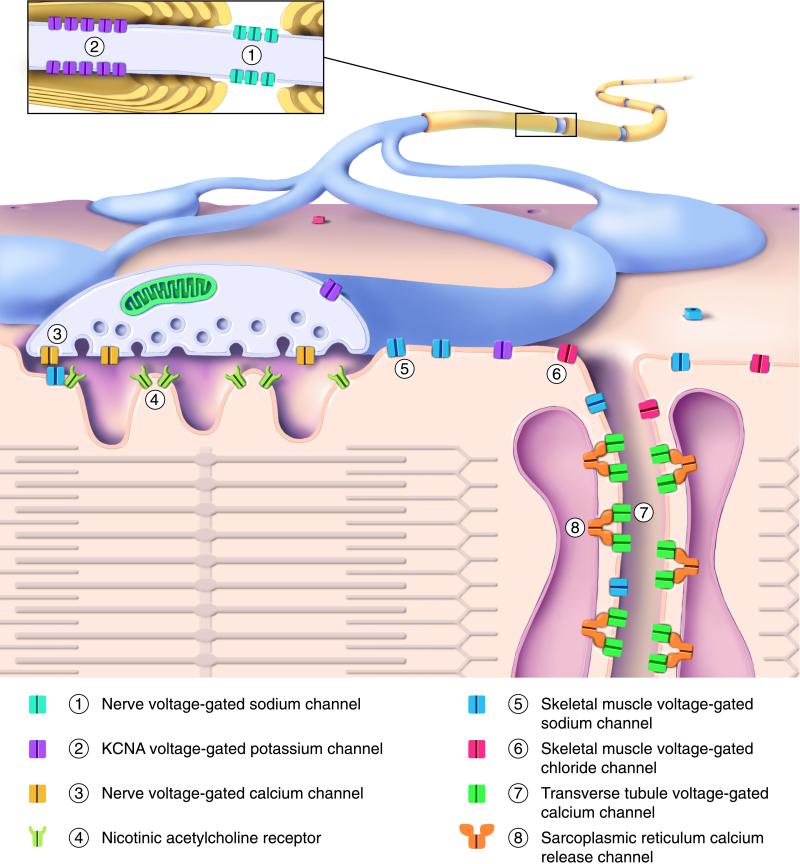
The importance of the specific properties of individual channel types for cellular excitability and behavior is best understood for the neuromuscular synapse that transmits signals from the motor nerve to the skeletal muscle. The axons of spinal motor neurons exit the spinal column and make their way to the muscle, where they form contacts with individual muscle fibers. Ion channels play crucial roles in the passage of signals along the nerve and from the muscle surface to the contractile machinery within (Fig. 1). The signal that travels down the axon, the action potential, results from the sequential opening of voltage-gated sodium channels (that allow external sodium to enter and depolarize the membrane potential) and potassium channels (that allow potassium to exit the cell, causing repolarization). Arrival of the action potential at the nerve terminal activates voltage-gated calcium channels, allowing calcium to enter the terminal, causing neurotransmitter release into the synaptic cleft. The released neurotransmitter, acetylcholine, binds to nicotinic acetylcholine receptors, which are ligand-gated cation channels, causing sodium to enter and depolarize the muscle cell membrane at synaptic sites where these acetylcholine receptors are clustered. This local depolarization leads to the activation of nearby voltage-gated sodium channels, which spread action potentials across the surface of the muscle fiber and into special invaginations of the plasma membrane, the transverse tubules, where calcium channels are present at high density. T-tubule calcium channels are physically associated with the calcium release channels embedded in the closely apposed membranes of the sarcoplasmic reticulum. The invading action potential causes the T-tubule calcium channels to undergo conformational changes that are relayed to the associated sarcoplasmic reticulum calcium channels, causing them to open. Calcium ions from within the sarcoplasmic reticulum flow into the cytoplasm, leading to muscle contraction. Chloride channels located on the T-tubule and muscle surface then return the muscle membrane potential to its resting level.
Figure 1.
Ion channels of the motor nerve and skeletal muscle involved in human disease. The drawing shows a myelinated axon branching to form synaptic contacts with a muscle fiber. (Upper) The outer surfaces of the axon and muscle. (Lower) The nerve presynaptic terminal and muscle fiber in cut section. (Inset) A magnified view of the nerve fiber, cut lengthwise near a node of Ranvier. Different channel types are identified by number and color, as indicated in the key below the drawing. The contractile proteins of the sarcomere are depicted schematically within the muscle. See text for additional details.
Remarkably, mutations of most of these key channel molecules have been found to cause human neuromuscular disorders. The distinct clinical syndromes can in many cases be understood based on the specific alterations the mutations produce in channel activity [Fig. 1, see recent reviews (55–59) for additional details and references]. Reducing the activity of potassium channels in the nerve fiber delays action potential repolarization and lowers the amount of excitation needed to produce action potentials. Potassium channel mutations with these effects underlie hereditary forms of myokymia, a spontaneous, involuntary rippling movement of skeletal muscle based on abnormal spontaneous action potential generation within the peripheral nerve. A large number of different mutations in genes encoding subunits of the acetylcholine receptor cause congenital myesthenic syndromes, disorders associated with muscle weakness and fatigue. Electrophysiological study has revealed that some of these acetylcholine receptor mutations reduce the number of channels at the cell surface, whereas others alter the rate channels open and close, cause spontaneous openings in the absense of neurotransmitter, or reduce the affinity of receptors for acetylcholine.
Mutations in the pore-forming subunits of sodium channels and chloride channels cause myotonia, a muscle abnormality in which relaxation after voluntary contraction is markedly delayed. Myotonic muscles recorded in vivo exhibit abnormal spontaneous oscillations in membrane potential, called myotonic discharges. What underlies these spontaneous oscillations? In normal muscle, depolarization of the postsynaptic membrane causes brief openings of sodium channels that take place within the first few msec after membrane depolarization. As these sodium channel openings subside, chloride enters the cell through more slowly opening chloride channels, promptly returning the membrane potential to its resting level. Sodium channels harboring mutations causing myotonia exhibit an abnormal tendency to open later or more persistently after membrane depolarization. Residual sodium entry through these abnormal channels repeatedly reinitiates the cycle of membrane depolarization. Chloride channel mutations causing myotonia reduce the amount of chloride ion that can enter the cell to repolarize the membrane, leading to oscillations.
