Abstract
Over the last five decades, progress in neural recording techniques has allowed the number of simultaneously recorded neurons to double approximately every 7 years, mimicking Moore’s law. Such exponential growth motivates us to ask how data analysis techniques are affected by progressively larger numbers of recorded neurons. Traditionally, neurons are analyzed independently on the basis of their tuning to stimuli or movement. Although tuning curve approaches are unaffected by growing numbers of simultaneously recorded neurons, newly developed techniques that analyze interactions between neurons become more accurate and more complex as the number of recorded neurons increases. Emerging data analysis techniques should consider both the computational costs and the potential for more accurate models associated with this exponential growth of the number of recorded neurons.
Since computers were introduced, their processing speed has grown exponentially, doubling approximately every 2 years, as formalized by Moore’s law1. This growth means that the time it takes to process a given amount of data is halved every 2 years. However, although processing speeds grow exponentially, datasets are also growing. For data processing to be feasible, it is essential that algorithms scale well with the amount of data, and scaling analysis is one of the central tools of theoretical computer science2. As neuroscience fundamentally aims at understanding the processing of huge numbers of neurons, we want to understand how recording and analysis techniques scale. Specifically, we examined how the number of simultaneously recorded neurons grows over time, what computational challenges this growth introduces and how well analysis techniques can take advantage of this growth to improve the prediction of neural activity.
Growth in the number of simultaneously recorded neurons
Since the advent of multi-electrode recordings in the 1950s, there has been tremendous growth in the number of simultaneously recorded single neurons3. With current multiple-electrode technology, signals from hundreds of individual neurons can be recorded simultaneously4,5. Using an in-depth search of the literature, we identified the studies with the highest numbers of simultaneously recorded neurons since the development of multi-electrode recording (see Supplementary Table 1 and Supplementary Methods). We found that, in good approximation, the number of recorded neurons has grown exponentially since the 1950s, doubling every 7 years (Fig. 1a). Although this growth is slower than that of computer speeds, it may have important implications for methods used to analyze neural data.
Figure 1.
Exponential growth in the number of recorded neurons. (a) Examining 56 studies published over the last five decades, we found that the number of simultaneously recorded neurons doubled approximately every 7 years. (b) A timeline of recording technologies during this period shows the development from single-electrode recordings to multi-electrode arrays and in vivo imaging techniques. Images of recording techniques reprinted from refs. 40–43 with permission of Elsevier, Springer Science + Business Media, and Am. Physiol. Soc. Image of Utah array reprinted from ref. 42, © 1999 IEEE. Ca2+ imaging reprinted from ref. 33, © 2003 Natl. Acad. Sci. USA.
Growth in the number of simultaneously recorded neurons has been driven by a number of innovations in the production, implementation and wiring of electrodes (Fig. 1b). For example, initially electrodes were made one-by-one, by hand; later, they were made by bundling hand-made wires. Recently developed silicon processing techniques allow many electrodes to be fabricated as arrays in a fully automated process3. Advances in neural recording techniques have also been facilitated by progress in computer hardware, such as data transfer speeds and storage capacity. Many innovations have jointly driven the exponential growth in neural recordings and many of today’s systems would have seemed impossible 30 years ago.
The pace of technological change is easy to underestimate. Soon after Moore’s law was formulated it was argued that computer processing speed or, more precisely, the number of components that could be placed on an integrated circuit would have to plateau in a few years6. Although there are certainly physical limits to the density of transistors that can be placed in a finite amount of space, computer speeds continue to grow rapidly. Similarly, as neuroscientists, it is difficult to imagine neural recordings doubling every 7 years. If this exponential growth were to continue, future electrophysiologists would be able to record from all of the approximately 100 billion neurons in the human brain in 220 years.
This prediction, extrapolated from the past 50 years of growth, seems absurd given today’s technology. Tissue displacement, for instance, may fundamentally limit the density with which electrodes can be implanted and bleaching and toxicity may limit the effectiveness of many optical techniques. Although experimental tools7, as well as improvements in automated spike-sorting techniques8, are beginning to lessen the need for human intervention, manual spike sorting may also be a substantial bottleneck for large-scale multi-electrode recordings. Despite these limitations, whole-brain spike recordings may not be beyond the realm of possibility. For example, one might imagine a system in which each neuron records spike times onto RNA molecules that could then by read-out by sequencing the results, one neuron at a time. Just as microchip fabrication technology has evolved drastically since the introduction of Moore’s law, progress in neural recording technology may allow growth beyond our current expectations.
