Abstract
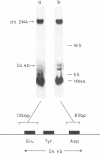
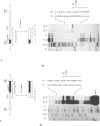
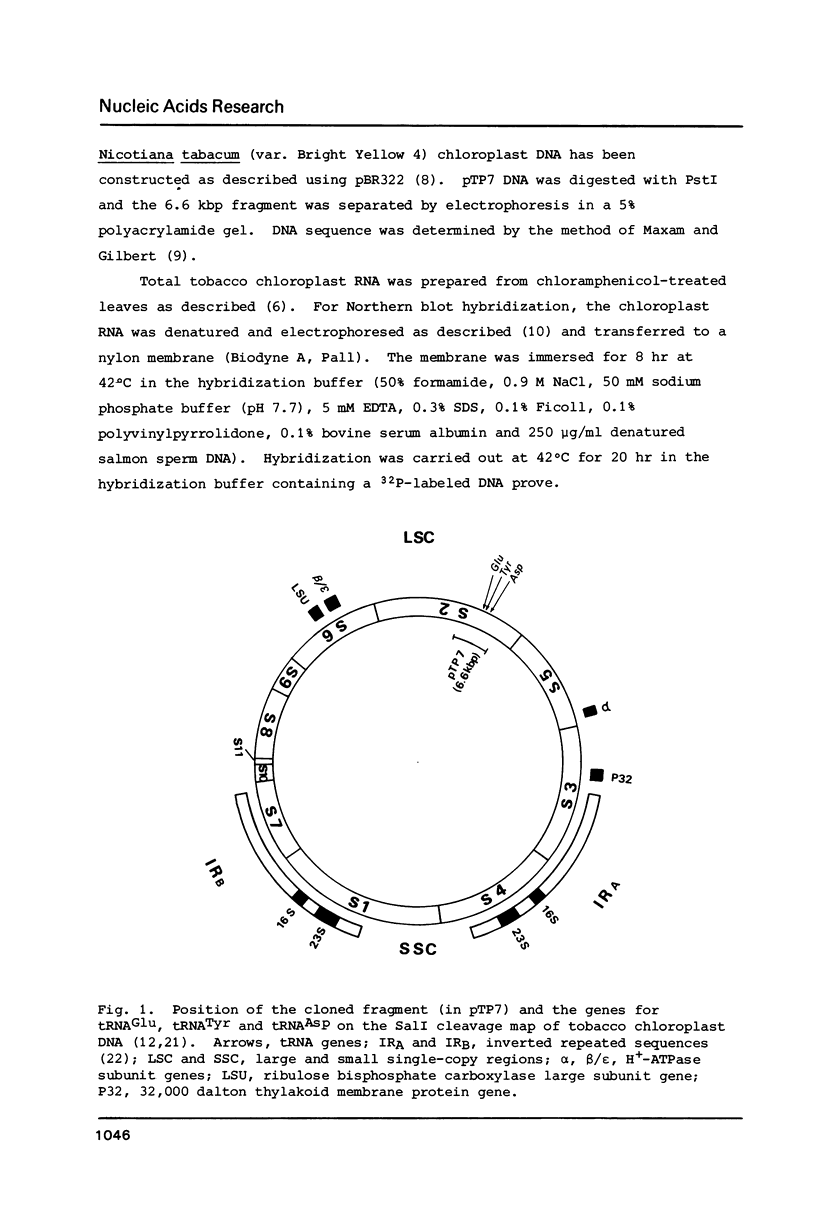
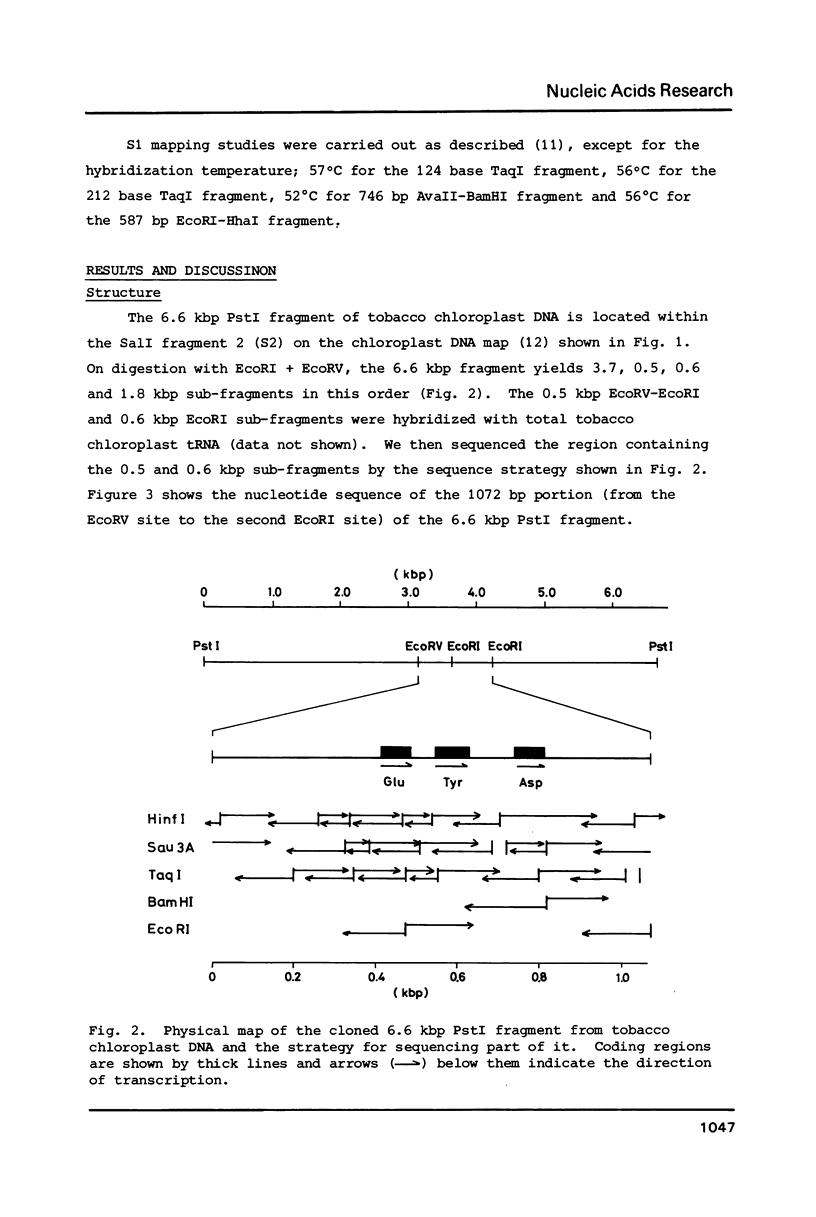
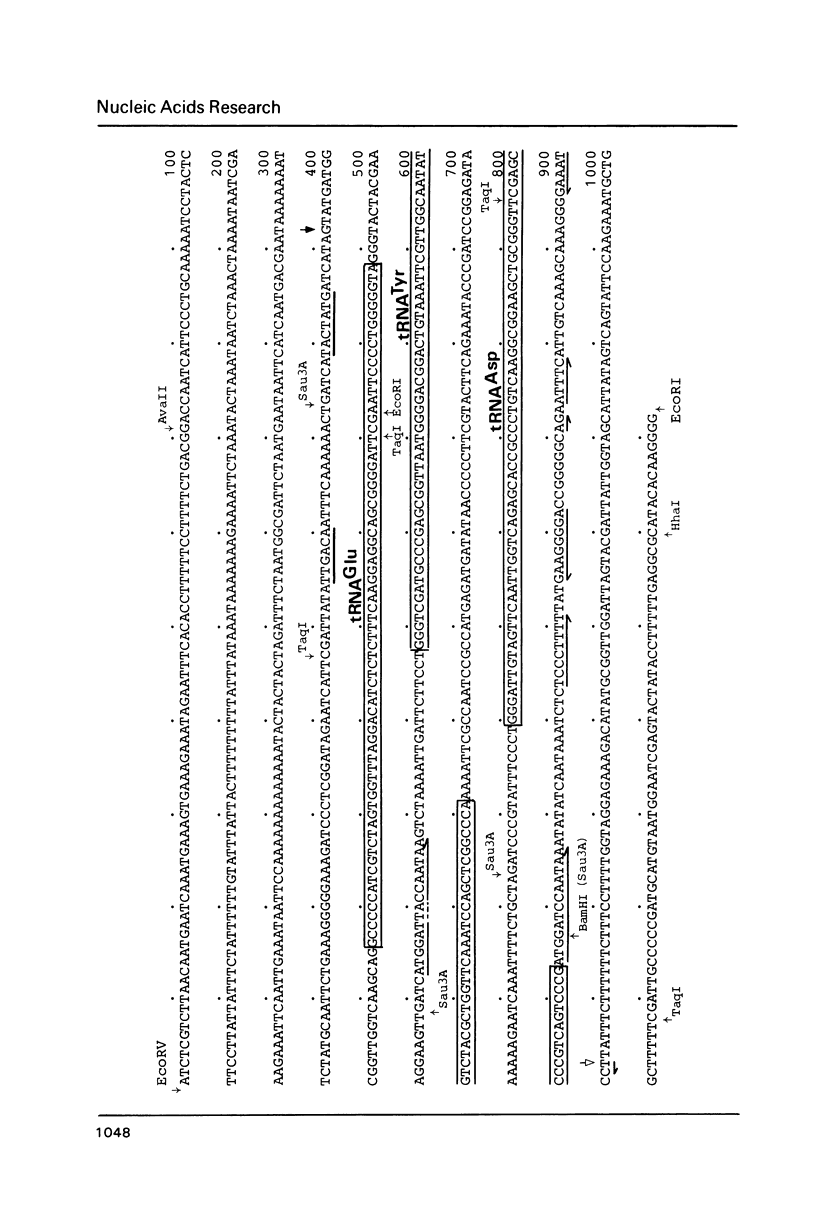
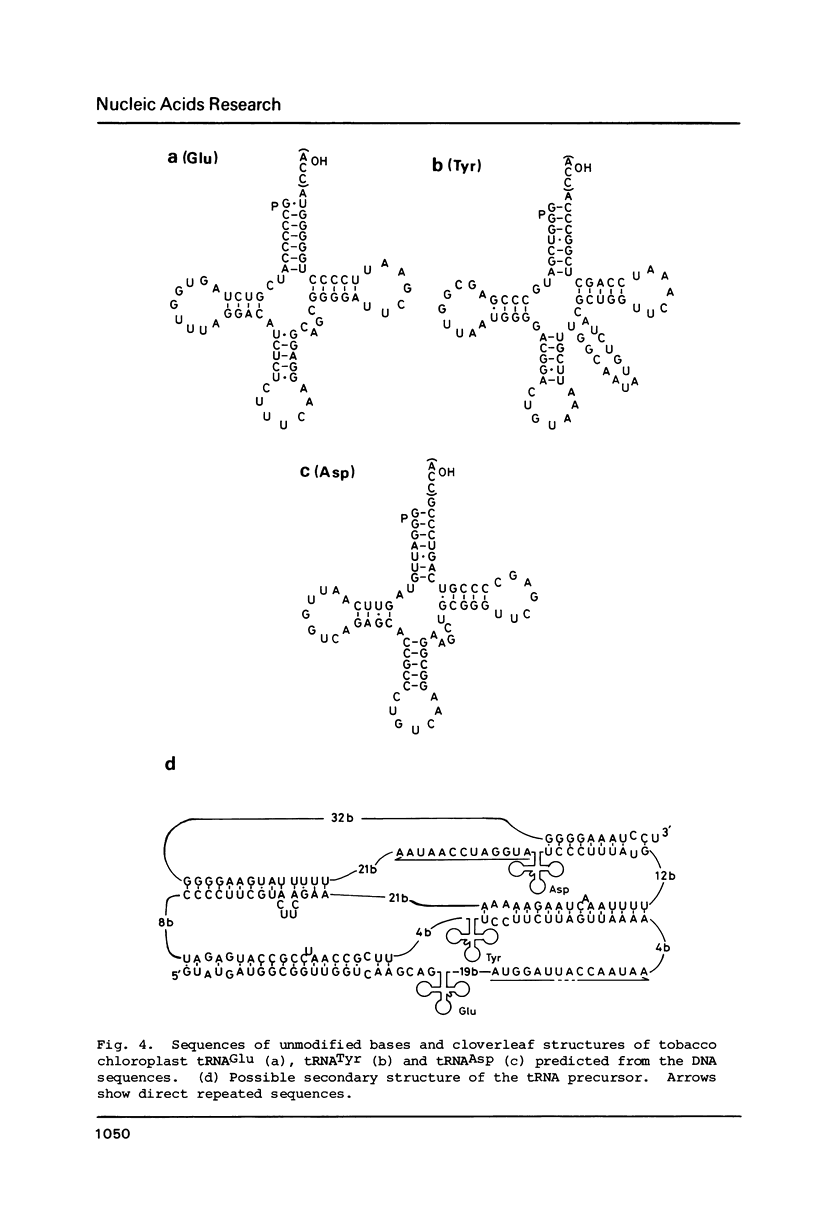
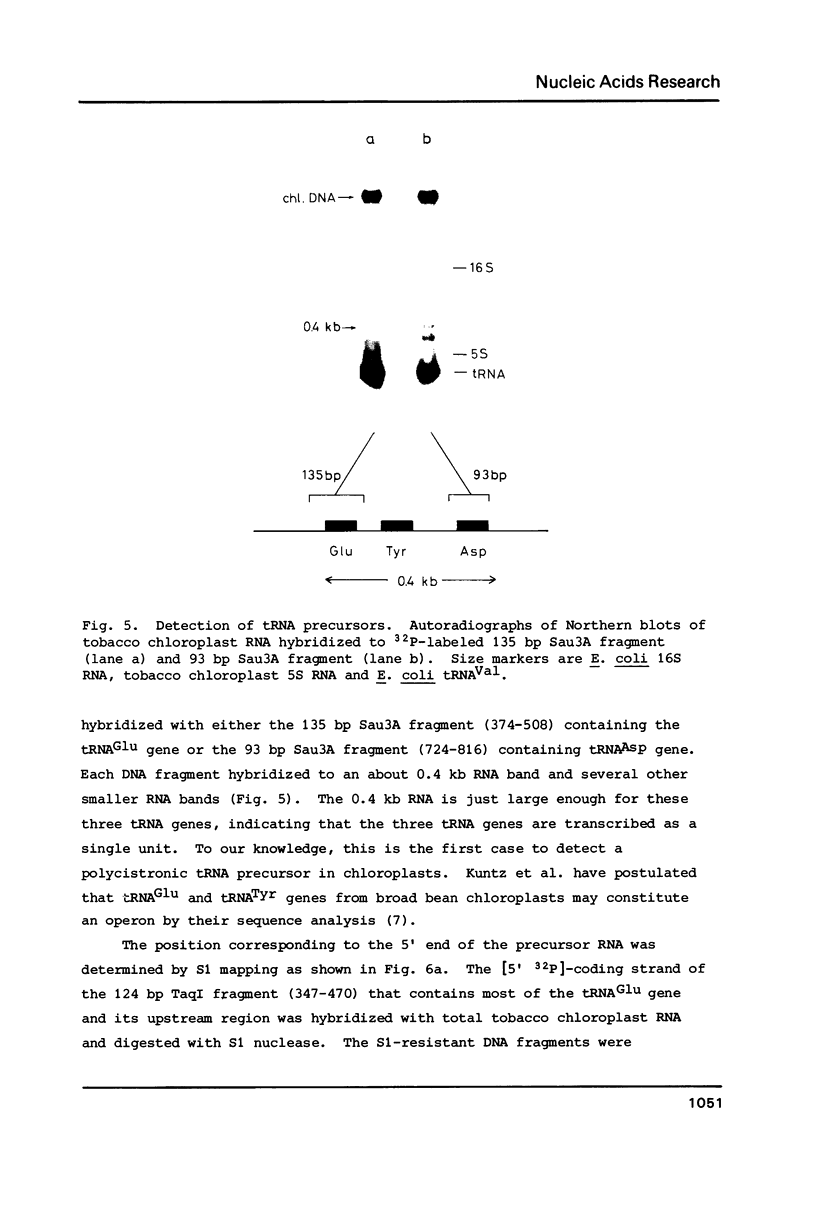
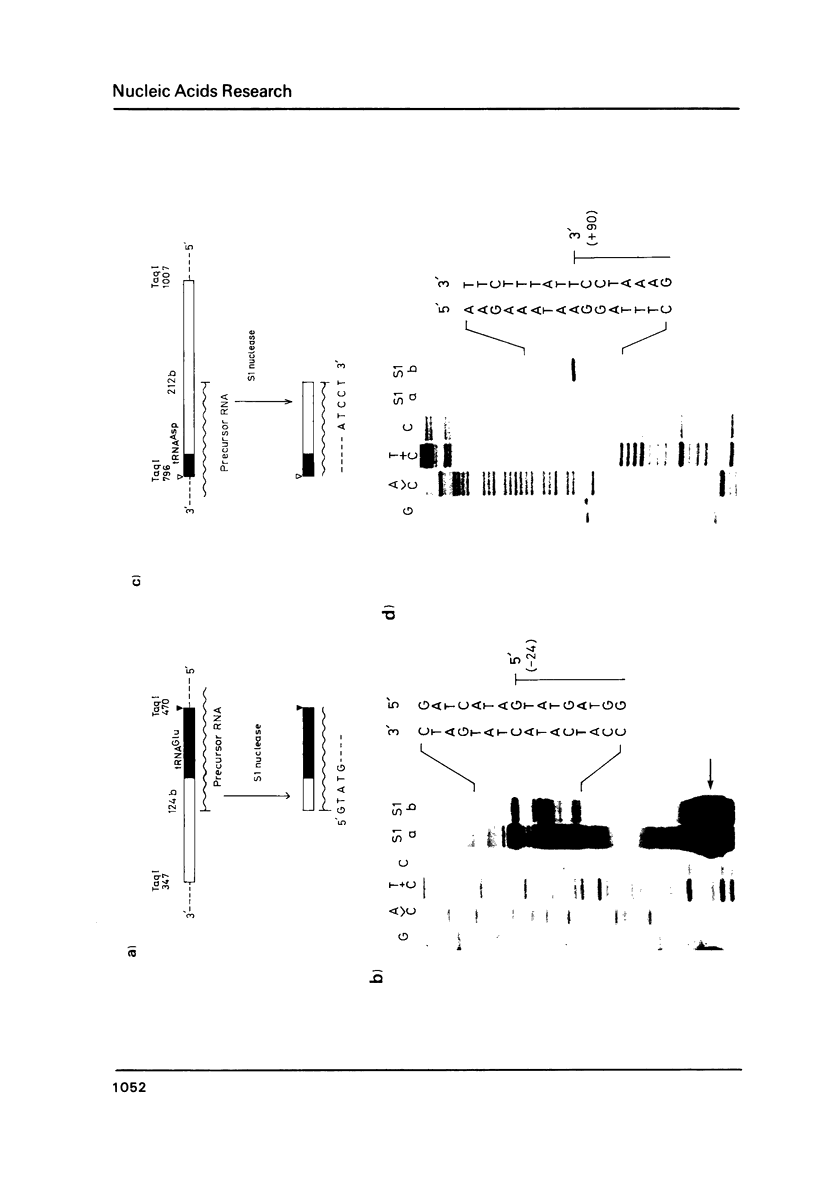
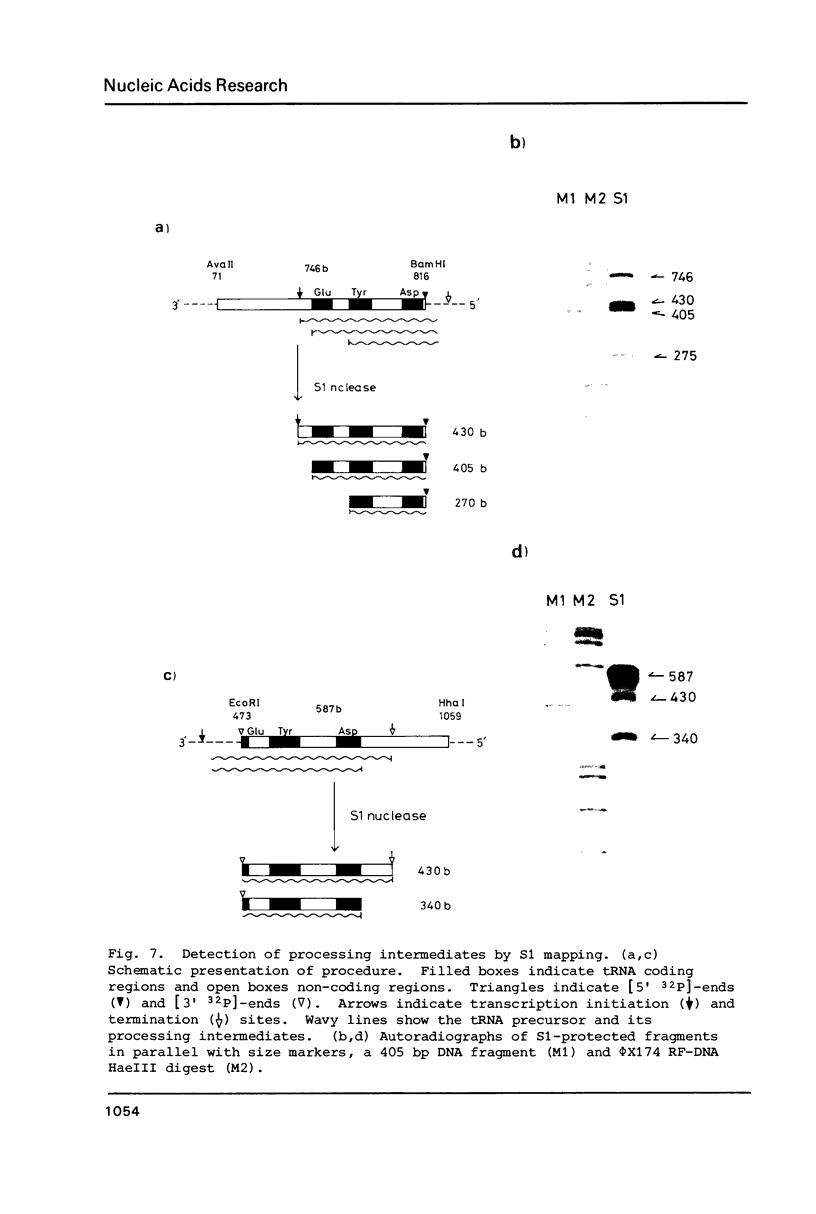
The location and nucleotide sequences of tobacco chloroplast genes for tRNAGlu(UUC), tRNATyr(GUA) and tRNAAsp(GUC) have been determined. These genes lie midway between the genes for alpha and beta/epsilon subunits of H+-ATPase on the large single-copy