Abstract
Although the role of the 2-oxoglutarate dehydrogenase complex (2-OGDHC) has previously been demonstrated in plant heterotrophic tissues its role in photosynthetically active tissues remains poorly understood. By using a combination of metabolite and transcript profiles we here investigated the function of 2-OGDHC in leaves of Arabidopsis thaliana via use of specific phosphonate inhibitors of the enzyme. Incubation of leaf disks with the inhibitors revealed that they produced the anticipated effects on the in situ enzyme activity. In vitro experiments revealed that succinyl phosphonate (SP) and a carboxy ethyl ester of SP are slow-binding inhibitors of the 2-OGDHC. Our results indicate that the reduced respiration rates are associated with changes in the regulation of metabolic and signaling pathways leading to an imbalance in carbon-nitrogen metabolism and cell homeostasis. The inducible alteration of primary metabolism was associated with altered expression of genes belonging to networks of amino acids, plant respiration, and sugar metabolism. In addition, by using isothermal titration calorimetry we excluded the possibility that the changes in gene expression resulted from an effect on 2-oxoglutarate (2OG) binding to the carbon/ATP sensing protein PII. We also demonstrated that the 2OG degradation by the 2-oxoglutarate dehydrogenase strongly influences the distribution of intermediates of the tricarboxylic acid (TCA) cycle and the GABA shunt. Our results indicate that the TCA cycle activity is clearly working in a non-cyclic manner upon 2-OGDHC inhibition during the light period.
Keywords: 2-oxoglutarate dehydrogenase, phosphonate inhibitors, TCA cycle, PII, 2-oxoglutarate, GABA
Introduction
The metabolic intermediate 2-oxoglutarate (2OG) sits at the juncture of the tricarboxylic acid (TCA) cycle and the nitrate assimilatory pathway as well as being proposed as an important signaling molecule in several species including Azotobacter vinelandii, Escherichia coli, and cyanobacteria (Little et al., 2000; Forchhammer, 2004; Jiang and Ninfa, 2009) and plants (Templeton and Moorhead, 2004; Uhrig et al., 2009). The level of 2OG is perceived by the evolutionarily conserved PII protein which was discovered over 40 years ago in E. coli and has subsequently been found in almost every bacterial and archaeal genome examined (Templeton and Moorhead, 2004). PII was more recently discovered in plants (Hsieh et al., 1998; Smith et al., 2002; Moorhead and Smith, 2003) and is localized to the plastid. Plant PII function is highly conserved compared to its bacterial progenitor and binds ATP and 2OG in an analogous manner (Smith et al., 2003). Unlike E. coli or Synechococcus elongatus, however, the Arabidopsis thaliana PII appears to lack post-translational modification (Smith et al., 2004; Uhrig et al., 2009). Given that ATP binding constants are well below endogenous plastidial levels and binding of 2OG is contingent upon a PII-ATP complex it has previously been suggested that plant PII must be a highly sensitive carbon sensor (Uhrig et al., 2009). PII is regulated at the transcriptional level (Ferrario-Mery et al., 2005) and the intriguing observation was made that unlike in prokaryotes defined PII regulatory targets in plants are restricted merely to two proteins N-acetyl-l-glutamate kinase and heteromeric acetyl-CoA carboxylase (Feria Bourrellier et al., 2009; Baud et al., 2010; Bourrellier et al., 2010). The regulatory circuits that control nitrogen metabolism are relatively well characterized in several bacterial model systems. However, much less is understood about how both the nitrogen and carbon status of the cell is perceived in vivo as well as the sensing and signaling systems that operate in illuminated leaves (Nunes-Nesi et al., 2010). Accordingly in illuminated leaves, TCA cycle decarboxylations can be reduced by 80% and the decarboxylation reaction of the pyruvate dehydrogenase is decreased by 30% when compared to dark respiration (Tcherkez et al., 2008). It is possible that this activity is not sufficient to produce all 2OG needed for nitrogen assimilation. However, in photosynthetic tissues, the activity of a complete TCA cycle appears to be reduced and a non-cyclic TCA pathway is likely more important in illuminated leaves due to the transport of organic acids out of the mitochondria (Tcherkez et al., 2009; Sweetlove et al., 2010). Furthermore, although recent years have witnessed substantial research effort, the precise source of the 2OG necessary for glutamate and glutamine production particularly during nitrogen assimilation remains poorly understood (Bunik and Fernie, 2009). Nevertheless, compelling evidence has been provided to suggest that the operation of (at least a partial) TCA cycle in illuminated leaves is essential for the provision of 2OG (Hodges, 2002; Hodges et al., 2003; Dutilleul et al., 2005; Kolbe et al., 2006; Lemaitre et al., 2007). In addition, the nitrogen assimilatory process has been documented to influence respiration during the light period (Cousins and Bloom, 2004; Guo et al., 2005; Foyer et al., 2011). Additional evidence that perturbation of 2OG metabolism alters nitrogen metabolism have been provided by transgenic tomato (Solanum lycopersicum) lines with a minor inhibition of the cytosolic NADP-dependent isocitrate dehydrogenase which were characterized by an altered amino acid composition as well as a shift in cellular redox balance (Sulpice et al., 2010). Furthermore, we have recently demonstrated, by the inhibition of the 2-oxoglutarate dehydrogenase reaction in potato tubers via application of a chemical inhibitor, that this enzyme play an important role in nitrate assimilation as well as in amino acid metabolism (Araújo et al., 2008). Nevertheless its role in photosynthetically active tissues remains only poorly understood. Several efforts have been made in the last decade to identify and understand the components of the nitrogen signaling pathway. However, the mechanisms of nitrogen signaling of other organisms have not yet been extrapolated to plants (Vidal and Gutierrez, 2008). That said, considerable advances in our understanding of the control of mitochondrial metabolism and its interactions with nitrogen metabolism and associated carbon/nitrogen interactions have occurred in recent years, particularly highlighting important roles in cellular redox homeostasis (Foyer et al., 2011).
To reveal molecular mechanisms of the association between neuropathologies and 2-oxoglutarate dehydrogenase complex (2-OGDHC) function, Bunik et al. (2005) introduced specific inhibitors of 2-OGDHC, which have been successfully applied in recent years for cellular (Sá Santos et al., 2006; Bunik et al., 2009b; Kabysheva et al., 2009; Zündorf et al., 2009), plant tissue (Araújo et al., 2008), and animal (Graf et al., 2009; Trofimova et al., 2010) studies. Having the phosphonate residue instead of the leaving carboxyl group of 2OG, these synthetic inhibitors target the starting and rate-limiting E1o component of 2-oxoglutarate dehydrogenase complex in a highly specific manner, imitating the transition state of the E1o-catalyzed step (Bunik et al., 1992, 2005). Hence, application of such phosphonate analogs of 2OG mimics the state following a decrease in 2-OGDHC activity.
Given that it is very difficult to compare wild-type and mutant phenotypes to understand the short-term events resulting from changes in enzyme activity in plants since mutants and wild types represent distinct steady states separated by an unknown series of events and exhibit pleiotropic effects that do not necessarily portray direct roles of such enzyme or specific responses to enzyme inhibitions we decided to study the short-term events associated with the chemical inhibition of the 2-OGDHC. In this study, we investigated the in situ function of the 2-OGDHC using these phosphonate analogs of 2OG to specifically inhibit the reaction within photosynthetically active cells. Combinatorial metabolite and transcript profiling were used to explore the alterations in primary metabolism and genome-wide gene expression programs of the illuminated leaf which are associated with the inhibition of 2-OGDHC. Our results revealed a positive association between 2OG metabolism and the GABA shunt cycle during the early time-course of inhibition. They also suggest that the metabolic perturbation resulting from the alteration of TCA cycle metabolism invokes the induction of expression of a specific set of genes associated with amino acid mobilization, as well as with those directly linked to alternative pathways of plant respiration. The consequences of this short-term inhibition were additionally monitored at the steady-state metabolite and flux levels, with the combined data discussed in the context of current models of the metabolic regulation of respiration and amino acid metabolism in the illuminated leaf.
Materials and Methods
Materials
All biochemicals, substrates, co-factors, and ion-exchange resins were from Sigma-Aldrich (Taufkirchen, Germany), with the exception of Lipoamide Dehydrogenase from Porcine Heart which was from Calzyme Laboratories (UK) Ltd. Radiochemicals were from Amersham International1. The phosphonate inhibitors were synthesized according to Bunik et al. (2005), with their identity and purity proven by NMR spectroscopy.
Plant material
Arabidopsis thaliana plants used in this study were of the Columbia ecotype (Col-0). Plants were sown on standard greenhouse soil (Stender AG; Schermbeck, Germany) in plastic pots of a 0.1-L capacity. The trays containing the pots were placed under a 16/8 h day/night (22/16°C) with 60/75% relative humidity and 180 μmol m−2 s−1 light intensity. The fully expanded 9th to 12th rosette leaves of 4-week-old plants were harvested for subsequent analysis. With the exception of enzyme assays and dark respiration measurements, all incubations in presence of inhibitors were done exactly in similar growth conditions in order to obtain a clear picture of changes in metabolism.
Enzyme assays
Enzymes were extracted exactly as described previously (Gibon et al., 2004) except that Triton X-100 was used at the concentration of 1% and glycerol at 20%. Enzymes were assayed exactly as described in the literature (Gibon et al., 2004; Nunes-Nesi et al., 2005; Studart-Guimarães et al., 2007). Briefly, pyruvate dehydrogenase was assayed as described previously (Randall and Miernyk, 1990), glutamate dehydrogenase as in Purnell et al. (2005), aconitase as detailed in Carrari et al. (2003), alcohol dehydrogenase as described previously (Chung and Ferl, 1999), alanine amino transferase as in Rocha et al. (2010), citrate synthase as in Gibon et al. (2004), isocitrate dehydrogenase as in Jenner et al. (2001), fumarase as in Nunes-Nesi et al. (2007a), and malate dehydrogenase as in Nunes-Nesi et al. (2005). The maximum activity of 2-oxoglutarate dehydrogenase was determined essentially as described previously (Araújo et al., 2008) with a few modifications. In brief, 2-oxoglutarate dehydrogenase activity was measured by determining NADH formation at 340 nm in a reaction medium containing 75 mM TES-KOH (pH 7.5), 0.05% (w/v) Triton X-100, 1.5 mM MgCl2, 2 mM NAD+, 0.12 mM lithium CoA, 0.2 mM TPP, 2.5 mM cysteine-HCl, 1 mM AMP, 2 mM sodium-2OG, and 5 units of lipoamide dehydrogenase. If not specified otherwise, the reaction was started by 2OG after the enzyme was preincubated in the reaction medium with or without inhibitors for 15 min. The initial linear part of the product accumulation curves (within 10 min of the reaction) was used to calculate the reaction rates.
Metabolic profiling
Metabolite extraction was carried out exactly as described previously (Lisec et al., 2006). As described above, leaf material were cut directly from an Arabidopsis plant, washed three times with 10 mM MES-KOH (pH 6.5), and then incubated (eight disks; 10 mm in diameter) in 2 mL of 10 mM MES-KOH buffer (pH 6.5) in a 100-mL Erlenmeyer flask shaken at 90 rpm in the absence or presence of 100 μM of SP or CESP at similar plant growth conditions. Leaves were sampled during the course of the experiment at the time points indicated in the text, washed briefly with 10 mM MES-KOH (pH 6.5) and immediately frozen in liquid nitrogen. The GC-MS system was comprised of a CTC CombiPAL autosampler, an Agilent 6890N gas chromatograph and a LECO Pegasus III TOF-MS running in EI+ mode. Metabolites were identified in comparison to database entries of authentic standards (Kopka et al., 2005; Schauer et al., 2005). Metabolite profiling data are reported following recent recommendations (Fernie et al., 2011).