Advances in neural recording and models of neural coding
Just as Moore’s law has an influence on the design of algorithms in computer science, advances in neural recording can and should influence the design of techniques for analyzing neural data. Ideally, data analysis techniques should be able to leverage larger numbers of simultaneously recorded neurons to better understand how the brain represents and processes information while avoiding the necessity for massive supercomputers. We first asked how the spike prediction accuracy of two commonly used neural data analysis methods scales with the number of simultaneously recorded neurons.
Understanding what makes neurons fire is a central question in neuroscience and being able to accurately predict neural activity is at the heart of many neural data analysis techniques9. These techniques generally ask how information about the external world is encoded in the spiking of neurons10. On the other hand, a number of applications, such as brain-machine interfaces, aim to use neural firing to predict behavior or estimate what stimuli are present in the external world. These two issues are together referred to as the neural coding problem. We want to understand how neurons encode information about the external world and we want to understand how neural signals can be decoded to provide information about the external world. In most cases, encoding and decoding models are tightly linked; leading decoding models are usually based on explicit models of encoding11–13.
We focused on models of neural encoding and two general approaches to the neural coding problem. Many methods focus on describing how neural firing relates to stimuli or the movement produced by an animal, using tuning curves or receptive fields. For example, in motor cortex, the firing of the majority of neurons appears to depend sinusoidally on the direction of the animal’s hand movement. A second class of methods focuses on describing how neurons interact and influence one another14–20 and assume that each neuron’s spiking may influence the spiking probability of other neurons. We fitted typical versions of both model classes to multi-electrode data recorded from the cortices of awake, behaving (motor task) or anesthetized (visual task) monkeys and determined how spike prediction accuracy scaled with the number of recorded neurons.
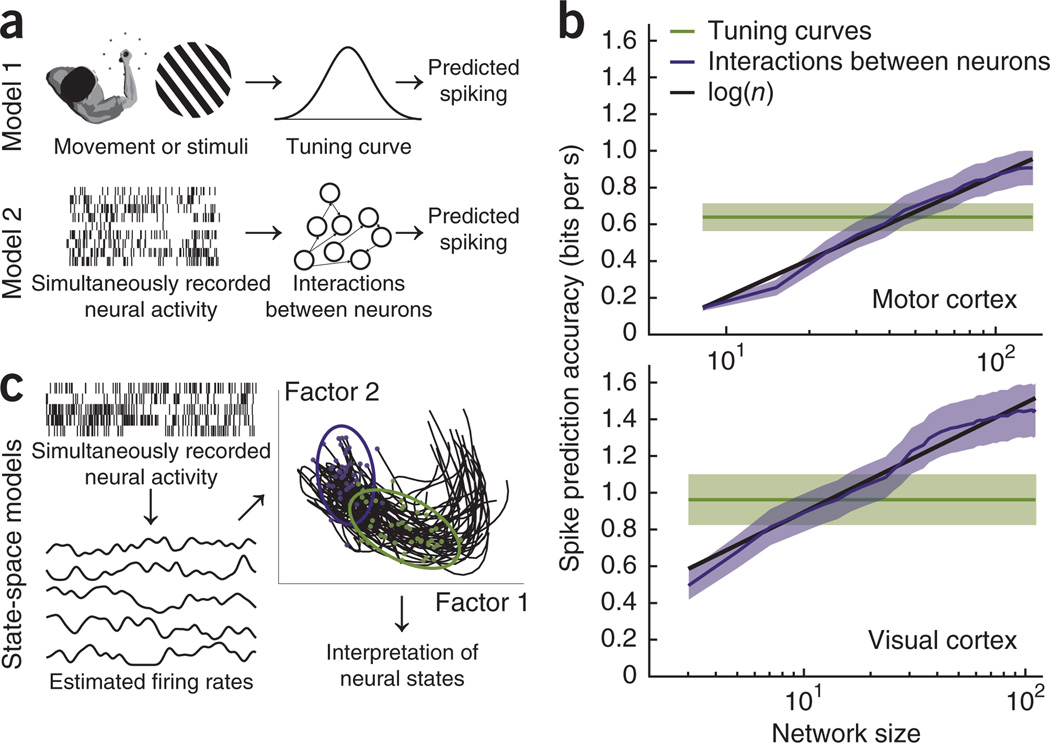
We analyzed datasets of recorded spikes using two models that both aim at predicting trial-by-trial spike counts: a tuning curve model that makes predictions based on external stimuli and a pair-wise interactions model that makes predictions based on the activity of the other simultaneously recorded neurons (Fig. 2a). In both models, we assumed that spike counts on a given trial were generated by a linear nonlinear Poisson model21, where the firing rate is determined either by a tuning curve or by coupling with the other recorded neurons. We estimated the parameters of these two models using maximum a posteriori estimation and assessed the spike prediction accuracy on trials that were not used during the estimation (Supplementary Methods). We were particularly interested in how the number of simultaneously recorded neurons affects spike prediction accuracy. For the interaction model, we varied the ‘network size’ by using a random subsample of the other recorded neurons and examined how prediction accuracy varies with the number of neurons used in the model.
Figure 2.