Mutations in muscle sodium and calcium channels cause forms of periodic paralysis, disorders associated with episodes of weakness lasting hours to days at a time. The sodium channel mutants exhibit abnormal, long-lasting currents. The resulting prolonged membrane depolarization causes a portion of the sodium channels to enter a desensitized or inactivated state, leading to membrane inexcitability. The molecular mechanism underlying calcium channel-associated periodic paralysis is less clear and awaits additional study.
Mutations in the sarcoplasmic calcium release channel cause malignant hyperthermia. Attacks often are provoked by general anesthesia. During the attack, the calcium release channel opens persistently, causing sustained muscle contraction, fever, and muscle injury. Mutations in T-tubule calcium channel and muscle sodium channel are less common causes of malignant hyperthermia.
Insight into the molecular basis of these disorders has allowed for a more rational approach to therapy. In forms of the myesthenic syndrome where acetylcholine receptor openings are prolonged, receptor blockers are useful; where mutations cause reductions in the opening probability of the receptors, acetylcholinesterase inhibitors that increase the lifetime of released acetylcholine and potassium channel blockers that promote transmitter release are of benefit. Sodium channel myotonias can be treated with “use-dependent” blockers of these channels that selectively inhibit the abnormal late openings of these mutant channels.
Ion Channel Disorders in the Central Nervous System
From this brief overview of channel mutations that affect signaling from motor nerve to muscle, a model system for synaptic transmission, we can appreciate how different ion channels work in a pathway to execute a command from the nervous system. Similar principles are used by neurons in the brain to process and integrate signals, although the network of neurons is far more complex. One lesson likely to be of general validity is that prolonged or excessive excitation may result from sodium channel mutations that cause recurrent channel openings, or potassium or chloride channel mutations that reduce channel activity.
We will discuss three areas where very recent progress has linked human central nervous system disorders with mutations in voltage-gated sodium and potassium channels: (i) the association of the epileptic syndrome, generalized epilepsy with febrile seizures plus, with a mutation in a neuronal sodium channel, (ii) the identification of genes causing benign neonatal familial convulsions, a generalized epilepsy, as the genes underlying an extensively studied potassium channel (the M-channel), and (iii) the characterization of possible cellular mechanisms underlying episodic ataxia with myokymia, another potassium channel disorder, by using animal models. Recent reviews (58, 60) and citations included in Table 1 provide additional discussion and references concerning other central nervous system channel diseases.
Altered Sodium Channel Function Is a Cause of Epilepsy
Epileptic seizures are behavioral attacks resulting from the overly synchronized and excessive activity of large groups of brain neurons. Symptoms vary widely, depending on the region and extent of the brain that participates in the abnormal electrical activity, but may include alterations or loss of consciousness, sustained or rhythmic muscle contraction, stereotyped gestural movements, and visual or somatosensory hallucinations. Epilepsy is present in about 1% of the general population and is more frequent during childhood (61). About 5–7% of children will have a seizure during the first 5 years of life; about half of these will be provoked by acute febrile illness. It has remained unclear why fever provokes seizures in some individuals during childhood, but not later in life. The febrile seizure trait exhibits a complex pattern of inheritance, suggesting the involvement of multiple genetic factors.
Sheaffer and Berkovic (4) recently described an extraordinary family in which numerous members experienced both febrile and nonfebrile seizures, a syndrome they termed generalized epilepsy with febrile seizures plus. In one branch of the family, consisting of 60 individuals in five generations, 23 were epileptic. The family was descended from early English settlers of a remote area of Australia. In the first years after immigration, marriage between close relatives sometimes occurred. Among the direct ancestors of this family, such consanguinous marriage occurred in three successive generations. This unusually homogeneous genetic background left later generations at risk for disorders that may result from the activity of weakly penetrant disease alleles. Mulley and colleagues (5) obtained evidence linking epilepsy in this family to a region on chromosome 19. They evaluated candidate genes previously localized to this region, including SCN1B, the β1 auxilliary subunit of the voltage-gated sodium channel (5). The SCN1B genes of epileptic individuals in the family contained a single base- pair substitution, causing a single amino acid change in the protein sequence.
Previous structural and functional studies of the sodium channel are helpful in assessing the significance of this mutation (49, 62–65). Sodium channels in mammalian brain, heart, and skeletal muscle contain pore-forming α subunits and β1 subunits associated in 1:1 stoichiometry. The β1 subunit has important effects on the gating of the channel, hastening the rates at which it opens and closes (49, 62–64). The β1 subunit is a membrane protein, with a small intracellular domain, a single transmembrane segment, and a large extracellular domain. The extracellular domain has a structure that is homologous to members of the Ig fold superfamily. Mutant β1 subunits with alanine substitutions at positions expected to disrupt the Ig fold lack the ability to modify channel gating properties (65). The mutation discovered by Mulley and colleagues (5) substitutes a tryptophan residue for a conserved cysteine that normally forms a disulfide bridge to stabilize the Ig fold domain. This mutation appears to result in loss of function—cells expressing the mutant exhibited slow channel openings and closings indistinguishable from those expressing α subunits alone (5).
As discussed above, the mutations in skeletal muscle sodium channel α subunits causing myotonia and periodic paralysis also result in the expression of channels that exhibit delayed and prolonged openings (57). Furthermore, mutations in the α subunit of the heart sodium channel gene, linked to the long QT syndrome and idiopathic ventricular fibrillation (forms of cardiac arrhythmia that cause sudden death), are associated with similar changes in channel gating (33, 34). The basis for these gating abnormalities has been revealed by single channel studies. It appears that normal sodium channels possess two distinct gating modes. The predominant mode is associated with brief openings occurring within the first few milliseconds after the membrane is depolarized. In the other, rarely occupied mode, channel openings are prolonged and may occur after many milliseconds of delay. In the sodium channel diseases so far studied, channels exhibit an increased tendency to enter the second gating mode associated with long, late openings. The sodium channel diseases, of muscle, heart, and now, brain, suggest one way in which ion channel mutations may result in episodic symptoms—-through selective changes in the characteristics of an infrequently occupied gating mode.