The levels of starch were determined exactly as described previously (Fernie et al., 2001). The levels of nitrate, total amino acids, and protein were measured as described previously (Sienkiewicz-Porzucek et al., 2008), while NAD(H) and NADP(H) were determined as described by Schippers et al. (2008).
Measurement of redistribution of stable isotope
The fate of 13C-labeled glucose was traced following incubation (at similar light growth conditions) of leaf disks in 20 mM labeled substrate in 10 mM MES-KOH (pH 6.5) for 1, 2, and 4 h. Fractional enrichment of metabolite pools was determined and label redistribution was expressed exactly as described previously (Roessner-Tunali et al., 2004; Tieman et al., 2006). From the above analysis and for sake of simplicity we decided to show only the 4 h incubation period since the majority of pools were still into a linear phase of labeling, as previously demonstrated (Roessner-Tunali et al., 2004).
Respiration measurements
Dark respiration was measured in an oxygen electrode following the protocol detailed previously (Geigenberger et al., 2000).
Transcript profiling
Transcript profiling was performed using the Arabidopsis Affymetrix ATH1 array as previously described (Redman et al., 2004). Three independent hybridizations were performed for each treatment (control samples and SP-treated samples). Data were processed using Robin software (Lohse et al., 2010) and are displayed following visualization in MapMan (Usadel et al., 2005) or PageMan (Usadel et al., 2006).
Cloning and overexpression of A. thaliana PII
The high-level expression vector encoding the A. thaliana PII gene product (GLB1) minus the transit peptide region was constructed and recombinant A. thaliana PII minus the transit peptide (H6PII-tp) was purified as described in Smith et al. (2002). The concentration of H6PII-tp was determined spectrophotometrically by the method described previously (Sienkiewicz-Porzucek et al., 2008) using BSA as a standard.
Microcalorimetric determination of A. thaliana PII-ligand binding constants
The binding of metabolites to the recombinant protein H6PII-tp was measured by isothermal titration calorimetry (ITC) using a VP-isothermal titration calorimeter from MicroCal, Inc. (Northampton, MA, USA). All titrations were performed at 28°C using a 0.298-μL injection syringe while stirring at 430 rpm exactly as described previously (Smith et al., 2003). All chemicals used were of the highest available grade and purchased from Sigma-Aldrich (Oakville, ON, Canada), except for the adenine nucleotides and 2OG, which were from Roche (Laval, QC, Canada). Heats of dilution isotherms for each experiment were collected in a blank run under conditions corresponding to the experimental run, except with the absence of protein. The ligand heat of dilution was selected as the blank run, since it was larger than the heat of dilution of the protein sample in all cases. The calorimetric data were analyzed using the Origin software, Version 5.0 (MicroCal). The dilution heats from the blank run were subtracted from the experimental calorimetric data using the Origin programs. The independent variables, the association binding constant Ka, and the heat of binding ΔH, were generated from the software by de-convoluting and curve-fitting the binding data using standard methods (Origin User’s Manual Version 5). The dependent variables, the dissociation constants (Kd = 1/Ka), Gibbs free energy changes (ΔG = −RT ln Ka), and entropy changes (ΔS = ΔH/T - ΔG/T), were calculated using ΔH and Ka values.
Statistical analysis
Where two observations are described in the text as different, this means that they were determined to be statistically different (P < 0.05) by the performance of Student’s t-tests.
Results
The inhibitory effects of phosphonate analogs of 2OG on the in vitro activity of the Arabidopsis 2-oxoglutarate dehydrogenase complex
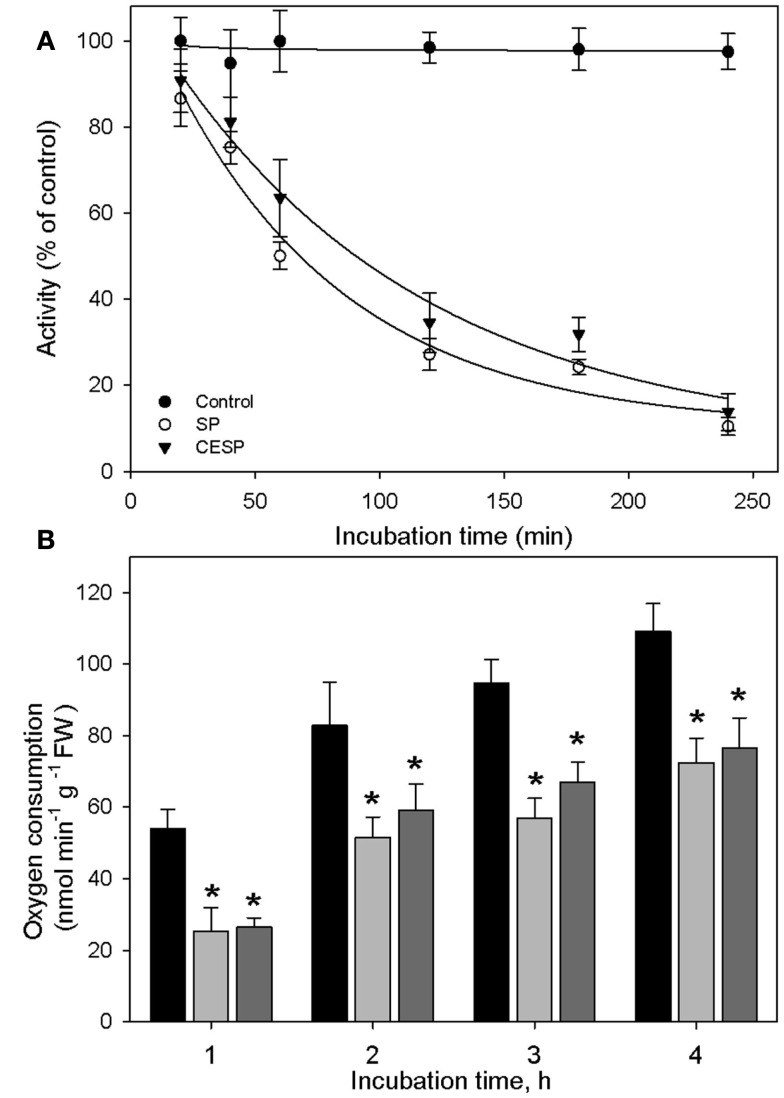
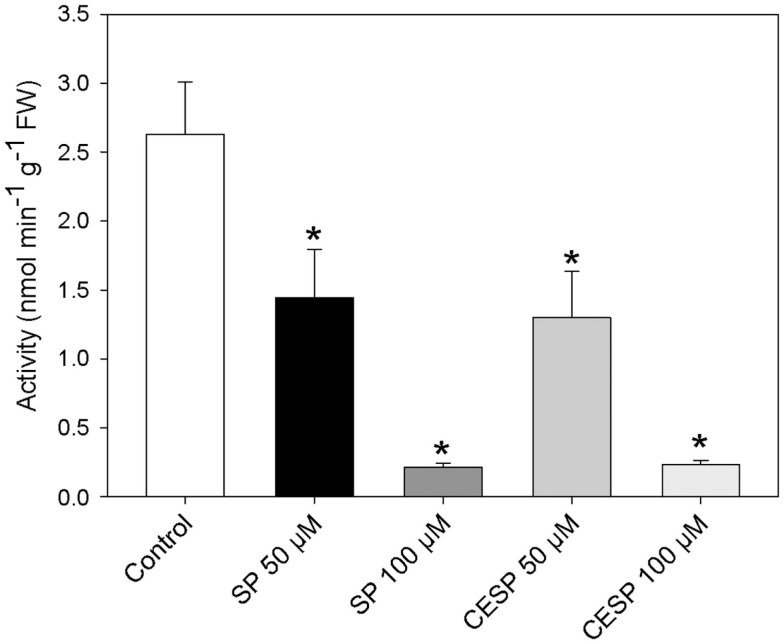
We have previously demonstrated that the phosphonate analogs succinylphosphonate (SP) and carboxy ethyl ester of SP (CESP) inhibited the 2-OGDHC activity markedly in a concentration-dependent manner, whereas the phosphono ethyl ester of SP (PESP) and triethyl ester of SP (TESP) gave less efficient inhibition in plant tissues (Araújo et al., 2008) which is a common feature of the inhibition, observed also in animal tissues (Bunik et al., 1992, 2005, 2009a). On the basis of these experiments we decided to concentrate our studies in illuminated leaf using the former two analogs. Following the protocol of Millar et al. (1999) we assayed the 2-OGDHC activity in crude extracts from A. thaliana leaf material in the presence of varying concentrations (50–100 μM) of the inhibitors SP and CESP. These experiments revealed a similar pattern of inhibition with an almost complete inhibition of the 2-OGDHC with the addition of 100 μM of inhibitors (Figure A1 in Appendix). It is important to note that the activity determined in leaf material is only around 10–20% of that observed in potato tubers (Araújo et al., 2008), however, this is consistent with other studies on photosynthetic and heterotrophic tissues (Hanning and Heldt, 1993; van der Merwe et al., 2010), reflecting a lower content of mitochondria in such tissues compared to heterotrophic ones. The fact that the above in vitro studies revealed that SP and CESP were potent inhibitors of the 2-OGDHC activity suggests that they have the potential to be valuable tools for studying the metabolic impact of the function of this enzyme in folio. We next performed feeding experiments in which leaf disks were incubated in similar light growth conditions (see Materials and Methods) in the presence or absence of 100 μM SP or CESP over a period of 4 h to evaluate whether we could effectively apply inhibitors to this tissue. For this purpose, we used disks isolated from 4-week-old A. thaliana Col-0 plants. Leaf disks were incubated in similar light growth conditions (see Materials and Methods), sampled, rapidly washed, and snap frozen at time points during their incubation in phosphonate-supplemented medium, and the effect of the inhibitors was assessed by measuring the activity of the 2-OGDHC in extracts of these samples. These studies revealed that incubation of the disks with either SP or CESP effectively decreased the activity of the 2-OGDHC. After 1 h, the activity in the SP-fed disks was half of the control (2.63 nmol min−1 g−1) activity (Figure 1A), while that in CESP-fed disks was reduced to approximately 60% of the control level (Figure 1A); after 4 h both inhibitors lead to a decrease in activity to near 10% of control levels.
Figure 1.
Inhibition of the 2-oxoglutarate dehydrogenase complex activity by SP and CESP. (A) Incubation of leaf disks with phosphonate analogs inhibit the activity of the 2-OGDHC. Arabidopsis thaliana leaf disks were incubated in 10 mM MES-KOH (pH 6.5) with 100 μM SP (white circle) or CESP (black triangle) for up to 4 h in similar light growth conditions. The control (black circles) was incubated in the absence of inhibitor. At the times indicated, the leaf disks were washed with 10 mM MES-KOH (pH 6.5) to remove excess inhibitors and then homogenized. 2-OGDHC activity of the extracts was measured in the standard assay medium without the inhibitors. Each value is the mean ± SE of six biological replicates. (B) Respiration of leaf disks incubated in the absence (black bars) or presence of 100 μM SP (gray bars) or CESP (dark gray bars). Freshly prepared leaf disks were transferred into the temperature controlled measuring chamber of an oxygen electrode containing 1 mL of 10 mM MES-KOH, pH 6.5. Each determination was performed in six biological replicates, and data presented are means ± SE of these replicates. The asterisks demarcate values that were judged to be significantly different from the control (P < 0.05) following the performance of Student’s t-tests. FW, fresh weight.