Approaches to neural data analysis and the scaling of spike prediction accuracy. (a) There are two main approaches to modeling multi-electrode data: mapping tuning properties to describe how neurons relate to stimuli or movement and mapping interactions between neurons. These techniques aim to predict spiking based on either external variables or other neural signals. (b) In data recorded from motor cortex (top) and visual cortex (bottom), spike prediction accuracy grows when modeling interactions between neurons, but is constant when modeling tuning curves. Shaded regions denote ± s.e.m. across neurons. (c) An alternative approach is to consider simultaneously recorded neural activity as an expression of a latent, low-dimensional state space. These spaces can be extracted by first estimating smooth firing rates for each neuron and then using a dimensionality reduction technique such as factor analysis. Features of these state spaces can then be used to predict reaction times or reach targets on a trial-by-trial basis or to describe neural variability. Purple and green ellipses represent neural variability at target onset and movement onset, respectively.
Spike data from 143 primary and pre-motor cortical neurons were recorded while a monkey performed a center-out reaching task22. In addition, spike data from 106 primary visual cortical neurons were recorded while an anesthetized monkey viewed oriented gratings23. In data from motor cortex, we considered sinusoidal tuning to the direction of hand movement, while in the data from visual cortex we considered tuning to the movement direction of an oriented grating. As the tuning curve model describes each neuron independently, spike prediction accuracy is constant as a function of the number of recorded neurons. For the interaction model, however, it is possible for spike prediction accuracy to vary as a function of the number of neurons (Fig. 2b). We found that spike prediction accuracy under the interaction model grows with the number of recorded neurons in both motor and visual cortex (as approximately log N). This result implies that accurately modeling interactions between neurons should become more important as the number of simultaneously recorded neurons continues to grow over time. Models that allow nonlinear interactions between neurons or that take into account higher-order interactions such as triplets may allow even more rapid growth in spike prediction accuracy, and developing such data analysis approaches is an essential topic for future research.
Although modeling larger numbers of neurons can certainly allow for more accurate prediction of spikes, the growth in the number of simultaneously recorded neurons is not without computational challenges. One issue in modeling these large, complex datasets is computer runtime. Modeling larger numbers of neurons requires more tuning curves (linear in the number of neurons) or more pair-wise interactions (quadratic in the number of neurons), which increases the computational complexity of these models. Models of higher-order interactions such as triplets require even more parameters. The increases in runtime associated with increases in the number of parameters are, at least partially, offset by increases in computer speed and Moore’s law, but it is still essential for the run-time of algorithms to scale well with the number of recorded neurons and recording lengths. Improving the efficiency of data analysis algorithms and developing hardware to accelerate them are currently active areas of research.