An Epilepsy Gene Hunt Leads to Genes Encoding the M-Channel
As the year 1998 drew to its end, two independent, decades-long searches converged: one by geneticists seeking the cause of another remarkable epileptic syndrome, and the second by channel specialists seeking to identify a key mechanism underlying cholinergic excitation in the central and autonomic nervous system. In the 1960s and 1970s, investigators reported several large families in which neonatal convulsions appeared to be dominantly inherited with high penetrance. This epileptic syndrome, benign neonatal familial convulsions (BNFC) (66) is an often-cited model for what neurologists term “idiopathic” epilepsy: affected individuals are epileptic, but have no other neurological deficits or other organ involvement. Recurrent, brief, generalized seizures begin on about the fourth day of life and cease after 1–3 months. Infants otherwise grow and develop normally. However, affected persons carry a 10–16% risk of developing epilepsy again later in life. When seizures occur after infancy, they are in many instances provoked by sudden emotional stress or startle (66). In 1989, Leppert and colleagues (67) found linkage of the disease gene to chromosome 20. Later, Lewis et al. (68) and Steinlein and colleagues (69) identified two families in which the BNFC syndrome was linked not to chromosome 20 but to chromosome 8.
A score of years ago, electrophysiologists recording from sympathetic ganglion neurons discovered the M-channel, a voltage-gated potassium channel that is suppressed by muscarinic acetylcholine receptor activity (70). M-channels open occasionally at the resting membrane potential and are slowly activated by membrane depolarization. Because of these slow kinetics, M-channel activation causes a delayed membrane hyperpolarization after a cell receives excitatory input. As a result, sympathetic neurons expressing large amounts of M-channels tend to fire one action potential after receiving excitatory input, then become transiently quiescent. Inhibition of the M-channel causes these neurons to become slightly depolarized and to fire multiple action potentials rhythmically after receiving excitatory inputs.
M-channels were later found in central neurons in the brain and the spinal cord (71). Besides acetylcholine, peptide transmitters such as luteinizing hormone-releasing hormone, substance P, and bradykinin also inhibit the M-channel, thereby increasing neuronal excitability (71). Although receptors that mediate the action of these transmitters are known to activate the GTP binding protein Gq/11 (72), the intracellular second messenger mobilized by the muscarinic acetylcholine receptor to suppress the M-channel has remained elusive (73, 74).
The action of acetylcholine on central neurons is of considerable interest because of its importance in cognition. Cholinergic neurons in the mammalian brain integrate inputs from subcortical regions such as the hypothalamus and midbrain and modulate neurons in the cortex, hippocampus, and other brain regions. Lesion of these cholinergic neurons impairs attention, alertness, learning, and memory; these defects can be reduced by cholinergic drugs (75). The loss of cognitive function in Alzheimer’s disease is correlated with a marked reduction of central cholinergic neurons. Acetylcholinesterase inhibitors, which increase the lifetime of acetylcholine that is released from remaining cholinergic neurons, improve the cognitive performance of patients in early stages of the disease (76). In the course of developing other pharmacological reagents as potential treatments of Alzheimer’s disease, researchers in the DuPont Merck Research Laboratories found that linopirdine both improves learning and memory in laboratory animals and enhances acetylcholine release because of its action on a potassium channel (77, 78). The most potent action of this drug in hippocampal and sympathetic neurons is the inhibition of the M-channel (79, 80).
The involvement of acetylcholine and the M-channel in controlling neuronal excitability is further underscored in an rodent model of epilepsy. Systemic administration of the muscarinic agonist pilocarpine potently causes acute status epilepticus and induces a chronic seizure phenotype reminiscent of human temporal lobe epilepsy (81). These effects are abolished in heterozygous or homozygous mutant mice lacking half or all of the forebrain m1 muscarinic acetylcholine receptors (82). Mutant mice lacking m1 receptors also fail to exhibit suppression of the M-channel when their sympathetic neurons are exposed to cholinergic agonists that activate muscarinic acetylcholine receptors (82), raising the question whether excessive muscarinic acetylcholine receptor-mediated suppression of the M-channel in central neurons might be a cause of epileptic seizures. Because of the M-channel’s key importance in controlling repetitive firing in many neurons (71), its extensive and potentially novel mechanisms of regulation (72–74), and its possible contributions to normal cognitive function, dementia, and epilepsy, the identification of genes for the M-channel has been eagerly awaited.
The searches for genes for benign neonatal familial convulsions (BNFC) and the M-channel ended with a surprise. The two BNFC genes on chromosome 20 and 8 encode highly homologous potassium channel subunits, KCNQ2 and KCNQ3 (1–3). Expression of either gene alone in Xenopus oocytes results in only small currents, but expression of the two genes together results in currents that are 10–50 times larger (7, 83). The channels resulting from KCNQ2 and KCNQ3 coexpression exhibit gating properties and linopirdine sensitivity similar to those of neuronal M-channels (6). In situ hybridization and Northern blotting experiments show that KCNQ2 and KCNQ3 mRNA are expressed in overlapping patterns in brain and sympathetic ganglia (6, 7). These data suggest that KCNQ2 and KCNQ3 coassemble in vivo to form the M-channel. Disease-causing missense mutations in KCNQ2 and KCNQ3 are associated with only modest reductions (20–30%) in current magnitude (7). This finding suggests that symptoms are not the result of toxic gain-of-function or dominant-negative activity, but rather, of a critical dependence of neuronal excitability on the absolute magnitude of KCNQ2/KCNQ3 potassium channel activity.