In order to estimate the impact of the inhibition of the 2-OGDHC on the physiology of the plant, we assayed the rate of dark respiration. For this end we incubated leaf disks in an oxygen electrode in the presence and absence of the inhibitors. The inhibitors decreased respiration rate over the entire observation period. However, following 1 h of incubation both the 2-OGDHC activity (Figure 1A) and respiration (Figure 1B) showed about 50% reduction. In contrast, after 4 h of incubation the respiration decrease was much less pronounced (approximating 30%), compared to the 2-OGDHC inhibition (about 80%), suggesting an induction of compensatory mechanisms in response to the 2-OGDHC inhibition.
Having established the inhibitory effects of these phosphonate analogs on the in vitro and in situ activity of the 2-OGDHC, we next performed a broader screen of enzymes associated with the TCA cycle in order to evaluate if the inhibitors affected other enzymes of this pathway. We chose to evaluate the effect of high concentrations (100 μM) of the inhibitors on activities of the mitochondrial enzymes transforming organic acids that are structural analogs of 2OG: namely aconitase, alanine amino transferase, alcohol dehydrogenase, citrate synthase, fumarase, isocitrate dehydrogenase, malate dehydrogenase, pyruvate dehydrogenase complex, pyruvate kinase, and succinyl-CoA ligase, as well as the important enzymes of nitrate metabolism nitrate reductase, glutamine synthetase (GS), glutamate synthase (Fd-GOGAT), and glutamate dehydrogenase. Neither SP nor CESP affected the activities of any of these enzymes (Table 1). We additionally assayed all of the enzymes in the absence of their described substrates to verify that they were incapable of utilizing either of the phosphonate analogs as a substrate. In no instance was a significant transformation of the phosphonate detected (data not shown).
Table 1.
Assays of the key enzymes related to the TCA cycle in the absence (Control) and presence of 100 μM of SP or CESP.
| Control | SP | CESP | |
|---|---|---|---|
| nmol min−1 g−1 FW | |||
| Aconitase | 41.5 ± 4.6 | 39.9 ± 5.4 | 40.4 ± 6.3 |
| Citrate synthase | 165.8 ± 12.6 | 160.1 ± 14.5 | 173.4 ± 10.1 |
| Fd-GOGAT | 1157.5 ± 94.4 | 1203.9 ± 111.3 | 1099.9 ± 102.7 |
| Glutamate dehydrogenase | 612.8 ± 72.5 | 597.8 ± 86.7 | 614.6 ± 84.7 |
| Glutamine synthetase | 981.9 ± 40.3 | 918.5 ± 59.3 | 938.6 ± 62.6 |
| NAD-isocitrate dehydrogenase | 262.9 ± 25.6 | 246.6 ± 26.2 | 253.2 ± 37.9 |
| Pyruvate kinase | 847.6 ± 64.4 | 873.4 ± 81.4 | 851.6 ± 58.3 |
| Nitrate reductase | 537.5 ± 30.3 | 551.7 ± 41.1 | 541.1 ± 29.9 |
| Succinyl-CoA ligase | 114.5 ± 12.9 | 124.1 ± 12.8 | 115.5 ± 13.7 |
| Pyruvate dehydrogenase | 27.4 ± 2.4 | 24.6 ± 4.4 | 25.7 ± 3.8 |
| μmol min−1 g−1 FW | |||
| Alcohol dehydrogenase | 90.8 ± 3.7 | 89.9 ± 3.3 | 88.1 ± 2.5 |
| Alanine amino transferase | 116.9 ± 5.7 | 103.8 ± 6.7 | 104.7 ± 5.4 |
| Fumarase | 5.6 ± 1.1 | 5.4 ± 0.8 | 4.9 ± 0.9 |
| NAD-malate dehydrogenase | 70.3 ± 8.1 | 72.6 ± 9.8 | 68.2 ± 9.2 |
The in vitro activity of the enzymes were determined in Arabidopsis thaliana leaves extract of fully expanded leaves of 4-week-old plants were harvested at the middle of the light period. Values are presented as the mean ± SE of four replicates.
Consequences of inhibition of the 2-oxoglutarate dehydrogenase complex on metabolism in the illuminated leaf
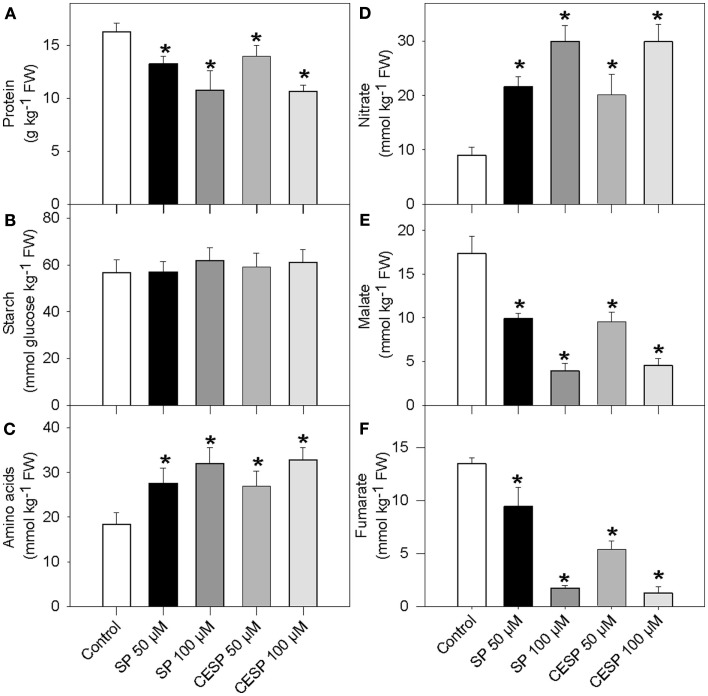
Given the considerable changes in respiration rates (Figure 1) and the recognized link between mitochondrial metabolism and associated carbon/nitrogen interactions (Foyer et al., 2011) we next analyzed the content of total protein and amino acid levels as well as starch, nitrate, malate, and fumarate following incubation of leaf disks in the presence of the inhibitors in exactly similar light growth conditions (Figure 2). The presence of inhibitor had similar consequences at each time point evaluated, so for the sake of simplicity we only present the data obtained from the 4-h treatment. Whilst the levels of amino acids and nitrate were significantly increased in the presence of increasing concentrations of both inhibitors (Figures 2C,D), the levels of protein, malate, and fumarate were significantly reduced (Figures 2A,E, F). Given that the levels of nitrate increased during the treatment, it seems likely that nitrate is taken up from the incubation medium, since MES buffer is a rich source of nitrate (Araújo et al., 2008). The starch content was unchanged following 2 h inhibitor incubation (Figure 2B).
Figure 2.
Effects of the inhibition of the 2-oxoglutarate dehydrogenase complex activity on metabolite levels. Following 4 h incubation of Arabidopsis leaf disks in exactly similar light growth conditions in the absence (white bars) or presence of 50 μM SP (dark bars), 100 μM SP (dark gray bars), 50 μM CESP (gray bars), or 100 μM CESP (light gray bars) levels of protein (A), starch (B), total amino acids (C), nitrate (D), malate (E), and fumarate (F) were measured. Values are means ± SE of six independent samplings. Asterisk indicates values that were determined by the Student’s t-test to be significantly different (P < 0.05) from the control treatment. FW, fresh weight.
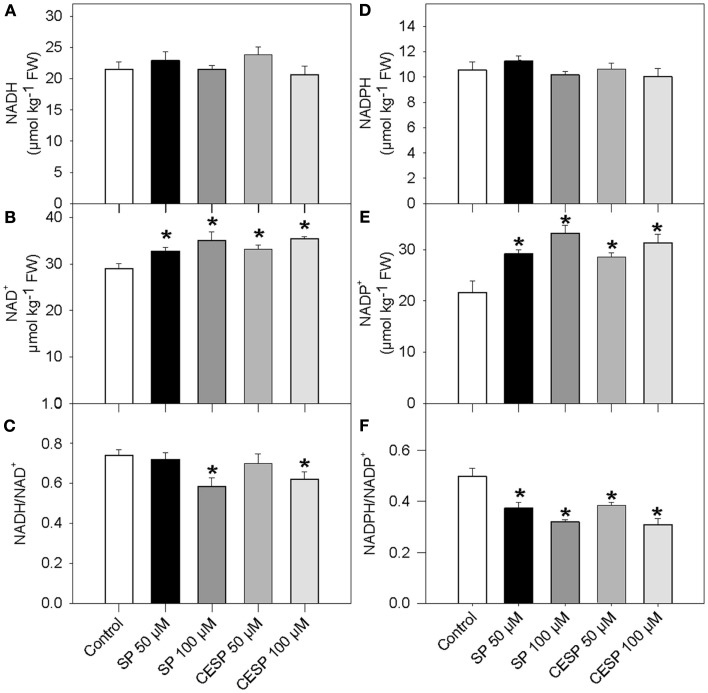
Since the oxidative decarboxylation of 2OG to succinyl-CoA catalyzed by the 2-oxoglutarate dehydrogenase also results in the production of reduced coenzyme NADH, it might be expected that inhibition in the activity of this enzyme would also affect the redox poise of the illuminated leaf. We therefore assayed the levels of pyridine dinucleotides in the phosphonate analog treated samples following incubation of leave disks in exactly similar growth conditions (Figure 3). Again the presence of inhibitor had similar consequences at each time point evaluated, so for the sake of simplicity we only present the data obtained from the 4-h treatment. Interestingly, whilst both NADH and NADPH remained constant (Figures 3A,D), NAD+ and NADP+ levels were significantly higher following the inhibition of the 2-OGDHC (Figures 3B,E). As a result, a trend toward a decrease in both the NADH/NAD+ and NADPH/NADP+ ratios, in a concentration-dependent manner, was observed (Figures 3C,F)
Figure 3.
Pyridine nucleotide levels and ratios in Arabidopsis leaves following inhibition of 2-OGDHC by phosphonate analogous. Following 4 h incubation of Arabidopsis leaf disks in exactly similar light growth conditions in the absence (white bars) or presence of 50 μM SP (dark bars), 100 μM (dark gray bars), 50 μM CESP (gray bars) or 100 μM CESP (light gray bars) levels of pyridine nucleotide [NADH (A), NAD+ (B), NADPH (D), and NADP+ (E)] and ratios [NADH/NAD+ (C) and NADPH/NADP+ (D)] were measured. Values are means ± SE of six independent samplings. Asterisk indicates values that were determined by the Student’s t-test to be significantly different (P < 0.05) from the control treatment. FW, fresh weight.