A second computational challenge is the curse of dimensionality. As the number of free parameters in a model increases, precisely estimating the parameters from a finite amount of data becomes more and more difficult. Modeling the pair-wise interactions between 100 neurons, for instance, requires ~10,000 parameters, and precisely estimating these parameters from a few hundred trials becomes difficult. Collecting more data is one solution to this problem, but we can also use modeling approaches that constrain the number of free parameters. For example, assuming that interactions between neurons are weak or rare markedly reduces the number of free parameters15,16. Alternatively, we may assume that neural activity is inherently low-dimensional and that only a few patterns of interactions exist or that interactions exist only between nearest-neighbors. Both of these techniques, regularization and dimensionality reduction, are active areas of research in machine learning. Ultimately, knowledge from anatomy and other physiological experiments can provide powerful constraints, and constraints that are tailored to neural data will be necessary to keep data analysis methods feasible in the face of growing numbers of recorded neurons. Although modeling interactions between neurons does introduce computational issues, statistical approaches to neural data analysis are being developed to address these problems.
Understanding massive neural populations
Understanding the high-dimensional datasets generated by modern recording techniques seems outstandingly complicated. After all, computational neuroscientists face the problem of condensing these massive datasets into simplifying principles about population activity. Ultimately, data from simultaneously recorded neurons promises to yield insight into the structure of the nervous system, hierarchical and modular information processing, neuronal microcircuits, as well as adaptation and learning at the network level. At the moment, many of these questions have not yet been formulated in a way that would allow data analysis to produce clear and concise answers. It may be argued that an important emerging objective of computational neuroscience is to find order in rich multi-neuron data.
Modeling the interactions, or functional connections, between neurons with multi-electrode recordings is beginning to shed light on the function and organization of the nervous system. Recent efforts modeling interactions between retinal ganglion cells, for instance, have revealed strong local neighborhood structure in addition to traditional ON/OFF receptive fields15. Similar models applied to cortical data have revealed modularity in primary motor cortex and dorsal pre-motor cortex24, as well as weak, functional interactions across cortex25. These methods have also been used to clarify the role of feedback in the thalamus26, the relationship between spikes and local field potentials27, and, on a small scale, the effects of spike timing–dependent plasticity28.
Considering interactions between neurons may offer some insight into the principles underlying neural activity, but there are also a number of recently developed methods that aim at providing simpler models of neural activity by assuming that the nervous system is inherently low dimensional29,30 (Fig. 2c). Such state-space methods allow the extraction of a small number of factors, much fewer than the number of neurons, which can be used to visualize and denoise multi-unit spike train data. Although interpreting these low-dimensional factors may present another set of challenges, these approaches have already led to insights into the activity of populations of neurons. Notably, features of the trajectories in state space can be correlated with a number of behavioral variables, such as reaction times30, and results using a state-space approach have shown that stimulus onset reduces neural variability across cortex in a wide range of areas31.
An important aspect of both state-space models and models of interactions between neurons is that they do not necessarily require modeling how individual neurons represent the external world. Although tuning curves and receptive fields have been enormously successful as models of neural encoding, they make it easy to overlook the importance of correlations between neurons and the fact that, excepting peripheral neurons, the functional properties of each neuron are caused by the inputs it receives from other neurons. By attempting to model the interactions and correlated activity of populations of simultaneously recorded neurons, state-space models and functional connectivity models may be able to shed light on how networks of neurons represent and process information.
Discussion
We observed that the number of simultaneously recorded single neurons has been growing rapidly, doubling approximately every 7 years. The trend described here predicts that in 15 years physiologists should be able to record from approximately 1,000 neurons. This seems feasible with a range of techniques. First, standard recording techniques using micro-wire arrays have been used with up to ~700 electrodes and recordings using on the order of 1,000 electrodes should appear in the near future. Second, population, two-photon calcium imaging using neuron-targeted scanning techniques have been used to record from hundreds of neurons. Advances in dyes and scanning methods, as well as statistical methods for extracting spikes from fluorescence signals promise to make this approach feasible for thousands of neurons as well32,33. Although prediction is notoriously difficult, especially about the future, it seems very likely that a 7-year doubling in the number of simultaneously recorded neurons will continue over the next couple of decades.