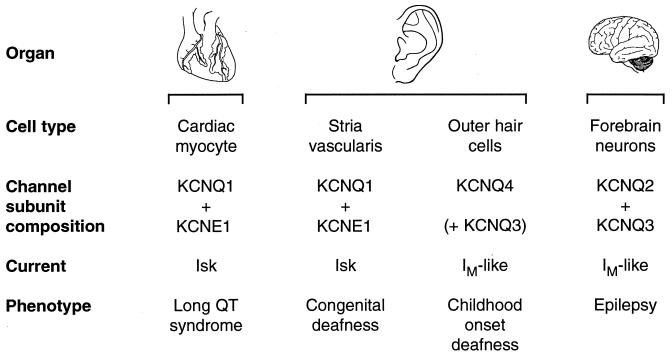
KCNQ2 and KCNQ3 are relatives of another channel, KCNQ1, which previously was shown to be mutated in long QT syndrome and in Jervell and Lange-Neilson syndrome, a recessive disorder characterized by a congenital bilateral deafness associated with the long QT syndrome (18, 31, 84). Recently, another member of the KCNQ potassium channel subfamily, KCNQ4, was identified as a cause of a dominantly inherited form of childhood-onset deafness (8). KCNQ1 and KCNQ4 both are expressed in the inner ear, but in different cell types. KCNQ1 is expressed, as a complex with a small auxiliary subunit called KCNE1, in the cells of a specialized endothelium, the stria vascularis. The stria is responsible for secretion of endolymph, the potassium-rich fluid that fills the middle chamber of the cochlea and baths the sound-receptive hair cells. In Jervell and Lange-Neilson syndrome, endolymph secretion does not occur normally, the middle chamber of the cochlea collapses, and hair cells degenerate. In contrast, KCNQ4 is expressed in outer hair cells. The outer hair cells function to increase the sensitivity of sound perception by mechanically amplifying sound vibrations within the cochlea. The specific cellular roles of KCNQ1 and KCNQ4 channels in the stria and the outer hair cells remain to be determined. It is noteworthy that KCNQ4 appears to form heterooligomeric channels when coexpressed with KCNQ3, but not with KCNQ2, KCNQ1, or KCNE1. Thus, three different heterooligomeric KCNQ channels are likely expressed in different cell types. Mutations in each of the genes contributing to these channels have been shown to cause human disease (Fig. 2).
Figure 2.
The KCNQ family of voltage-gated potassium channels. Four pore-forming subunit genes (KCNQ1–4) and one small auxiliary subunit gene (KCNE1) are known.
Episodic Ataxia Type 1 (EA-1), Shaker Flies, and Epileptic Mice
How might the identification of culprit channel genes bearing mutations lead to mechanistic understanding of the resulting neurological diseases? Recent studies related to the pathogenesis of EA-1 are instructive. EA-1 is a dominantly inherited disorder involving both the brain and peripheral nerves (85, 86). Patients experience recurrent attacks of unsteady gait and loss of limb coordination lasting minutes to hours. This phenotype suggests an intermittent derangement of cerebellar function. In some cases, transient cognitive deficits accompany the motor symptoms. Attacks may be provoked by a sudden stress or startle. Myokymia, discussed above, is continuously present in many EA-1 patients. This myokymia is unaffected by pharmacological block at the proximal portion of peripheral nerves, but is reduced by distal nerve block and abolished by inhibition of neuromuscular transmission. This observation suggests that, unlike myotonia or epilepsy, myokymia results from abnormal hyperactivity in the peripheral nerve.
In 1994, Litt and colleagues (12) linked EA-1 to missense mutations in KCNA1, a voltage-gated potassium channel gene. KCNA1 is a mammalian homologue of the fly gene Shaker. Adult Shaker flies exhibit abnormal limb shaking when anesthetized with ether. Shaker larvae showed abnormal hyperexcitability of the motor nerve terminals (87). Shaker and its mammalian homologues are among the most extensively studied of cloned ion channels (88).
Extensive further work, including physiological studies of the mutant channels, anatomical studies of KCNA channel localization in the central nervous system and peripheral nerve, and studies of kcna1 knockout mice, gives useful clues for understanding the basis of the EA-1 phenotype. By using specific antibodies, researchers found that kcna1 is localized to a key site regulating cerebellar output—the axons and nerve terminals of the basket cells that make inhibitory synaptic contacts on the proximal segment of the purkinje cell axon (44, 89). Patch-clamp recordings of basket cell terminals made in cerebellar slices revealed an extraordinarily high potassium channel density, in excess of 1,000 channels per μm2 (90). Block of these kcna channels with α-dendrotoxin causes dramatic increases in the spontaneous inhibitory postsynaptic potentials recorded in purkinje cells, suggesting that kcna regulates the excitability of the basket cell presynaptic terminals (90). Kcna channels also are concentrated at the juxtaparanodal regions of myelinated axons. In these regions that flank the nodes of Ranvier (see Fig. 1, Inset) the kcna channel appears to stabilize the resting membrane potential, possibly preventing aberrant action potential generation that may result in myokymia (89, 91, 92).
Because studies in Xenopus oocytes indicate that the mutations causing EA-1 result in reductions in KCNA1 expression and current magnitude (93, 94), knockout mice lacking the murine homologue of KCNA1 may provide a useful animal model for EA-1. Tempel and colleagues (11) have generated kcna1 knockout mice. Interestingly, mice heterozygous for the null allele appear normal; homozygous mutants have very frequent generalized epileptic seizures (11). The homozygous mutant mice also exhibit a mild loss of coordination under normal conditions and attacks of tremulousness and marked ataxia after cold-temperature stress (9). Physiological studies of these mice reveal presynaptic hyperexcitability at both the neuromuscular and cerebellar basket cell synapses (9, 10).
Prospects
Molecular studies of ion channels, begun a generation ago, have revealed a remarkable diversity of channel genes. The impending completion of genome projects in several model organisms as well as humans probably signals the end of the cloning era as previously practiced. However, the characterization of the products of the channel genes, and of the channel mutations that cause brain diseases, has in many ways only begun. As we have discussed, the channel disorders of the neuromuscular synapse illustrate how a large variety of disease phenotypes may result from mutations in channels functioning together at a single anatomical site. In the brain, a far greater variety of channels are expressed, and the roles played by specific channels are, for the most part, poorly understood. Yet, studies of the first few brain channel disorders indicate that subtle changes in the electrophysiological properties of a single channel type can have important effects on behavior.
A challenge that awaits us is to more comprehensively analyze the contributions made by a large number of the channel genes at the cellular and neuronal network level. The importance of mouse genetic models for this work is widely appreciated and will grow. It is noteworthy that studies of inbred lines of mice with epilepsy, cerebellar deficits, and other neurological abnormalities, also have resulted in the cloning of channel alleles (60, 95). Besides these spontaneous mutations, targeted mutation of several different channel genes also have been shown to cause neurological phenotypes, including epilepsy (96). As illustrated by the studies of kcna1 knockout mice already completed, these animal models make possible efforts to unravel the functions of particular channel genes through parallel studies at the molecular, cellular, neuronal network, and behavioral levels.