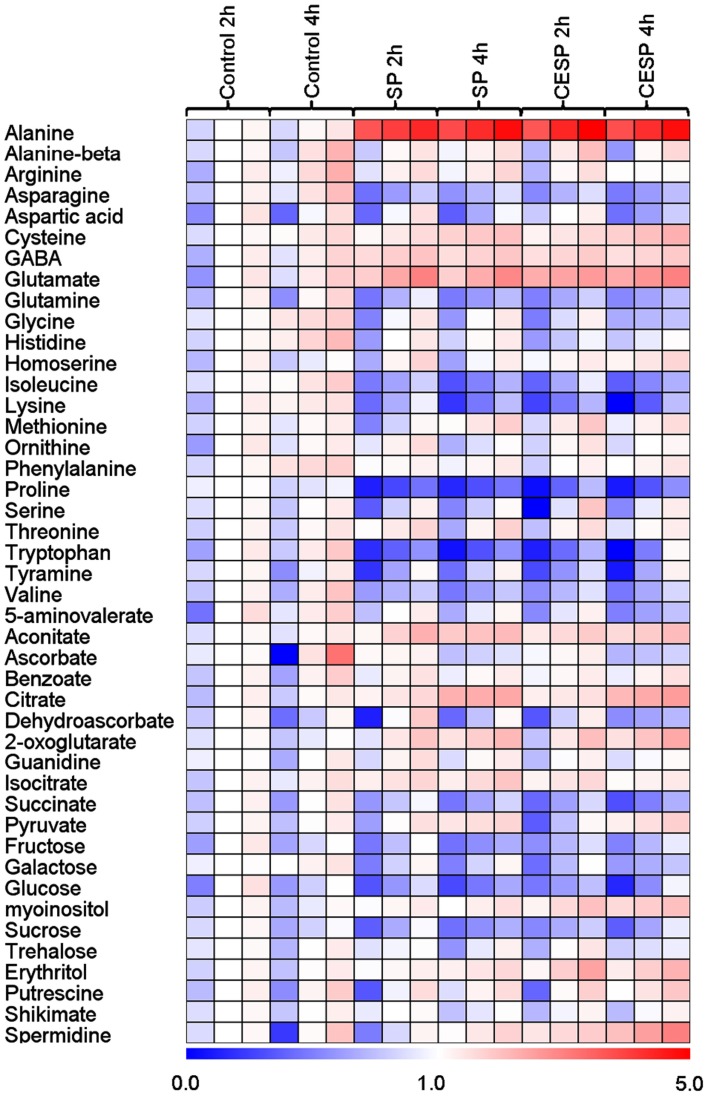
In order to obtain a clearer picture of the changes in the primary metabolism we next utilized a gas chromatography-mass spectrometry (GC-MS)-based metabolic profiling method (Fernie et al., 2004; Lisec et al., 2006) to quantify the relative metabolite levels following incubation in buffer in the presence or absence of 100 μM SP or CESP for 2 or 4 h under the exact same growth conditions. The data obtained are displayed in false color in the heat map of Figure 4 in order to provide an easy overview (the full data set is additionally available as Table A1 in Appendix). From this display, it is noticeable that there were changes in the levels of metabolites even in the absence of inhibitor. However, these were relatively minor and very few of these changes were statistically significant. The changes in metabolite profiles were, by and large, qualitatively and quantitatively similar between the inhibitors, which is in good accordance with the similar level of enzyme inhibition displayed by the phosphonates (Figure A1 in Appendix). Furthermore, inhibition of the 2-OGDHC activity is evident from the fact that 2OG was significantly elevated at all time points following incubation with SP or CESP. We observed large increases in glutamate and alanine (more than two and fourfold, respectively) and the significant increase in γ-aminobutyrate (GABA). Within the TCA cycle, the levels of the intermediates pyruvate, citrate, isocitrate, and aconitate were generally increased, whereas the levels of fumarate and malate were reduced (Figure 2) indicating a new steady-state of the flux through the TCA cycle with the inhibition of 2-oxoglutarate dehydrogenase and suggesting that the TCA cycle is working in a non-cyclic manner (Sweetlove et al., 2010). Interestingly the levels of succinate were also reduced after inhibition of the 2-OGDHC, albeit only statistically significant following 4-h incubation. This corresponds to the observed increase in respiration (Figure 1), with both findings consistent with a possible compensatory upregulation of other pathways of succinate production, and an increased control of the respiration beyond the 2-oxoglutarate dehydrogenase. Some amino acids were decreased most notably those derived from 2OG (proline and glutamine) but the pyruvate-derived alanine was strongly increased. Surprisingly the oxaloacetate-derived amino acids homoserine, threonine, and methionine were generally unaltered as were the 3-phosphoglycerate-derived serine and glycine. By contrast, tryptophan, asparagine, and the branched-chain amino acids isoleucine, lysine, and valine were significantly decreased. Furthermore the levels of the sugars (fructose, glucose, and sucrose) were significantly reduced following incubation with the phosphonate analogs (Figure 4). Another change of note in the metabolite profile was the significant increase in the levels of the polyamine spermidine following 4-h incubation. Intriguingly, the most prominent changes in the data set were observed for the levels of alanine, which increased more than fourfold following incubation in both SP and CESP, most likely reflecting an increased anaerobic fermentation in the treated samples.
Figure 4.
Heat map representing the changes in relative metabolite contents of treated and control Arabidopsis leaf disks. Leaf disks were cut directly from 4-week-old plants, washed three times with 10 mM MES-KOH (pH 6.5), and then incubated during up to 4 h in 10 mM MES-KOH buffer (pH 6.5) containing 2.0 mM glucose and 100 μM of SP or CESP. Metabolites were determined as described in the Section “Materials and Methods.” Data are normalized with respect to the mean response calculated for the control at 2 h. The full data sets from these metabolic profiling studies are available as Table A1 in Appendix including statistical treatment. Green and red represent a decrease and an increase of metabolite content, respectively, in the SP-treated samples with respect to the control samples. The color scale used is reproduced in the Figure.
In order to provide corroborative evidence for some of the changes highlighted above, we performed one further experiment. Namely, in order to gain further evidence for an augmentation of anaerobic fermentation in treated samples we measured the activity of two enzymes (alanine amino transferase and alcohol dehydrogenase) known to be directly involved in fermentation following incubation for up to 4 h in the phosphonate analogs. In contrast to the suggestion observed with our metabolite profile, following incubation with both inhibitors, the maximal catalytic activities of these two enzymes were not different than that of the control (Table A2 in Appendix) suggesting that these changes were the metabolic consequence of inhibiting the 2-OGDHC rather than a direct effect of the phosphonate analogs on the fermentative enzymes themselves.
The inhibition of the 2-oxoglutarate dehydrogenase is associated with the up regulation of alternative respiratory pathways
In order to provide corroborative evidence for some of the changes highlighted above, we assessed the fermentative and GABA shunt fluxes by following the metabolic fate of 13C-labeled glucose and evaluated the redistribution of 13C to TCA cycle, GABA shunt, and photorespiratory pathways after 4 h incubation in similar growth conditions in presence or absence of SP and CESP. The changes in isotope redistribution profiles were, by and large, qualitatively and quantitatively similar between the inhibitors. Not surprisingly the isotope redistribution to 2OG was significantly increased with both inhibitors as well as that of glutamate and GABA while a reduced redistribution was observed for glutamine, confirming an up regulation of the GABA shunt in treated samples (Table 2). Given that the GABA shunt can bypass the reaction catalyzed by succinyl-CoA ligase and sustain succinate provision to the TCA cycle we would expected no impact on the metabolites of the second part of the TCA cycle. Intriguingly, however, the label redistribution to malate and fumarate was consistently reduced in the presence of the inhibitors (Table 2) indicating that the up regulation of the GABA shunt was not enough to rescue the TCA cycle function at normal levels. Interestingly, a reduced redistribution to branched-chain amino acids, lysine, and valine, was also observed, perhaps indicative of an increased usage of these amino acids as alternative respiratory substrates (Araújo et al., 2010). Taken together, these data and the respiration measurements described above indicates that following 2-OGDHC inhibition a metabolic rearrangement is reconstructed generating clear imbalance of the TCA cycle coupled with an up regulation of the GABA shunt.
Table 2.
Redistribution of heavy label following glucose feeding of control and treated Arabidopsis leaves.
| Control | SP | CESP | |
|---|---|---|---|
| μmol C1 equivalent g−1 FW h−1 | |||
| Alanine | 4.837 ± 1.248 | 3.989 ± 0.578 | 4.417 ± 0.707 |
| Asparagine | 0.129 ± 0.042 | 0.117 ± 0.004 | 0.124 ± 0.014 |
| Aspartate | 0.085 ± 0.020 | 0.060 ± 0.014 | 0.075 ± 0.015 |
| Fructose | 0.479 ± 0.032 | 0.388 ± 0.107 | 0.613 ± 0.074 |
| Fumarate | 0.231 ± 0.068 | 0.114 ± 0.013 | 0.143 ± 0.066 |
| GABA | 0.077 ± 0.006 | 0.115 ± 0.006 | 0.971 ± 0.007 |
| Glutamate | 1.199 ± 0.172 | 1.712 ± 0.163 | 1.591 ± 0.134 |
| Glutamine | 0.931 ± 0.134 | 0.679 ± 0.097 | 0.663 ± 0.180 |
| Histidine | 0.992 ± 0.139 | 0.859 ± 0.045 | 0.887 ± 0.118 |
| Isoleucine | 0.018 ± 0.005 | 0.015 ± 0.003 | 0.012 ± 0.003 |
| Lysine | 1.477 ± 0.221 | 1.036 ± 0.128 | 1.167 ± 0.136 |
| Malate | 2.174 ± 0.166 | 1.522 ± 0.158 | 1.408 ± 0.206 |
| Methionine | 0.044 ± 0.001 | 0.046 ± 0.001 | 0.049 ± 0.001 |
| Ornithine | 0.364 ± 0.108 | 0.073 ± 0.015 | 0.063 ± 0.020 |
| 2-Oxoglutarate | 0.320 ± 0.168 | 0.391 ± 0.035 | 0.383 ± 0.126 |
| Proline | 0.029 ± 0.012 | 0.001 ± 0.001 | 0.002 ± 0.001 |
| Serine | 0.115 ± 0.022 | 0.087 ± 0.014 | 0.088 ± 0.018 |
| Sucrose | 1.323 ± 0.134 | 1.184 ± 0.326 | 1.197 ± 0.255 |
| Valine | 0.449 ± 0.109 | 0.325 ± 0.054 | 0.341 ± 0.061 |
Fully expanded leaves of 4-week-old plants were harvested at the middle of the light period, washed three times with 10 mM MES-KOH (pH 6.5), and then incubated during 4 h in similar light growth conditions in 10 mM MES-KOH buffer (pH 6.5) containing 20.0 [U-13C] mM glucose and 100 μM of SP or CESP. Values represent absolute redistribution of the label and are given as means ± SE of determinations on six independent samples. Those set in bold type were determined by the Student’s t-test to be significantly different (P < 0.05) from the control.
Influence of the inhibition of the 2-oxoglutarate dehydrogenase complex on gene expression
We were also interested in studying the impact of the 2-OGDHC inhibition in a broader context which also includes alterations of gene expression. For this purpose, we performed microarray analysis of samples incubated for 4 h in similar growth conditions in presence and absence of SP using Affymetrix ATH1 chips. The entire dataset of this microarray analysis is provided in Data Set S1 in Supplementary Material.