These advances in neural recording are an important consideration for emerging data analysis techniques. We examined how growth in the number of recorded neurons affects spike prediction accuracy in two approaches to neural encoding. The spike prediction accuracy of tuning curve models is unaffected by growth in the number of recorded neurons, as neurons are treated independently. However, in both primary visual cortex and motor cortex, modeling interactions between neurons allows spike prediction accuracy to scale with the number of recorded neurons. It is important to note that the log N scaling that we observed likely depends on a number of factors. These data are from an incomplete, highly under-sampled set of neurons in cortex. In more complete recordings, spike prediction accuracy is expected to saturate as more and more of the relevant inputs are observed18,34. Even in cortex, there is evidence to suggest that the strength of correlations between neurons depends strongly on the spatial scale23 and that very nearby neurons may be relatively independent35. In a given dataset, the spatial distribution of the recorded neurons, the strength of the interactions and the completeness of the recordings are all important considerations for our understanding of how spike prediction accuracy scales with the number of recorded neurons.
Advances in neural recording will undoubtedly affect many other areas of computational neuroscience. As the number of simultaneously recorded neurons grows, models that have traditionally only been tested using large-scale neural simulations will be able to access large, comparable datasets36,37. Models of network dynamics and population coding38,39 will be able to draw from increasingly complete neural data. However, making these links will likely require more sophisticated tools for statistical inference and data analysis.
Unlike Moore’s law, which is driven by consumer demand, advances in neural recording are ultimately driven by scientific questions. Functional connectivity methods that describe the interactions between neurons have the potential to provide increasingly accurate spike prediction as the number of simultaneously recorded neurons grows. However, understanding the activity of large populations of neurons will require even better data analysis tools and computational techniques that allow simplifying conclusions to be drawn from complex, high-dimensional data. Exponential growth in the number of simultaneously recorded neurons introduces additional computational challenges both in terms of computer run-time and the dimensionality of models. However, new models can also leverage this growth to improve prediction accuracy and better understand the representation and processing of information in populations of interacting neurons. The trends described here suggest that advances in neural recording should be a standard consideration when designing these new data analysis methods. Techniques such as regularization and dimensionality reduction that are explicitly aimed at improving scaling behavior and are tailored to neural data will be important tools for understanding growing neural datasets.
Supplementary Material
ACKNOWLEDGMENTS
Thanks to A. Kohn and members of the Kohn laboratory for providing data from visual cortex (US National Institutes of Health EY016774) and N. Hatsopoulos and J. Reimer for providing data from motor cortex. All animal use procedures were approved by the institutional animal care and use committees at Albert Einstein College of Medicine and the University of Chicago, respectively. Thanks to B. Yu and J. Cunningham for providing the GPFA code and B. Yu for insightful discussions. This work was supported by the Chicago Community Trust and US National Institutes of Health grants 1R01NS063399 and 2P01NS044393.
Footnotes
Note: Supplementary information is available on the Nature Neuroscience website.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprintsandpermissions/.