While neurobiologists illuminate the links between individual channel genes and behavior, structural biologists and pharmacologists will characterize in new detail the molecular contours of the different channel proteins and identify compounds that modify the activity of individual channel types with greater specificity. In 1998, this effort entered a period of accelerating progress. The structure of a bacterial potassium channel was solved by x-ray crystallography (97), and the structures of components of eukaryotic channels responsible for inactivation and deactivation gating (98, 99), subunit association (100), and ligand binding (101) also were determined.
As genomic tools become more powerful, it also may be possible to identify the important channel gene polymorphisms in human populations and to correlate particular alleles with more common, genetically complex forms of epilepsy and other traits. In the nematode C. elegans, a naturally occurring polymorphism in a G protein-coupled receptor has been shown to underlie the preference of different individuals for solitary or social living (102). A comprehensive analysis of the properties of naturally occurring human channel variants and their influence over behavior likely will deepen our understanding of nervous system function.
Acknowledgments
We thank members of the Jan laboratory, PNAS reviewers, and Drs. B. Schwappach, R. B. Layzer and J. C. Jen for helpful comments, and Wendy Hiller Gee for illustrations. E.C.C. was supported by The Parke–Davis Young Investigator Award in Epilepsy Research from the American Academy of Neurology Education and Research Foundation and by the National Institutes of Health. L.Y.J. was supported by the National Institutes of Health and is an Investigator of the Howard Hughes Medical Institute.
ABBREVIATION
- EA-1
episodic ataxia type 1
References
- 1.Charlier C, Singh N A, Ryan S G, Lewis T B, Reus B E, Leach R J, Leppert M. Nat Genet. 1998;18:53–55. doi: 10.1038/ng0198-53. [DOI] [PubMed] [Google Scholar]
- 2.Singh N A, Charlier C, Stauffer D, DuPont B R, Leach R J, Melis R, Ronen G M, Bjerre I, Quattlebaum T, Murphy J V, et al. Nat Genet. 1998;18:25–29. doi: 10.1038/ng0198-25. [DOI] [PubMed] [Google Scholar]
- 3.Biervert C, Schroeder B C, Kubisch C, Berkovic S F, Propping P, Jentsch T J, Steinlein O K. Science. 1998;279:403–406. doi: 10.1126/science.279.5349.403. [DOI] [PubMed] [Google Scholar]
- 4.Scheffer I E, Berkovic S F. Brain. 1997;120:479–490. doi: 10.1093/brain/120.3.479. [DOI] [PubMed] [Google Scholar]
- 5.Wallace R H, Wang D W, Singh R, Scheffer I E, George A L, Jr, Phillips H A, Saar K, Reis A, Johnson E W, Sutherland G R, et al. Nat Genet. 1998;19:366–370. doi: 10.1038/1252. [DOI] [PubMed] [Google Scholar]
- 6.Wang H-S, Pan Z, Shi W, Brown B S, Wymore R S, Cohen I S, Dixon J E, McKinnon D. Science. 1998;282:1890–1893. doi: 10.1126/science.282.5395.1890. [DOI] [PubMed] [Google Scholar]
- 7.Schroeder B C, Kubisch C, Stein V, Jentsch T J. Nature (London) 1998;396:687–690. doi: 10.1038/25367. [DOI] [PubMed] [Google Scholar]
- 8.Kubisch C, Schroeder B C, Friedrich T, Lutjohann B, El-Amraoui A, Marlin S, Petit C, Jentsch T J. Cell. 1999;96:437–446. doi: 10.1016/s0092-8674(00)80556-5. [DOI] [PubMed] [Google Scholar]
- 9.Zhou L, Zhang C-L, Messing A, Chiu S Y. J Neurosci. 1998;18:7200–7215. doi: 10.1523/JNEUROSCI.18-18-07200.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang, C.-L., Messing, A. & Chiu, S. Y. (1999) J. Neurosci., in press.
- 11.Smart S, Lopantsev V, Zhang C L, Robbins C A, Wang H, Chiu S Y, Schwartzkroin P A, Messing A, Tempel B. Neuron. 1998;20:809–820. doi: 10.1016/s0896-6273(00)81018-1. [DOI] [PubMed] [Google Scholar]
- 12.Browne D L, Gancher S T, Nutt J G, Brunt E R, Smith E A, Kramer P, Litt M. Nat Genet. 1994;8:136–140. doi: 10.1038/ng1094-136. [DOI] [PubMed] [Google Scholar]
- 13.Ophoff R A, Terwindt G M, Vergouwe M N, van Eijk R, Oefner P J, Hoffman S M, Lamerdin J E, Mohrenweiser H W, Bulman D E, Ferrari M, et al. Cell. 1996;87:543–552. doi: 10.1016/s0092-8674(00)81373-2. [DOI] [PubMed] [Google Scholar]
- 14.Zhuchenko O, Bailey J, Bonnen P, Ashizawa T, Stockton D W, Amos C, Dobyns W B, Subramony S H, Zoghbi H Y, Lee C C. Nat Genet. 1997;15:62–69. doi: 10.1038/ng0197-62. [DOI] [PubMed] [Google Scholar]
- 15.Yue Q, Jen J C, Thwe M M, Nelson S F, Baloh R W. Am J Med Genet. 1998;77:298–301. doi: 10.1002/(sici)1096-8628(19980526)77:4<298::aid-ajmg9>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 16.Steinlein O K, Mulley J C, Propping P, Wallace R H, Phillips H A, Sutherland G R, Scheffer I E, Berkovic S F. Nat Genet. 1995;11:201–203. doi: 10.1038/ng1095-201. [DOI] [PubMed] [Google Scholar]