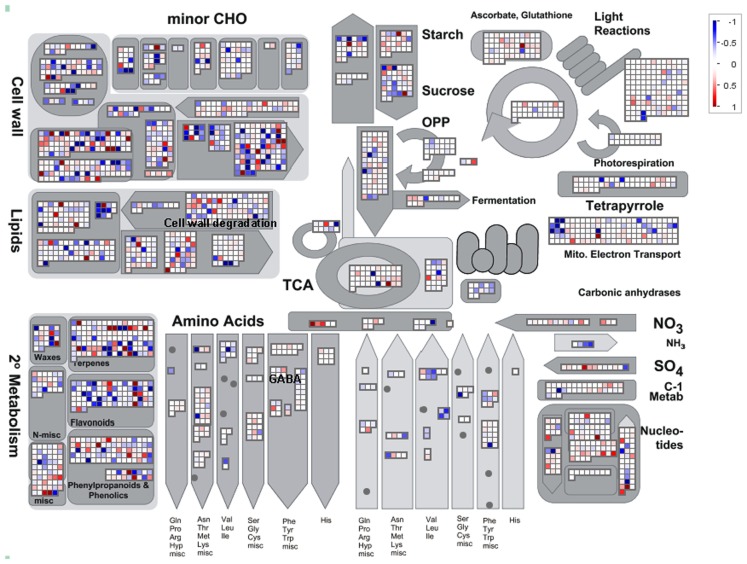
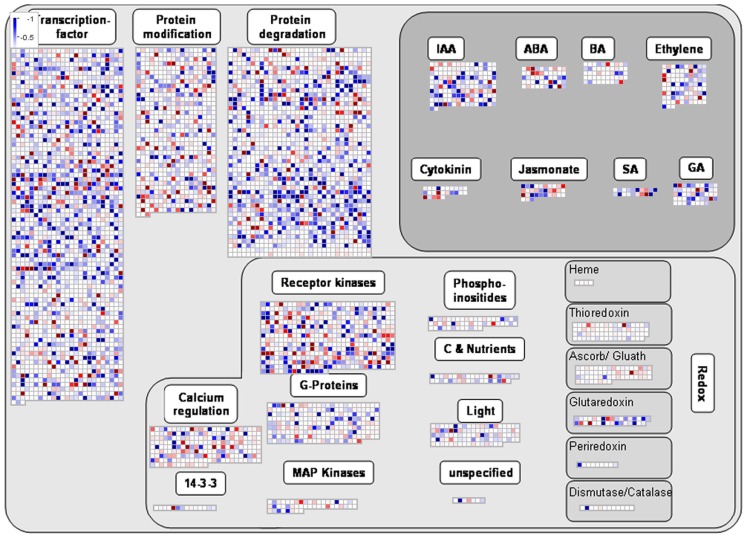
ANOVA of the microarray results indicated that the expression of 76 genes showed a significant and at least 1.5-fold change between SP treated and control samples, with detection P-value of a false discovery rate <0.005 in all replicates (Tables A3 and A4 in Appendix). We further identified the genes that were consistently up-regulated (33, Table A3 in Appendix) and down regulated (43; Table A4 in Appendix) across all samples and subjected them to overrepresentation analysis using the tools embedded in the MapMan software tools2. The major effects of the phosphonate treatment included the significant repression of the expression of 43 genes belonging to functional categories associated with anabolic processes (Table A4 in Appendix and Figure 5). Among these we observed (i) genes associated with protein synthesis (including genes encoding ribosomal proteins and genes encoding transcription factors); (ii) genes encoding enzymes associated with amino acid metabolism, from which genes interestingly belong to the branched-chain amino acid metabolism (AT1G10070, branched-chain amino acid transaminases; AT3G45300, isovaleryl CoA dehydrogenase; AT4G33150, saccharopine dehydrogenase); (iii) genes encoding enzymes associated with nitrogen metabolism (AT2G38400, AGT3; alanine glyoxylate aminotransferase; At5g18170, GDH1, glutamate dehydrogenase; At5g07440, GDH2, glutamate dehydrogenase); (iv) two methyl transferase genes; and (v) genes associated with RNA processing. Among the genes that were up-regulated (Table A3 in Appendix) changes were observed in chloroplast ribosomal proteins which might be associated to perturbed protein synthesis, adenylosuccinate lyase, an enzyme of the purine nucleotide cycle in which fumarate is generated from aspartate (Siehl et al., 1996) most likely in order to increase or maintain the concentration of TCA cycle intermediates. Since we previously documented that inhibition of the TCA cycle have a direct and immediate effect on cell wall biosynthesis (Araújo et al., 2008; Sweetlove et al., 2010; van der Merwe et al., 2010) we also looked whether changes in 2OG metabolism were associated with alteration in the expression pattern of genes involved in cell wall metabolism and only a significant up regulation of XTR6 (Xyloglucan endotransglycosylase 6) was observed. In addition to the repression of expression of genes associated with the above processes, the SP treatment also seems to have mixed effects (induction/repression) on small sets of genes involved in several metabolic processes as well as those associated with RNA transcription and processing, post-translational modifications, and protein turnover (Figure 5; Tables A3 and A4, Figure A2 in Appendix).
Figure 5.
Overview of the MapMan visualization of differences in transcript levels between control samples and SP-treated samples. Genes associated with metabolic pathways were analyzed by the MapMan software (http://mapman.gabipd.org/web/guest/mapman). Blue and red represent a decrease and an increase of expression, respectively, in the SP-treated samples with respect to the control samples. The color scale used is reproduced in the Figure. Fully expanded leaves of 4-week-old plants were harvested at the middle of the light period, washed three times with 10 mM MES-KOH (pH 6.5), and then incubated in presence or absence of 100 μM of SP during 4 h in 10 mM MES-KOH buffer (pH 6.5) containing 2.0 mM glucose.
The presence of phosphonate analogs does not compromise the binding of 2OG to the putative regulatory PII protein
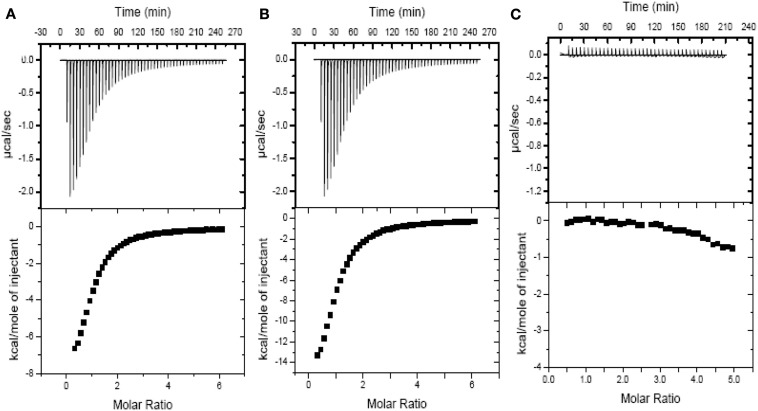
Due to the fundamental role of the bacterial PII as an allosteric sensor and coordinator of responses to carbon and nitrogen levels (Smith et al., 2003) and in light of the multiple changes in gene expression following the 2-OGDHC inhibitor incubation we chose to investigate whether the phosphonate analogs are also able to affect the binding of PII to 2OG. The binding of the phosphonates to recombinant PII was determined by measuring the heat flow (μcal s−1) as a function of time, using ITC. 2OG is an effector molecule that binds to PII and is suggested to mediate signaling of carbon status in the chloroplast (Smith et al., 2003; Ninfa and Jiang, 2005). Additionally it has been demonstrated that 2OG binds PII only when the ATP binding site in PII is occupied by ATP (Smith et al., 2003). The binding of 2OG to PII was observed exactly as previously described (Smith et al., 2003), however, the addition of either SP or CESP did not affect the thermodynamic parameters of this ligation (Figure 6; Table 3). Nevertheless, in the absence of 2OG no binding of ATP to the Arabidopsis PII could be detected in the presence of the inhibitors (Figure 6C) suggesting that the phosphonates neither inhibited the 2OG binding to PII protein, nor bound to the PII binding site in the absence of 2OG. This implies that the observed changes in gene expression observed occurred most likely independently of the action of PII.
Figure 6.
Isothermal titration profiles comparing binding of 2OG to H6PII-tp in the absence and presence of phosphonate analogs or ATP. The upper panel shows the raw data in the form of the heat effect during titration of 120 μm PII with 2OG (5.0–500 μm) and ATP in the presence of SP (A), CESP (B) and in presence of SP but without ATP (C). The lower panel shows the experimental data for the titration of PII plotted in the derivative format, i.e., the binding isotherm, and the best fit curve.
Table 3.
Thermodynamic parameters of 2OG binding to Arabidopsis thaliana PII in the presence of ATP, SP, or CESP.
| Binding molecule | Kd (μM)* | ΔH (kcal mol-1) | ΔG (kcal mol-1) | TΔS (kcal mol-1) |
|---|---|---|---|---|
| 2OG | ||||
| PII: ATP | 110.7 ± 3.4 | −10.5 ± 0.3 | −6.8 ± 0.4 | −3.6 ± 0.9 |
| PII: ATP + SP | 105.5 ± 2.7 | −9.9 ± 0.4 | −5.9 ± 0.3 | −4.1 ± 1.3 |
| PII: ATP + CESP | 114.1 ± 3.2 | −10.7 ± 0.2 | −7.1 ± 0.6 | −3.7 ± 1.5 |
*Values are means ± SE of six independent samplings. Ligand-complexed PII was in the presence of excess ligand, either 1.0 mm ATP or 2OG. ΔG values were calculated using ΔG = −RT ln Ka = ΔH − TΔS. Values for thermodynamic parameters could not be determined without either 2OG or ATP in presence of each phosphonate analog due to weak binding suggesting that the those compounds cannot replace 2OG in the PII binding site.
Discussion
Here we used phosphonate inhibitors of 2-OGDHC to inducibly alter the metabolic flux of 2OG degradation in order to investigate both the role of 2OG degradation within primary metabolism and genome-wide transcriptional programs in a short-term response in the illuminated leaf. We were able to demonstrate that these analogs neither affected the expression or activities of a wide range of TCA cycle enzymes (Table 1) nor the binding of 2OG to PII (Figure 6; Table 3) suggesting that they are highly appropriate to address the role of 2-OGDHC also in the illuminated leaf. A number of studies have pointed toward an essential role of the 2-OGDHC reaction in overall cellular metabolic activity (for a review see Bunik and Fernie, 2009). Additionally we have previously, via use of the same specific chemical inhibitors used here, demonstrated that 2-OGDHC is limiting for respiration and plays an important role in nitrogen assimilation in potato tubers (Araújo et al., 2008). Interestingly, the metabolic perturbation following its inhibition in heterotrophic plant tissues is similar to that observed in species of other kingdoms (Bunik and Fernie, 2009). Additional evidence that perturbation of 2OG metabolism alters nitrogen metabolism have been provided by showing that transgenic tomato (Solanum lycopersicum) lines with a minor inhibition of the cytosolic NADP-dependent isocitrate dehydrogenase were characterized by an altered leaf nitrate and amino acid composition (Sulpice et al., 2010).
Our current knowledge indicates that plants possess an intricate regulatory machinery that coordinates the capacity of nitrogen assimilation with carbon metabolism, nutrient availability, and other environmental factors linking the demands needed for plant growth and development (Nunes-Nesi et al., 2010). The accumulation of 2OG in leaves, which occurs as a consequence of the chemical blockage of 2-OGDHC, not only leads to significant shifts in the levels of glutamate and GABA, but also affects the pool of other amino acids (Figure 4; Table A1 in Appendix). This finding is broadly similar to that observed in potato tubers and is highly consistent with results from other studies suggesting that the diverse pathways of amino acid metabolism are highly interconnected (Less and Galili, 2008; Gu et al., 2010; Angelovici et al., 2011). The results presented here also indicate an upregulation of the GABA shunt, as previously observed in SP-treated potato tubers (Araújo et al., 2008) and neurons (Sá Santos et al., 2006) as well as upon the inhibition of the reaction catalyzed by succinyl-CoA ligase (Studart-Guimarães et al., 2007). Interestingly the chemical inhibition of the 2-OGDHC is associated with a down regulation of alternative respiratory pathways such as the involvement of branched-chain amino acids. The levels of branched-chain amino acids were strongly reduced (Figure 4) and this was associated with a down regulation of a set of genes associated with their degradation (Table 3). Whilst the mechanism behind such response is currently unclear it is tempting to speculate that one (or more) breakdown compounds generated during the catabolism of branched-chain amino acids might be responsible for this feedback regulation. Here we provide evidence that the inhibition of the 2-OGDHC leads to alteration in respiratory and nitrogen metabolism despite being characterized by a compensatory increased flux of the GABA shunt to maintain operation of the TCA cycle. This finding, when taken together with those from previous studies, highlights the metabolic importance of the GABA shunt in plants (Fait et al., 2008) and is similar to findings following neuronal 2-OGDHC inhibition (Sá Santos et al., 2006; Shi et al., 2009). Interestingly, GABA-transaminase activity appears to be the limiting step under stress conditions, resulting in the accumulation of GABA, which could in turn be used to rapidly generate succinate and energy via the TCA cycle upon removal of the stress (Simpson et al., 2010; Shelp et al., 2012). It is important to mention that the increase in both NAD+ and NADP+ levels upon 2-OGDHC inhibition (Figure 3) is in accordance with the key role these coenzymes plays in cellular redox reactions, including those of the TCA cycle (Schippers et al., 2008). In good agreement with previous studies our results also demonstrated that alterations in NAD+/NADH ratios (Figure 3), as observed here, were coupled with dramatic consequences on cellular metabolism (Dutilleul et al., 2005; Shen et al., 2006; Schippers et al., 2008).