References
- 1.Moore GE. Cramming more components onto integrated circuits. Electronics. 1965;38 [Google Scholar]
- 2.Papadimitriou CH. Computational Complexity. John Wiley and Sons; 2003. [Google Scholar]
- 3.Nicolelis M. Methods for Neural Ensemble Recordings. 2nd edn. CRC Press; 2007. [PubMed] [Google Scholar]
- 4.Nicolelis M, et al. Chronic, multisite, multielectrode recordings in macaque monkeys. Proc. Natl. Acad. Sci. USA. 2003;100:11041–11046. doi: 10.1073/pnas.1934665100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelly R, et al. Comparison of recordings from microelectrode arrays and single electrodes in the visual cortex. J. Neurosci. 2007;27:261–264. doi: 10.1523/JNEUROSCI.4906-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore G. In: Understanding Moore’s Law: Four Decades of Innovation. Brock DC, editor. Chemical Heritage Foundation; 2006. Ch. 7. [Google Scholar]
- 7.Harris K, Henze D, Csicsvari J, Hirase H, Buzsaki G. Accuracy of tetrode spike separation as determined by simultaneous intracellular and extracellular measurements. J. Neurophysiol. 2000;84:401–414. doi: 10.1152/jn.2000.84.1.401. [DOI] [PubMed] [Google Scholar]
- 8.Lewicki M. A review of methods for spike sorting: the detection and classification of neural action potentials. Network. 1998;9:R53–R78. [PubMed] [Google Scholar]
- 9.Brown EN, Kass RE, Mitra PP. Multiple neural spike train data analysis: state-of-the-art and future challenges. Nat. Neurosci. 2004;7:456–461. doi: 10.1038/nn1228. [DOI] [PubMed] [Google Scholar]
- 10.Kass R, Ventura V, Brown E. Statistical issues in the analysis of neuronal data. J. Neurophysiol. 2005;94:8–25. doi: 10.1152/jn.00648.2004. [DOI] [PubMed] [Google Scholar]
- 11.Paninski L, et al. A new look at state-space models for neural data. J. Comput. Neurosci. 2009;29:1–20. doi: 10.1007/s10827-009-0179-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paninski L, Pillow J, Lewi J. Statistical models for neural encoding, decoding, and optimal stimulus design. Prog. Brain Res. 2007;165:493–507. doi: 10.1016/S0079-6123(06)65031-0. [DOI] [PubMed] [Google Scholar]
- 13.Brockwell A, Rojas A, Kass R. Recursive Bayesian decoding of motor cortical signals by particle filtering. J. Neurophysiol. 2004;91:1899–1907. doi: 10.1152/jn.00438.2003. [DOI] [PubMed] [Google Scholar]
- 14.Okatan M, Wilson MA, Brown EN. Analyzing functional connectivity using a network likelihood model of ensemble neural spiking activity. Neural Comput. 2005;17:1927–1961. doi: 10.1162/0899766054322973. [DOI] [PubMed] [Google Scholar]
- 15.Pillow JW, et al. Spatio-temporal correlations and visual signaling in a complete neuronal population. Nature. 2008;454:995–999. doi: 10.1038/nature07140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stevenson IH, Rebesco JM, Miller LE, Körding KP. Inferring functional connections between neurons. Curr. Opin. Neurobiol. 2008;18:582–588. doi: 10.1016/j.conb.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Truccolo W, Eden UT, Fellows MR, Donoghue JP, Brown EN. A point process framework for relating neural spiking activity to spiking history, neural ensemble and extrinsic covariate effects. J. Neurophysiol. 2005;93:1074–1089. doi: 10.1152/jn.00697.2004. [DOI] [PubMed] [Google Scholar]
- 18.Schneidman E, Berry MJ, II, Segev R, Bialek W. Weak pairwise correlations imply strongly correlated network states in a neural population. Nature. 2006;440:1007–1012. doi: 10.1038/nature04701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maynard E, et al. Neuronal interactions improve cortical population coding of movement direction. J. Neurosci. 1999;19:8083–8093. doi: 10.1523/JNEUROSCI.19-18-08083.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris K, Csicsvari J, Hirase H, Dragoi G, Buzsáki G. Organization of cell assemblies in the hippocampus. Nature. 2003;424:552–556. doi: 10.1038/nature01834. [DOI] [PubMed] [Google Scholar]
- 21.Paninski L. Maximum likelihood estimation of cascade point-process neural encoding models. Network. 2004;15:243–262. [PubMed] [Google Scholar]
- 22.Hatsopoulos N, Joshi J, O’Leary JG. Decoding continuous and discrete motor behaviors using motor and premotor cortical ensembles. J. Neurophysiol. 2004;92:1165–1174. doi: 10.1152/jn.01245.2003. [DOI] [PubMed] [Google Scholar]