- 17.Shiang R, Ryan S G, Zhu Y Z, Hahn A F, O’Connell P, Wasmuth J J. Nat Genet. 1993;5:351–358. doi: 10.1038/ng1293-351. [DOI] [PubMed] [Google Scholar]
- 18.Neyroud N, Tesson F, Denjoy I, Leibovici M, Donger C, Barhanin J, Faure S, Gary F, Coumel P, Petit C, et al. Nat Genet. 1997;15:186–189. doi: 10.1038/ng0297-186. [DOI] [PubMed] [Google Scholar]
- 19.Splawski I, Tristani-Firouzi M, Lehmann M H, Sanguinetti M C, Keating M T. Nat Genet. 1997;17:338–340. doi: 10.1038/ng1197-338. [DOI] [PubMed] [Google Scholar]
- 20.Tyson J, Tranebjaerg L, Bellman S, Wren C, Taylor J F, Bathen J, Aslaksen B, Sorland S J, Lund O, Malcolm S, et al. Hum Mol Genet. 1997;6:2179–2185. doi: 10.1093/hmg/6.12.2179. [DOI] [PubMed] [Google Scholar]
- 21.Fontaine B, Khurana T S, Hoffman E P, Bruns G A, Haines J L, Trofatter J A, Hanson M P, Rich J, McFarlane H, Yasek D M, et al. Science. 1990;250:1000–1002. doi: 10.1126/science.2173143. [DOI] [PubMed] [Google Scholar]
- 22.Ptacek L J, George A L, Jr, Griggs R C, Tawil R, Kallen R G, Barchi R L, Robertson M, Leppert M F. Cell. 1991;67:1021–1027. doi: 10.1016/0092-8674(91)90374-8. [DOI] [PubMed] [Google Scholar]
- 23.Ptacek L J, George A L, Jr, Barchi R L, Griggs R C, Riggs J E, Robertson M, Leppert M F. Neuron. 1992;8:891–897. doi: 10.1016/0896-6273(92)90203-p. [DOI] [PubMed] [Google Scholar]
- 24.McClatchey A I, Van den Bergh P, Pericak-Vance M A, Raskind W, Verellen C, McKenna-Yasek D, Rao K, Haines J L, Bird T, Brown R H, Jr, et al. Cell. 1992;68:769–774. doi: 10.1016/0092-8674(92)90151-2. [DOI] [PubMed] [Google Scholar]
- 25.Ptacek L J, Tawil R, Griggs R C, Engel A G, Layzer R B, Kwiecinski H, McManis P G, Santiago L, Moore M, Fouad G, et al. Cell. 1994;77:863–868. doi: 10.1016/0092-8674(94)90135-x. [DOI] [PubMed] [Google Scholar]
- 26.Koch M C, Steinmeyer K, Lorenz C, Ricker K, Wolf F, Otto M, Zoll B, Lehmann-Horn F, Grzeschik K H, Jentsch T J. Science. 1992;257:797–800. doi: 10.1126/science.1379744. [DOI] [PubMed] [Google Scholar]
- 27.George A L, Jr, Crackower M A, Abdalla J A, Hudson A J, Ebers G C. Nat Genet. 1993;3:305–310. doi: 10.1038/ng0493-305. [DOI] [PubMed] [Google Scholar]
- 28.Quane K A, Healy J M, Keating K E, Manning B M, Couch F J, Palmucci L M, Doriguzzi C, Fagerlund T H, Berg K, Ording H, et al. Nat Genet. 1993;5:51–55. doi: 10.1038/ng0993-51. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Chen H S, Khanna V K, De Leon S, Phillips M S, Schappert K, Britt B A, Browell A K, MacLennan D H. Nat Genet. 1993;5:46–50. doi: 10.1038/ng0993-46. [DOI] [PubMed] [Google Scholar]
- 30.Sine S M, Ohno K, Bouzat C, Auerbach A, Milone M, Pruitt J N, Engel A G. Neuron. 1995;15:229–239. doi: 10.1016/0896-6273(95)90080-2. [DOI] [PubMed] [Google Scholar]
- 31.Wang Q, Curran M E, Splawski I, Burn T C, Millholland J M, VanRaay T J, Shen J, Timothy K W, Vincent G M, de Jager T, et al. Nat Genet. 1996;12:17–23. doi: 10.1038/ng0196-17. [DOI] [PubMed] [Google Scholar]
- 32.Curran M E, Splawski I, Timothy K W, Vincent G M, Green E D, Keating M T. Cell. 1995;80:795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 33.Bennett P B, Yazawa K, Makita N, George A L., Jr Nature (London) 1995;376:683–685. doi: 10.1038/376683a0. [DOI] [PubMed] [Google Scholar]
- 34.Chen Q, Kirsch G E, Zhang D, Brugada R, Brugada J, Brugada P, Potenza D, Moya A, Borggrefe M, Breithardt G, et al. Nature (London) 1998;392:293–296. doi: 10.1038/32675. [DOI] [PubMed] [Google Scholar]
- 35.Hille B. Ionic Channels of Excitable Membranes. Sunderland, MA: Sinauer; 1992. [Google Scholar]
- 36.Sakmann B, Neher E. Single-Channel Recording. New York: Plenum; 1995. [Google Scholar]
- 37.Stuart G, Sakmann B. Nature (London) 1994;367:69–72. doi: 10.1038/367069a0. [DOI] [PubMed] [Google Scholar]
- 38.Yuste R, Tank D. Neuron. 1996;16:701–716. doi: 10.1016/s0896-6273(00)80091-4. [DOI] [PubMed] [Google Scholar]
- 39.Hoffman D A, Magee J C, Colbert C M, Johnston D. Nature (London) 1997;387:869–875. doi: 10.1038/43119. [DOI] [PubMed] [Google Scholar]
- 40.Debanne D, Guerineau N C, Gahwiler B H, Thompson S M. Nature (London) 1997;389:286–289. doi: 10.1038/38502. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi T, Forsythe I D, Tsujimoto T, Barnes-Davies M, Onodera K. Science. 1996;274:594–597. doi: 10.1126/science.274.5287.594. [DOI] [PubMed] [Google Scholar]