region of the chloroplast DNA. The gene organization is tRNAGlu - 59bp spacer - tRNATyr - 108bp spacer - tRNAAsp on the same DNA strand. Northern blot hybridization studies revealed that these three tRNA genes are cotranscribed. The transcription initiation site was localized at 24 bp upstream from the tRNAGlu coding region and its termination site at 90 bp downstream from the tRNAAsp coding region by S1 mapping. The tricistronic tRNA precursor is thus calculated to be 512 bases long. Its processing was also studied by S1 mapping.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Deno H., Kato A., Shinozaki K., Sugiura M. Nucleotide sequences of tobacco chloroplast genes for elongator tRNAMet and tRNAVal (UAC): the tRNAVal (UAC) gene contains a long intron. Nucleic Acids Res. 1982 Dec 11;10(23):7511–7520. doi: 10.1093/nar/10.23.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deno H., Shinozaki K., Sugiura M. Nucleotide sequence of tobacco chloroplast gene for the alpha subunit of proton-translocating ATPase. Nucleic Acids Res. 1983 Apr 11;11(7):2185–2191. doi: 10.1093/nar/11.7.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruissem W., Prescott D. M., Greenberg B. M., Hallick R. B. Transcription of E. coli and Euglena chloroplast tRNA gene clusters and processing of polycistronic transcripts in a HeLa cell-free system. Cell. 1982 Aug;30(1):81–92. doi: 10.1016/0092-8674(82)90014-9. [DOI] [PubMed] [Google Scholar]
- Hollingsworth M. J., Hallick R. B. Euglena gracilis chloroplast transfer RNA transcription units. Nucleotide sequence analysis of a tRNATyr-tRNAHis-tRNAMet-tRNATrp-tRNAGlu-tRNAGly gene cluster. J Biol Chem. 1982 Nov 10;257(21):12795–12799. [PubMed] [Google Scholar]
- Holschuh K., Bottomley W., Whitfeld P. R. Sequence of the genes for tRNACys and tRNAAsp from spinach chloroplasts. Nucleic Acids Res. 1983 Dec 