- 23.Smith M, Kohn A. Spatial and temporal scales of neuronal correlation in primary visual cortex. J. Neurosci. 2008;28:12591–12603. doi: 10.1523/JNEUROSCI.2929-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevenson IH, et al. Bayesian inference of functional connectivity and network structure from spikes. IEEE Trans. Neural Syst. Rehabil. Eng. 2009;17:203–213. doi: 10.1109/TNSRE.2008.2010471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Truccolo W, Hochberg L, Donoghue J. Collective dynamics in human and monkey sensorimotor cortex: predicting single neuron spikes. Nat. Neurosci. 2009;13:105–111. doi: 10.1038/nn.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Babadi B, Casti A, Xiao Y, Kaplan E, Paninski L. A generalized linear model of the impact of direct and indirect inputs to the lateral geniculate nucleus. J. Vis. 2010;10:22. doi: 10.1167/10.10.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly R, Smith M, Kass R, Lee T. Local field potentials indicate network state and account for neuronal response variability. J. Comput. Neurosci. 2010;29:567–579. doi: 10.1007/s10827-009-0208-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rebesco JM, Stevenson IH, Koerding K, Solla SA, Miller LE. Rewiring neural interactions by micro-stimulation. Front. Syst. Neurosci. 2010;4:39. doi: 10.3389/fnsys.2010.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu B, et al. Gaussian-process factor analysis for low-dimensional single-trial analysis of neural population activity. J. Neurophysiol. 2009;102:614–635. doi: 10.1152/jn.90941.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Churchland M, Yu B, Sahani M, Shenoy K. Techniques for extracting singletrial activity patterns from large-scale neural recordings. Curr. Opin. Neurobiol. 2007;17:609–618. doi: 10.1016/j.conb.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Churchland M, et al. Stimulus onset quenches neural variability: a widespread cortical phenomenon. Nat. Neurosci. 2010;13:369–378. doi: 10.1038/nn.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogelstein J, et al. Spike inference from calcium imaging using sequential monte carlo methods. Biophys. J. 2009;97:636–655. doi: 10.1016/j.bpj.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stosiek C, Garaschuk O, Holthoff K, Konnerth A. In vivo two-photon calcium imaging of neuronal networks. Proc. Natl. Acad. Sci. USA. 2003;100:7319–7324. doi: 10.1073/pnas.1232232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shlens J, et al. The structure of multi-neuron firing patterns in primate retina. J. Neurosci. 2006;26:8254–8266. doi: 10.1523/JNEUROSCI.1282-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ecker A, et al. Decorrelated neuronal firing in cortical microcircuits. Science. 2010;327:584–587. doi: 10.1126/science.1179867. [DOI] [PubMed] [Google Scholar]
- 36.Vogels T, Rajan K, Abbott L. Neural network dynamics. Annu. Rev. Neurosci. 2005;28:357–376. doi: 10.1146/annurev.neuro.28.061604.135637. [DOI] [PubMed] [Google Scholar]
- 37.Brette R, et al. Simulation of networks of spiking neurons: a review of tools and strategies. J. Comput. Neurosci. 2007;23:349–398. doi: 10.1007/s10827-007-0038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Averbeck B, Latham P, Pouget A. Neural correlations, population coding and computation. Nat. Rev. Neurosci. 2006;7:358–366. doi: 10.1038/nrn1888. [DOI] [PubMed] [Google Scholar]
- 39.Pouget A, Dayan P, Zemel R. Information processing with population codes. Nat. Rev. Neurosci. 2000;1:125–132. doi: 10.1038/35039062. [DOI] [PubMed] [Google Scholar]
- 40.Barna J, Arezzo J, Vaughan H., Jr A new multielectrode array for the simultaneous recording of field potentials and unit activity. Electroencephalogr. Clin. Neurophysiol. 1981;52:494–496. doi: 10.1016/0013-4694(81)90035-3. [DOI] [PubMed] [Google Scholar]
- 41.Krüger J, Bach M. Simultaneous recording with 30 microelectrodes in monkey visual cortex. Exp. Brain Res. 1981;41:191–194. doi: 10.1007/BF00236609. [DOI] [PubMed] [Google Scholar]
- 42.Rousche P, Normann R. Chronic intracortical microstimulation (ICMS) of cat sensory cortex using the Utah Intracortical Electrode Array. IEEE Trans. Rehabil. Eng. 2002;7:56–68. doi: 10.1109/86.750552. [DOI] [PubMed] [Google Scholar]
- 43.Blanche T, Spacek M, Hetke J, Swindale N. Polytrodes: high-density silicon electrode arrays for large-scale multiunit recording. J. Neurophysiol. 2005;93:2987–3000. doi: 10.1152/jn.01023.2004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.