- 42.Wu L G, Westenbroek R E, Borst J G G, Catterall W A, Sakmann B. J Neurosci. 1999;19:726–736. doi: 10.1523/JNEUROSCI.19-02-00726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shigemoto R, Kulik A, Roberts J D, Ohishi H, Nusser Z, Kaneko T, Somogyi P. Nature (London) 1996;381:523–525. doi: 10.1038/381523a0. [DOI] [PubMed] [Google Scholar]
- 44.Laube G, Roper J, Pitt J C, Sewing S, Kistner U, Garner C C, Pongs O, Veh R W. Brain Res Mol Brain Res. 1996;42:51–61. doi: 10.1016/s0169-328x(96)00120-9. [DOI] [PubMed] [Google Scholar]
- 45.Otterson O P, Chaudhry F A, Danbolt N C, Laake J H, Nagelhus E A, Storm-Mathisen J, Torp R. Progr Brain Res. 1997;114:97–107. doi: 10.1016/s0079-6123(08)63360-9. [DOI] [PubMed] [Google Scholar]
- 46.Rubio M E, Wenthold R J. Neuron. 1997;18:939–950. doi: 10.1016/s0896-6273(00)80333-5. [DOI] [PubMed] [Google Scholar]
- 47.Sheng M, Kim E. Curr Opin Neurobiol. 1996;6:602–608. doi: 10.1016/s0959-4388(96)80091-2. [DOI] [PubMed] [Google Scholar]
- 48.Bargmann C. Science. 1998;282:2028–2033. doi: 10.1126/science.282.5396.2028. [DOI] [PubMed] [Google Scholar]
- 49.Isom L L, De Jongh K S, Patton D E, Reber B F, Offord J, Charbonneau H, Walsh K, Goldin A L, Catterall W A. Science. 1992;256:839–842. doi: 10.1126/science.1375395. [DOI] [PubMed] [Google Scholar]
- 50.Trimmer J S. Curr Opin Neurobiol. 1998;8:370–374. doi: 10.1016/s0959-4388(98)80063-9. [DOI] [PubMed] [Google Scholar]
- 51.Hudspeth A J, Lewis R S. J Physiol. 1988;400:275–297. doi: 10.1113/jphysiol.1988.sp017120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramanathan K, Michael T H, Jiang G J, Hiel H, Fuchs P A. Science. 1999;283:215–217. doi: 10.1126/science.283.5399.215. [DOI] [PubMed] [Google Scholar]
- 53.Rosenblatt K P, Sun Z P, Heller S, Hudspeth A J. Neuron. 1997;19:1061–1075. doi: 10.1016/s0896-6273(00)80397-9. [DOI] [PubMed] [Google Scholar]
- 54.Navaratnam D S, Bell T J, Tu T D, Cohen E L, Oberholtzer J C. Neuron. 1997;19:1077–1085. doi: 10.1016/s0896-6273(00)80398-0. [DOI] [PubMed] [Google Scholar]
- 55.Kubisch C, Schmidt-Rose T, Fontaine B, Bretag A H, Jentsch T J. Hum Mol Genet. 1998;7:1753–1760. doi: 10.1093/hmg/7.11.1753. [DOI] [PubMed] [Google Scholar]
- 56.Ptacek L. Am J Med. 1998;105:58–70. doi: 10.1016/s0002-9343(98)00123-5. [DOI] [PubMed] [Google Scholar]
- 57.Cannon S C. Annu Rev Neurosci. 1996;19:141–164. doi: 10.1146/annurev.ne.19.030196.001041. [DOI] [PubMed] [Google Scholar]
- 58.Greenberg D A. Ann Neurol. 1997;42:275–282. doi: 10.1002/ana.410420302. [DOI] [PubMed] [Google Scholar]
- 59.Engel A G, Ohno K, Milone M, Sine S M. Ann NY Acad Sci. 1998;841:140–156. doi: 10.1111/j.1749-6632.1998.tb10921.x. [DOI] [PubMed] [Google Scholar]
- 60.Doyle J L, Stubb L. Trends Genet. 1998;14:92–98. doi: 10.1016/s0168-9525(97)01370-x. [DOI] [PubMed] [Google Scholar]
- 61.Engel J, Pedley T A. Epilepsy: A Comprehensive Textbook. Philadelphia: Raven; 1998. [Google Scholar]
- 62.Bennett P B, Jr, Makita N, George A L., Jr FEBS Lett. 1993;326:21–24. doi: 10.1016/0014-5793(93)81752-l. [DOI] [PubMed] [Google Scholar]
- 63.Patton D E, Isom L L, Catterall W A, Goldin A L. J Biol Chem. 1994;269:17649–17655. [PubMed] [Google Scholar]
- 64.Qu Y, Isom L L, Westenbroek R E, Rogers J C, Tanada T N, McCormick K A, Scheuer T, Catterall W A. J Biol Chem. 1995;270:25696–25701. doi: 10.1074/jbc.270.43.25696. [DOI] [PubMed] [Google Scholar]
- 65.McCormick K A, Isom L L, Ragsdale D, Smith D, Scheuer T, Catterall W A. J Biol Chem. 1998;273:3954–3962. doi: 10.1074/jbc.273.7.3954. [DOI] [PubMed] [Google Scholar]
- 66.Ronen G M, Rosales T O, Connolly M, Anderson V E, Leppert M. Neurology. 1993;43:1355–1360. doi: 10.1212/wnl.43.7.1355. [DOI] [PubMed] [Google Scholar]
- 67.Leppert M, Anderson V E, Quattlebaum T, Stauffer D, O’Connell P, Nakamura Y, Lalouel J M, White R. Nature (London) 1989;337:647–648. doi: 10.1038/337647a0. [DOI] [PubMed] [Google Scholar]
- 68.Lewis T B, Leach R J, Ward K, O’Connell P, Ryan S G. Am J Hum Genet. 1993;53:670–675. [PMC free article] [PubMed] [Google Scholar]
- 69.Steinlein O, Schuster V, Fischer C, Haussler M. Hum Genet. 1995;95:411–415. doi: 10.1007/BF00208966. [DOI] [PubMed] [Google Scholar]
- 70.Brown D A, Adams P R. Nature (London) 1980;283:673–676. doi: 10.1038/283673a0. [DOI] [PubMed] [Google Scholar]
- 71.Brown D. Trends Neurosci. 1988;11:294–299. doi: 10.1016/0166-2236(88)90089-6. [DOI] [PubMed] [Google Scholar]