Several recent studies have revealed that plants, like mammals, contain an alternative route of electron provision to the mitochondrial electron transport chain mediated by the Electron Transfer Flavoprotein (ETF) complex (Ishizaki et al., 2005, 2006). Branched-chain amino acid catabolism feeds into this pathway directly via the reactions catalyzed by isovaleryl dehydrogenase and 2-d-hydroxyglutarate dehydrogenase as well as indirectly by fueling the TCA cycle (Engqvist et al., 2009; Araújo et al., 2010, 2011, 2012). Whilst these pathways are particularly prominent during dark-induced senescence (Araújo et al., 2010), recent evidence suggests that they also operate in the dark-period of a normal light-dark cycle in the photosynthetic tissue of Arabidopsis (Caldana et al., 2011; Engqvist et al., 2011). In a similar vein, reduction of branched-chain amino acid catabolism in tomato results in an approximately 20% reduction in the rate of respiration in young fruits (Kochevenko et al., 2012). The fact that reduced expression of genes associated with these pathways results in reduced levels of branched-chain amino acids suggests that at least under certain conditions this pathway may also contribute to respiration in the illuminated leaf. However, the signal(s) which regulate(s) the expression of this alternative pathway is/are currently unknown. Moreover, the upregulation of these alternative pathways was not sufficient to entirely compensate for the lack of 2-OGDHC, highlighting its importance for maintenance of respiration.
The observation that a blockage in the reaction catalyzed by 2-OGDHC results in considerable changes in the expression of certain sets of genes (Figure 5; Table 3; Table A3, Figure A2 in Appendix) is highly intriguing particularly in light of the fact that we were able to demonstrate that this was not a direct consequence of an effect on the PII sensor protein (Figure 6; Table 3). In fact in higher plants the role of the PII protein remains much less clear than in prokaryotic organisms where it is a crucial integrator of cellular carbon, nitrogen, and energy levels (Uhrig et al., 2009). Several recent reports have suggested that plant mitochondria are part of signaling pathways known as mitochondrial retrograde regulation (MRR) pathway and participate in the response to oxidative stress (Alhagdow et al., 2007; Rhoads and Subbaiah, 2007; Sweetlove et al., 2007; Møller and Sweetlove, 2010). Whilst mitochondrial derived 2OG would, at least in theory, represent a good candidate signal, being downstream of the aconitase reaction which is a known to be particularly susceptible to oxidative stress (Lehmann et al., 2009), 2OG can also be produced in the cytosol. However, in blocking its consumption in the mitochondrion in our study, the primary accumulation of 2OG will most likely take place in the mitochondria.
Intriguingly, studying genes regarded as hallmarks of MRR including those involved in the response to dysfunctional mitochondrial function such as AOX and alternative NAD(P)H dehydrogenases as well as glutathione reductase, catalases, ascorbate peroxidases, and superoxide dismutases revealed that these were not affected following incubation in the phosphonate analogs. As mentioned above this manipulation did however (or did additionally) result in a co-ordinate regulation of many aspects of protein synthesis, amino acid metabolism, and nitrogen metabolism all of which would be anticipated to participate in the observed shifting of the cellular carbon:nitrogen balance as well as an increase in genes associated with RNA processing which indicates a highly complex integration of nitrogen metabolism and protein synthesis in the illuminated leaf (Nunes-Nesi et al., 2010). When taken together these changes are highly consistent with what would be expected at a global level following restriction in energy metabolism. They are, however, quite dramatic when considered alongside the current debate as to the relative activity of respiration in the illuminated leaf (Tcherkez et al., 2005, 2008, 2009, 2011; Nunes-Nesi et al., 2007b, 2008). Thus, while some studies indicate an almost complete inhibition of the TCA cycle in illuminated leaves (Tcherkez et al., 2005, 2008), others suggest significant operation of the TCA cycle in the light (Nunes-Nesi et al., 2007b). However, an inhibition of nearly 30% for the decarboxylation reaction catalyzed by the pyruvate dehydrogenase in the light compared with darkness was recently estimated (Tcherkez et al., 2009). The TCA cycle has completely different flux modes between light and dark respirations (Sweetlove et al., 2010), and is therefore likely to have a very different regulation during these two periods. One possible explanation is that the non-cyclic TCA might be operating in situations of 2-OGDHC inhibition in order to support the need of 2OG for nitrogen assimilation. Our results indicate that the TCA cycle activity is clearly working in a non-cyclic manner upon 2-OGDHC inhibition during the light period. This result is in good agreement with a recent report demonstrating that the 2-OGDHC activity is strongly inhibited in the light being more effectively involved in the remobilization of night-stored molecules and therefore most likely responsible for providing 2OG for glutamate synthesis in illuminated leaves (Gauthier et al., 2010). That said it is clear that the inhibition of mitochondrial consumption of 2OG results in a distinct reprogramming of gene expression and many further experiments will be required to elucidate the exact mechanism(s) by which this is achieved. Intriguingly, when transcripts encoding the two enzymes thus far characterized to be under the control of PII in plants – N-acetyl-l-glutamate kinase and heteromeric acetyl-CoA carboxylase (Feria Bourrellier et al., 2009; Baud et al., 2010; Bourrellier et al., 2010) are examined no differences were observed suggesting that their expression is not affected by PII following the inhibition of 2-OGDHC.
In summary, from our current study we can state that the inhibition of 2-OGDHC results in rapid changes both in gene expression and cellular metabolism. Despite the upregulation of several compensatory processes the effect of this inhibition could not be fully reversed and is in keeping with our hypothesis that this step harbors the most considerable portion of the metabolic control vested in the TCA cycle (Araújo et al., 2012). In addition, the fact that inhibiting the consumption of 2OG within the mitochondria resulted in reprogramming of gene expression suggests that a downstream metabolite thereof may act as an important signal metabolite which is capable of regulating the expression of multiple nuclear genes. The ITC experiments conducted here allow us to rule out a direct effect of the analogs on the putative regulatory PII proteins in regulating this function. Whilst the precise nature of the interaction between mitochondrially derived 2OG and changes in nuclear gene expression could not be fully resolved in this study it remains an exciting topic for future research.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Plant_Physiology/10.3389/fpls.2012.00114/abstract
Full transcriptome data set.
Acknowledgments
Discussions with Prof. Mark Stitt (same institute) were highly valuable in the development of this work. We gratefully acknowledge the synthesis and characterization of the phosphonate analogs of 2OG by Dr. N. K. Lukashev and Dr. A. V. Kazantsev from Chemistry Department of Moscow Lomonosov State University; we additionally thank K. Koehl and the Green Team of the MPI in producing high quality plant material for the experiments described here. Financial support was provided by the Max Planck Society (Wagner L. Araújo and Alisdair R. Fernie), the National Council for Scientific and Technological Development CNPq-Brazil (grant number 478261/2010-1 to Adriano Nunes-Nesi and 472787/2011-0 to Wagner L. Araújo), The Alberta Ingenuity Center for Carbohydrate Science (GBGM), Natural Sciences and Engineering Research Council of Canada (GBGM), a grant from Russian Foundation of Basic Research (grant 10-04-90007) and a travel grant of Alexander von Humboldt Foundation (program RUS/1003594; to Victoria I. Bunik). Danilo M. Daloso was supported by a PhD grand from the Foundation for Research Assistance of the Minas Gerais State, Brazil (FAPEMIG).
Appendix
Figure A1.
Inhibition of the 2-oxoglutarate dehydrogenase complex activity by SP and PESP.
Figure A2.
MapMan visualization of differences in transcript levels between control samples and SP-treated samples.
Table A1.
Relative metabolite content of treated and control Arabidopsis leaf disks.