20;11(24):8547–8554. doi: 10.1093/nar/11.24.8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch W., Edwards K., Kössel H. Sequencing of the 16S-23S spacer in a ribosomal RNA operon of Zea mays chloroplast DNA reveals two split tRNA genes. Cell. 1981 Jul;25(1):203–213. doi: 10.1016/0092-8674(81)90245-2. [DOI] [PubMed] [Google Scholar]
- Kuntz M., Weil J. H., Steinmetz A. Nucleotide sequence of a 2 kbp BamH I fragment of Vicia faba chloroplast DNA containing the genes for threonine, glutamic acid and tyrosine transfer RNAs. Nucleic Acids Res. 1984 Jun 25;12(12):5037–5047. doi: 10.1093/nar/12.12.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme M., Kamogashira T., Shinozoki K., Sugiura M. Locations and sequences of tobacco chloroplast genes for tRNAPro(UGG), tRNATrp, tRNAfMet and tRNAGly(GCC): the tRNAGly contains only two base-pairs in the D stem. Nucleic Acids Res. 1984 Sep 11;12(17):6741–6749. doi: 10.1093/nar/12.17.6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Deno H., Kato A., Sugiura M. Overlap and cotranscription of the genes for the beta and epsilon subunits of tobacco chloroplast ATPase. Gene. 1983 Oct;24(2-3):147–155. doi: 10.1016/0378-1119(83)90074-4. [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Sugiura M. The nucleotide sequence of the tobacco chloroplast gene for the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. Gene. 1982 Nov;20(1):91–102. doi: 10.1016/0378-1119(82)90090-7. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Gauss D. H. Compilation of tRNA sequences. Nucleic Acids Res. 1984;12 (Suppl):r1–57. [PMC free article] [PubMed] [Google Scholar]
- Takaiwa F., Sugiura M. Nucleotide sequence of the 16S - 23S spacer region in an rRNA gene cluster from tobacco chloroplast DNA. Nucleic Acids Res. 1982 Apr 24;10(8):2665–2676. doi: 10.1093/nar/10.8.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]