- 72.Caulfield M P, Jones S, Vallis Y, Buckley N J, Kim G-D, Milligan G, Brown D A. J Physiol. 1994;477:415–422. doi: 10.1113/jphysiol.1994.sp020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marrion N V. Annu Rev Physiol. 1997;59:483–504. doi: 10.1146/annurev.physiol.59.1.483. [DOI] [PubMed] [Google Scholar]
- 74.Cruzblanca H, Koh D-S, Hille B. Proc Natl Acad Sci USA. 1998;95:7151–7156. doi: 10.1073/pnas.95.12.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ridley R M, Murray T K, Johnson J A, Baker H F. Brain Res. 1986;376:108–116. doi: 10.1016/0006-8993(86)90904-2. [DOI] [PubMed] [Google Scholar]
- 76.Rogers S L, Farlow M R, Doody R S, Mohs R, Friedhoff L T. Neurology. 1998;50:136–145. doi: 10.1212/wnl.50.1.136. [DOI] [PubMed] [Google Scholar]
- 77.Cook L, Nickolson V J, Steinfels G F, Rohrbach K W, DeNoble V J. Drug Dev Res. 1990;19:301–314. [Google Scholar]
- 78.Nickolson V J, Tam S W, Myers M J, Cook L. Drug Dev Res. 1990;19:285–300. [Google Scholar]
- 79.Aiken S P, Lampe B J, Murphy P A, Brown B S. Br J Pharmacol. 1995;115:1163–1168. doi: 10.1111/j.1476-5381.1995.tb15019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lamas J A, Selyanko A A, Brown D A. Eur J Neurosci. 1997;9:605–616. doi: 10.1111/j.1460-9568.1997.tb01637.x. [DOI] [PubMed] [Google Scholar]
- 81.Cavelheiro E A. Epilepsia. 1996;37:1015–1019. doi: 10.1111/j.1528-1157.1996.tb00541.x. [DOI] [PubMed] [Google Scholar]
- 82.Hamilton S E, Loose M D, Qi M, Levey A I, Hille B, McKnight G S, Idzerda R L, Nathanson N M. Proc Natl Acad Sci USA. 1997;94:13311–13316. doi: 10.1073/pnas.94.24.13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang W P, Levesque P C, Little W A, Conder M L, Ramakrishnan P, Neubauer M G, Blanar M A. J Biol Chem. 1998;273:19419–19423. doi: 10.1074/jbc.273.31.19419. [DOI] [PubMed] [Google Scholar]
- 84.Wollnik B, Schroeder B C, Kubisch C, Esperer H D, Wieacker P, Jentsch T J. Hum Mol Genet. 1997;6:1943–1949. doi: 10.1093/hmg/6.11.1943. [DOI] [PubMed] [Google Scholar]
- 85.VanDyke D H, Griggs R C, Murphy M J, Goldstein M N. J Neurol Sci. 1975;25:109–118. doi: 10.1016/0022-510x(75)90191-4. [DOI] [PubMed] [Google Scholar]
- 86.Gancher S T, Nutt J G. Movement Disorders. 1986;1:239–253. doi: 10.1002/mds.870010404. [DOI] [PubMed] [Google Scholar]
- 87.Jan Y N, Jan L Y, Dennis M. Proc R Soc London Ser B. 1977;198:87–108. doi: 10.1098/rspb.1977.0087. [DOI] [PubMed] [Google Scholar]
- 88.Jan L Y, Jan Y N. Annu Rev Neurosci. 1997;20:91–123. doi: 10.1146/annurev.neuro.20.1.91. [DOI] [PubMed] [Google Scholar]
- 89.Wang H, Kunkel D D, Martin T M, Schwartzkroin P A, Tempel B L. Nature (London) 1993;365:75–79. doi: 10.1038/365075a0. [DOI] [PubMed] [Google Scholar]
- 90.Southan A P, Robertson B. J Neurosci. 1998;18:948–955. doi: 10.1523/JNEUROSCI.18-03-00948.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Koh D S, Vogel W. J Membr Biol. 1996;149:221–232. doi: 10.1007/s002329900022. [DOI] [PubMed] [Google Scholar]
- 92.Wang H, Allen M L, Grigg J J, Noebels J L, Tempel B L. Neuron. 1995;15:1337–1347. doi: 10.1016/0896-6273(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 93.Adelman J P, Bond C T, Pessia M, Maylie J. Neuron. 1995;15:1449–1454. doi: 10.1016/0896-6273(95)90022-5. [DOI] [PubMed] [Google Scholar]
- 94.Zerr P, Adelman J P, Maylie J. J Neurosci. 1998;18:2842–2848. doi: 10.1523/JNEUROSCI.18-08-02842.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Letts V A, Felix R, Biddlecome G H, Arikkath J, Mahaffey C L, Valenzuela A, Bartlett F S N, Mori Y, Campbell K P, Frankel W N. Nat Genet. 1998;19:340–347. doi: 10.1038/1228. [DOI] [PubMed] [Google Scholar]
- 96.Noebels J L. Neuron. 1996;16:241–244. doi: 10.1016/s0896-6273(00)80042-2. [DOI] [PubMed] [Google Scholar]
- 97.Doyle D A, Cabral J M, Pfuetzner R A, Kuo A, Gulbis J M, Cohen S L, Chait B T, MacKinnon R. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 98.Antz C, Geyer M, Fakler B, Schott M K, Guy H R, Frank R, Ruppersberg J P, Kalbitzer H R. Nature (London) 1997;385:272–275. doi: 10.1038/385272a0. [DOI] [PubMed] [Google Scholar]
- 99.Cabral J H M, Lee A, Cohen S L, Chait B T, Li M, MacKinnon R. Cell. 1998;95:649–655. doi: 10.1016/s0092-8674(00)81635-9. [DOI] [PubMed] [Google Scholar]
- 100.Kreusch A, Pfaffinger P, Stevens C F, Choe S. Nature (London) 1998;392:945–948. doi: 10.1038/31978. [DOI] [PubMed] [Google Scholar]
- 101.Armstrong N, Sun Y, Chen G Q, Gouaux E. Nature (London) 1998;395:913–917. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- 102.de Bono M, Bargmann C. Cell. 1998;94:679–689. doi: 10.1016/s0092-8674(00)81609-8. [DOI] [PubMed] [Google Scholar]