| Control 2 h | Control 4 h | SP 2 h | SP 4 h | CESP 2 h | CESP 4 h | |
|---|---|---|---|---|---|---|
| AMINO ACIDS | ||||||
| Alanine | 1 ± 0.17 | 1.13 ± 0.29 | 4.04 ± 0.35 | 4.34 ± 0.49 | 4.42 ± 0.74 | 4.29 ± 0.51 |
| Alanine, beta | 1 ± 0.15 | 1.5 ± 0.72 | 1.12 ± 0.33 | 1.25 ± 0.29 | 1.37 ± 0.65 | 1.11 ± 0.52 |
| Arginine | 1 ± 0.32 | 1.61 ± 0.67 | 1.24 ± 0.35 | 1.32 ± 0.36 | 1.12 ± 0.41 | 1.05 ± 0.02 |
| Asparagine | 1 ± 0.24 | 1.48 ± 0.58 | 0.61 ± 0.17 | 0.72 ± 0.15 | 0.7 ± 0.17 | 0.61 ± 0.13 |
| Aspartate | 1 ± 0.45 | 0.97 ± 0.58 | 0.97 ± 0.56 | 0.67 ± 0.3 | 1.03 ± 0.24 | 0.62 ± 0.18 |
| Cysteine | 1 ± 0.14 | 1.35 ± 0.28 | 1.36 ± 0.23 | 1.87 ± 0.13 | 1.42 ± 0.21 | 2.01 ± 0.25 |
| GABA | 1 ± 0.31 | 1.29 ± 0.41 | 1.77 ± 0.17 | 1.71 ± 0.17 | 1.67 ± 0.15 | 1.69 ± 0.16 |
| Glutamate | 1 ± 0.42 | 1.34 ± 0.47 | 2.39 ± 0.6 | 2.31 ± 0.56 | 2.45 ± 0.14 | 2.67 ± 0.33 |
| Glutamine | 1 ± 0.28 | 1.12 ± 0.56 | 0.7 ± 0.23 | 0.6 ± 0.13 | 0.66 ± 0.15 | 0.64 ± 0.11 |
| Glycine | 1 ± 0.1 | 1.58 ± 0.19 | 0.97 ± 0.45 | 0.99 ± 0.4 | 0.86 ± 0.38 | 0.72 ± 0.05 |
| Histidine | 1 ± 0.17 | 1.69 ± 0.42 | 1 ± 0.39 | 1.08 ± 0.26 | 0.78 ± 0.18 | 0.92 ± 0.15 |
| Homoserine | 1 ± 0.28 | 0.92 ± 0.13 | 1.18 ± 0.52 | 0.97 ± 0.34 | 1.15 ± 0.19 | 1.42 ± 0.24 |
| Isoleucine | 1 ± 0.13 | 1.44 ± 0.37 | 0.64 ± 0.16 | 0.51 ± 0.19 | 0.66 ± 0.27 | 0.53 ± 0.16 |
| Lysine | 1 ± 0.3 | 1.35 ± 0.15 | 0.68 ± 0.26 | 0.47 ± 0.26 | 0.48 ± 0.22 | 0.36 ± 0.38 |
| Methionine | 1 ± 0.18 | 1.11 ± 0.21 | 0.82 ± 0.31 | 1.42 ± 0.35 | 1.36 ± 0.52 | 1.24 ± 0.31 |
| Ornithine | 1 ± 0.39 | 1.11 ± 0.23 | 1.23 ± 0.33 | 0.87 ± 0.18 | 1.14 ± 0.32 | 1 ± 0.16 |
| Phenylalanine | 1 ± 0.16 | 1.61 ± 0.14 | 1.1 ± 0.11 | 1.15 ± 0.2 | 1.03 ± 0.23 | 1.2 ± 0.2 |
| Proline | 1 ± 0.06 | 0.89 ± 0.07 | 0.28 ± 0.16 | 0.31 ± 0.16 | 0.39 ± 0.34 | 0.33 ± 0.23 |
| Serine | 1 ± 0.13 | 1.06 ± 0.28 | 0.81 ± 0.44 | 0.8 ± 0.28 | 0.88 ± 1.04 | 0.92 ± 0.4 |
| Threonine | 1 ± 0.19 | 1.12 ± 0.33 | 1.35 ± 0.3 | 1.2 ± 0.53 | 1.16 ± 0.4 | 1.09 ± 0.21 |
| Tryptophan | 1 ± 0.37 | 1.33 ± 0.54 | 0.38 ± 0.19 | 0.32 ± 0.25 | 0.42 ± 0.3 | 0.49 ± 0.6 |
| Tyramine | 1 ± 0.16 | 0.95 ± 0.4 | 0.64 ± 0.44 | 0.81 ± 0.38 | 0.58 ± 0.29 | 0.66 ± 0.57 |
| Valine | 1 ± 0.22 | 1.32 ± 0.63 | 0.7 ± 0.09 | 0.63 ± 0.15 | 0.72 ± 0.16 | 0.66 ± 0.18 |
| 5-Aminovalerate | 1 ± 0.55 | 1.33 ± 0.44 | 1.03 ± 0.27 | 0.92 ± 0.25 | 0.9 ± 0.36 | 0.63 ± 0.13 |
| ORGANIC ACIDS | ||||||
| Aconitate | 1 ± 0.13 | 1.12 ± 0.23 | 1.71 ± 0.53 | 1.97 ± 0.12 | 1.6 ± 0.22 | 1.83 ± 0.24 |
| Ascorbate | 1 ± 0.08 | 1.48 ± 1.75 | 1.17 ± 0.06 | 0.82 ± 0.07 | 1.13 ± 0.16 | 0.76 ± 0.06 |
| Benzoate | 1 ± 0.23 | 1.23 ± 0.59 | 1.2 ± 0.28 | 1.12 ± 0.2 | 1.13 ± 0.17 | 1.21 ± 0.28 |
| Citrate | 1 ± 0.27 | 1.08 ± 0.3 | 1.41 ± 0.2 | 2.31 ± 0.07 | 1.38 ± 0.11 | 2.34 ± 0.22 |
| Dehydroascorbate | 1 ± 0.21 | 0.79 ± 0.36 | 0.99 ± 0.87 | 0.76 ± 0.36 | 0.81 ± 0.47 | 0.64 ± 0.08 |
| 2-Oxoglutarate | 1 ± 0.12 | 0.91 ± 0.14 | 1.4 ± 0.51 | 1.79 ± 0.33 | 1.39 ± 0.63 | 1.94 ± 0.44 |
| Guanidine | 1 ± 0.06 | 1.03 ± 0.36 | 1.2 ± 0.36 | 1.1 ± 0.23 | 0.99 ± 0.26 | 0.98 ± 0.12 |
| Isocitrate | 1 ± 0.23 | 1.23 ± 0.31 | 1.47 ± 0.2 | 1.6 ± 0.31 | 1.41 ± 0.21 | 1.22 ± 0.15 |
| Succinate | 1 ± 0.25 | 1.03 ± 0.43 | 0.78 ± 0.19 | 0.65 ± 0.19 | 0.63 ± 0.22 | 0.5 ± 0.19 |
| Pyruvate | 1 ± 0.19 | 1.04 ± 0.29 | 1.06 ± 0.44 | 1.54 ± 0.13 | 0.75 ± 0.39 | 1.52 ± 0.24 |
| SUGAR AND SUGAR-ALCOHOLS | ||||||
| Fructose | 1 ± 0.38 | 0.84 ± 0.19 | 0.75 ± 0.28 | 0.56 ± 0.11 | 0.72 ± 0.16 | 0.72 ± 0.2 |
| Galactose | 1 ± 0.06 | 1.23 ± 0.23 | 0.82 ± 0.34 | 0.83 ± 0.33 | 0.73 ± 0.3 | 0.68 ± 0.09 |
| Glucose | 1 ± 0.5 | 0.81 ± 0.22 | 0.59 ± 0.26 | 0.47 ± 0.19 | 0.61 ± 0.14 | 0.55 ± 0.4 |
| Myoinositol | 1 ± 0.2 | 0.92 ± 0.19 | 1.16 ± 0.17 | 1.27 ± 0.27 | 1.6 ± 0.4 | 1.81 ± 0.2 |
| Sucrose | 1 ± 0.15 | 0.82 ± 0.16 | 0.67 ± 0.31 | 0.56 ± 0.12 | 0.67 ± 0.13 | 0.65 ± 0.28 |
| Trehalose, alpha, alpha | 1 ± 0.1 | 1.02 ± 0.32 | 0.96 ± 0.08 | 0.91 ± 0.33 | 1.05 ± 0.36 | 0.87 ± 0.07 |
| OTHERS | ||||||
| Erythritol | 1 ± 0.18 | 1.05 ± 0.29 | 1.13 ± 0.14 | 1.43 ± 0.18 | 1.8 ± 0.65 | 1.77 ± 0.45 |
| Putrescine | 1 ± 0.27 | 1.19 ± 0.64 | 0.95 ± 0.61 | 1.21 ± 0.35 | 1.08 ± 0.68 | 1.45 ± 0.43 |
| Shikimate | 1 ± 0.13 | 1.03 ± 0.24 | 1 ± 0.08 | 0.9 ± 0.14 | 0.96 ± 0.24 | 0.98 ± 0.25 |
| Spermidine | 1 ± 0.14 | 1.08 ± 0.86 | 0.85 ± 0.36 | 1.38 ± 0.33 | 1.61 ± 0.2 | 2.51 ± 0.5 |
Leaf discs were cut directly from 4-week-old plants, washed three times with 10 mM MES-KOH (pH 6.5), and then incubated during up to 4 h in 10 mM MES-KOH buffer (pH 6.5) containing 2.0 mM glucose and 100 μM of SP or CESP. Metabolites were determined as described in the Section “Materials and Methods.” Data are normalized with respect to the mean response calculated for the control at 2 h. Values presented are the mean ± SE of six biological replicates; values set in bold and underline type were judged to be significantly different from the control (P < 0.05), following the performance of Student’s t-tests.
Table A2.
Assays of the key enzymes related to fermentation process following inhibition of 2-OGDHC by phosphonate analogous.
| Control | SP | CESP | |
|---|---|---|---|
| μmol min−1 g−1 FW | |||
| Alcohol dehydrogenase | 38.2 ± 4.1 | 35.9 ± 2.9 | 41.5 ± 4.1 |
| Alanine amino transferase | 98.7 ± 10.7 | 82.9 ± 17.4 | 97.5 ± 9.9 |
Fully expanded leaves of 4-week-old were harvested at the middle of the light period, washed three times with 10 mM MES-KOH (pH 6.5), and then incubated during 4 h in similar light growth conditions in 10 mM MES-KOH buffer (pH 6.5) containing 100 μM of SP or CESP. Values are presented as the mean ±SE of six biological replicates.
Table A3.
Expression values of up-regulated genes between control and SP-treated samples*.
| Proeb ID | AGI | FC | Gene annotation |
|---|---|---|---|
| 260804_at | At1g78410 | 2.50 | VQ motif-containing protein |
| 248676_at | At5g48850 | 1.90 | Male sterility MS5 family protein |
| 254343_at | At4g21990 | 1.89 | APR3 (APS REDUCTASE 3) |
| 249752_at | At5g24660 | 1.78 | Similar to unknown protein |
| 246884_at | At5g26220 | 1.77 | ChaC-like family protein |
| 256577_at | At3g28220 | 1.71 | Meprin and TRAF homology domain-containing protein |
| 245928_s_at | At5g24770 | 1.71 | VSP2 (VEGETATIVE STORAGE PROTEIN 2); acid phosphatase] |
| 248961_at | At5g45650 | 1.70 | Subtilase family protein |
| 245524_at | At4g15920 | 1.67 | Nodulin MtN3 family protein |
| 255622_at | At4g01070 | 1.67 | GT72B1; UDP-glucosyltransferase |
| 250502_at | At5g09590 | 1.66 | mtHSC70-2 (HEAT SHOCK PROTEIN 70) |
| 265530_at | At2g06050 | 1.65 | OPR3 (OPDA-REDUCTASE 3); 12-oxophytodienoate reductase |
| 256366_at | At1g66880 | 1.63 | Serine/threonine protein kinase family protein |
| 256745_at | At3g29360 | 1.62 | UDP-glucose 6-dehydrogenase, putative |
| 252496_at | At3g46790 | 1.60 | CRR2 (CHLORORESPIRATORY REDUCTION 2) |
| 256454_at | At1g75280 | 1.60 | Isoflavone reductase, putative |
| 254042_at | At4g25810 | 1.59 | XTR6 (XYLOGLUCAN ENDOTRANSGLYCOSYLASE 6) |
| 254432_at | AT4G20830 | 1.58 | FAD-binding domain-containing protein |
| 244982_at | Atcg00780 | 1.58 | Encodes a chloroplast ribosomal protein L14 |
| 247415_at | At5g63060 | 1.57 | Transporter |
| 261522_at | At1g71710 | 1.56 | Inositol polyphosphate 5-phosphatase, putative |
| 254789_at | At4g12880 | 1.56 | Plastocyanin-like domain-containing protein |
| 261177_at | At1g04770 | 1.55 | Male sterility MS5 family protein |
| 254563_at | At4g19120 | 1.54 | ERD3 (EARLY RESPONSIVE TO DEHYDRATION 3) |
| 253215_at | At4g34950 | 1.54 | Nodulin family protein |
| 245026_at | Atcg00140 | 1.54 | ATPase III subunit |
| 253129_at | At4g36020 | 1.54 | CSDP1 (COLD SHOCK DOMAIN PROTEIN 1) |
| 256461_s_at | At1g36280 | 1.54 | Adenylosuccinate lyase, putative |
| 249918_at | At5g19240 | 1.53 | Identical to uncharacterized GPI-anchored protein |
| 266295_at | At2g29550 | 1.52 | TUB7 (tubulin beta-7 chain) |
| 265058_s_at | At1g52030 | 1.52 | MBP2 (MYROSINASE-BINDING PROTEIN 2) |
| 248036_at | At5g55920 | 1.52 | Nucleolar protein, putative |
| 253687_at | At4g29520 | 1.51 | Unknown protein |
Fully expanded leaves of 4-week-old were harvested at the middle of the light period, washed three times with 10 mM MES-KOH (pH 6.5), and then incubated in presence or absence of 100 μM of SP during 4 h in 10 mM MES-KOH buffer (pH 6.5) containing 2.0 mM glucose. Transcriptome analysis was performed using Affymetrix ATH1 GeneChip array (22810 probes). Only statistically significant (P flag in all experiments and detection P-value ≤0.005) genes were presented. FC, fold change of SP/control.
*Genes were extracted from the list of genes exhibiting significant changes in expression between SP-treated and control samples, and were consistently down regulated in all samples. Average transcript levels were calculated from three independent replicates of Affymetrix ATH1 GeneChips. The full data set is additionally available as Data Set S1 in Supplementary Material.
Table A4.
Expression values of down regulated of genes between control and SP-treated samples*.
| Proeb ID | AGI | FC | Gene annotation |
|---|---|---|---|
| 264524_at | At1g10070 | 0.41 | ATBCAT-2; branched-chain amino acid transaminase |
| 252415_at | At3g47340 | 0.50 | ASN1; dark inducible 6 |
| 248050_at | At5g56100 | 0.50 | Glycine-rich protein |
| 267035_at | At2g38400 | 0.51 | AGT3; alanine glyoxylate aminotransferase 3 |
| 264261_at | At1g09240 | 0.54 | TNAS3; nicotianamine synthase 3 |
| 261135_at | At1g19610 | 0.55 | LCR78/PDF1.4; low-molecular-weight cysteine-rich 78 |
| 267138_s_at | At2g38230 | 0.55 | ATPDX1.1; pyridoxine biosynthesis 1.1 |
| 245349_at | At4g16690 | 0.58 | ATMES16; methyl esterase 16 |
| 264788_at | At2g17880 | 0.58 | DNAJ heat shock protein, putative |
| 255155_at | No match | 0.59 | No_match |
| 253754_at | At4g29020 | 0.59 | Glycine-rich protein |
| 251272_at | At3g61890 | 0.60 | ATHB-12; Arabidopsis homeobox protein 12 |
| 263118_at | At1g03090 | 0.60 | MCCA; 3-methylcrotonyl-CoA carboxylase 1 |
| 264782_at | At1g08810 | 0.60 | MYB60; Myb transcription factor |
| 262635_at | At1g06570 | 0.60 | PDS1; Phytoene desaturation 1 |
| 251539_at | At3g58690 | 0.60 | Protein kinase family protein |
| 256940_at | At3g30720 | 0.61 | Unknown protein |
| 250032_at | At5g18170 | 0.61 | GDH1; GLUTAMATE DEHYDROGENASE 1 |
| 266984_at | At2g39570 | 0.61 | ACT domain-containing protein |
| 252570_at | At3g45300 | 0.62 | IVD; ISOVALERYL COA DEHYDROGENASE |
| 251356_at | At3g61060 | 0.62 | ATPP2-A13; phloem protein 2-A13 |
| 262010_at | At1g35612 | 0.62 | Ulp1 protease family protein |
| 246051_at | At5g28840 | 0.62 | GME; GDP-d-mannose 3′,5′-epimerase |
| 258527_at | At3g06850 | 0.63 | DIN3/LTA1; dark inducible 3, alpha-ketoacid dehydrogenase |
| 263265_at | At2g38820 | 0.63 | Unknown protein |
| 248315_at | At5g52630 | 0.63 | Pentatricopeptide (PPR) repeat-containing protein |
| 253373_at | At4g33150 | 0.64 | LKR; Saccharopine dehydrogenase |
| 254877_at | At4g11640 | 0.64 | ATSR; Arabidopsis signal responsive, serine racemase |
| 251642_at | At3g57520 | 0.64 | ATSIP2; Arabidopsis seed imbibition 2, O-glycoside hydrolase |
| 246060_at | At5g08420 | 0.64 | RNA-binding KH domain-containing protein |
| 265387_at | At2g20670 | 0.64 | Unknown protein |
| 259976_at | At1g76560 | 0.64 | CP12-3; CP12 domain-containing protein 3 |
| 254561_at | At4g19160 | 0.64 | Unknown protein |
| 250482_at | At5g10320 | 0.65 | Unknown protein |
| 249133_at | At5g43130 | 0.65 | TAF4; TBP-associated factor 4 |
| 251650_at | At3g57360 | 0.65 | Unknown protein |
| 250580_at | At5g07440 | 0.65 | GDH2; glutamate dehydrogenase 2 |
| 248913_at | At5g45760 | 0.65 | WD-40 repeat family protein |
| 258095_at | At3g23610 | 0.65 | DsPTP1; dual specificity protein phosphatase |
| 247398_at | At5g62950 | 0.66 | RNA polymerase II, Rpb4 |
| 262761_at | At1g10720 | 0.66 | BSD domain-containing protein |
| 247783_at | At5g58800 | 0.66 | Quinone reductase family protein |
| 259934_at | At1g71340 | 0.66 | ATGDPD4; glycerophosphoryl diester phosphodiesterase 4 |
Fully expanded leaves of 4-week-old were harvested at the middle of the light period, washed three times with 10 mM MES-KOH (pH 6.5), and then incubated in presence or absence of 100 μM of SP during 4 h in 10 mM MES-KOH buffer (pH 6.5). Transcriptome analysis was performed using Affymetrix ATH1 GeneChip array (22810 probes). Only statistically significant (P flag in all experiments and detection P-value ≤0.005) genes were presented. FC, fold change of SP/control.
*Genes were extracted from the list of genes exhibiting significant changes in expression between SP-treated and control samples, and were consistently down regulated in all samples. Average transcript levels were calculated from three independent replicates of Affymetrix ATH1 GeneChips. The full data set is additionally available as Data Set S1 in Supplementary Material.
Footnotes
References
- Alhagdow M., Mounet F., Gilbert L., Nunes-Nesi A., Garcia V., Just D., Petit J., Beauvoit B., Fernie A. R., Rothan C., Baldet P. (2007). Silencing of the mitochondrial ascorbate synthesizing enzyme L-galactono-1,4-lactone dehydrogenase affects plant and fruit development in tomato. Plant Physiol. 145, 1408–1422 10.1104/pp.107.106500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelovici R., Fait A., Fernie A. R., Galili G. (2011). A seed high-lysine trait is negatively associated with the TCA cycle and slows down Arabidopsis seed germination. New Phytol. 189, 148–159 10.1111/j.1469-8137.2010.03478.x [DOI] [PubMed] [Google Scholar]
- Araújo W. L., Ishizaki K., Nunes-Nesi A., Larson T. R., Tohge T., Krahnert I., Witt S., Obata T., Schauer N., Graham I. A., Leaver C. J., Fernie A. R. (2010). Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22, 1549–1563 10.1105/tpc.110.075630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo W. L., Nunes-Nesi A., Nikoloski Z., Sweetlove L. J., Fernie A. R. (2012). Metabolic control and regulation of the tricarboxylic acid cycle in photosynthetic and heterotrophic plant tissues. Plant Cell Environ. 35, 1–21 10.1111/j.1365-3040.2011.02332.x [DOI] [PubMed] [Google Scholar]
- Araújo W. L., Nunes-Nesi A., Trenkamp S., Bunik V. I., Fernie A. R. (2008). Inhibition of 2-oxoglutarate dehydrogenase in potato tuber suggests the enzyme is limiting for respiration and confirms its importance in nitrogen assimilation. Plant Physiol. 148, 1782–1796 10.1104/pp.108.126219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo W. L., Tohge T., Ishizaki K., Leaver C. J., Fernie A. R. (2011). Protein degradation – an alternative respiratory substrate for stressed plants. Trends Plant Sci. 16, 489–498 [DOI] [PubMed] [Google Scholar]
- Baud S., Bourrellier A. B. F., Azzopardi M., Berger A., Dechorgnat J., Daniel-Vedele F., Lepiniec L., Miquel M., Rochat C., Hodges M., Ferrario-Mery S. (2010). PII is induced by WRINKLED1 and fine-tunes fatty acid composition in seeds of Arabidopsis thaliana. Plant J. 64, 291–303 10.1111/j.1365-313X.2010.04332.x [DOI] [PubMed] [Google Scholar]
- Bourrellier A. B. F., Valot B., Guillot A., Ambard-Bretteville F., Vidal J., Hodges M. (2010). Chloroplast acetyl-CoA carboxylase activity is 2-oxoglutarate-regulated by interaction of PII with the biotin carboxyl carrier subunit. Proc. Natl. Acad. Sci. U.S.A. 107, 502–507 10.1073/pnas.0910097107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunik V. I., Biryukov A. I., Zhukov Y. N. (1992). Inhibition of pigeon breast muscle alpha-ketoglutarate dehydrogenase by phosphonate analogues of alpha-ketoglutarate. FEBS Lett. 303, 197–201 10.1016/0014-5793(92)80518-L [DOI] [PubMed] [Google Scholar]
- Bunik V. I., Denton T. T., Xu H., Thompson C. M., Cooper A. J. L., Gibson G. E. (2005). Phosphonate analogs of α-ketoglutarate inhibit the activity of the α-ketoglutarate dehydrogenase complex isolated from brain and in cultured cells. Biochemistry 44, 10552–10561 10.1021/bi0503100 [DOI] [PubMed] [Google Scholar]
- Bunik V. I., Fernie A. R. (2009). Metabolic control exerted by the 2-oxoglutarate dehydrogenase reaction: a cross-kingdom comparison of the crossroad between energy production and nitrogen assimilation. Biochem. J. 422, 405–421 10.1042/BJ20090722 [DOI] [PubMed] [Google Scholar]
- Bunik V. I., Kabysheva M. S., Klimuk E. I., Storozhevykh T. P., Pinelis V. G. (2009a). Phosphono analogues of 2-oxoglutarate protect cerebellar granule neurons upon glutamate excitotoxicity. Ann. N. Y. Acad. Sci. 1171, 521–529 10.1111/j.1749-6632.2009.04709.x [DOI] [PubMed] [Google Scholar]
- Bunik V. I., Lovat M., Groznaya A., Graf A., Dunaeva T., Trofimova L., Sokolova N. (2009b). Succinyl phosphonate, a protector of the 2-oxoglutarate dehydrogenase complex, corrects behavioral impairments in rats exposed to hypoxia or ethanol. Alzheimers Dement. 5, P476–P477 10.1016/j.jalz.2009.04.718 [DOI] [Google Scholar]
- Caldana C., Degenkolbe T., Cuadros-Inostroza A., Klie S., Sulpice R., Leisse A., Steinhauser D., Fernie A. R., Willmitzer L., Hannah M. A. (2011). High-density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. Plant J. 67, 869–884 10.1111/j.1365-313X.2011.04640.x [DOI] [PubMed] [Google Scholar]
- Carrari F., Nunes-Nesi A., Gibon Y., Lytovchenko A., Loureiro M. E., Fernie A. R. (2003). Reduced expression of aconitase results in an enhanced rate of photosynthesis and marked shifts in carbon partitioning in illuminated leaves of wild species tomato. Plant Physiol. 133, 1322–1335 10.1104/pp.103.026716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.-J., Ferl R. J. (1999). Arabidopsis alcohol dehydrogenase expression in both shoots and roots is conditioned by root growth environment. Plant Physiol. 121, 429–436 10.1104/pp.121.2.429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousins A. B., Bloom A. J. (2004). Oxygen consumption during leaf nitrate assimilation in a C-3 and C-4 plant: the role of mitochondrial respiration. Plant Cell Environ. 27, 1537–1545 10.1111/j.1365-3040.2004.01257.x [DOI] [Google Scholar]
- Dutilleul C., Lelarge C., Prioul J.-L., De Paepe R., Foyer C. H., Noctor G. (2005). Mitochondria-driven changes in leaf NAD status exert a crucial influence on the control of nitrate assimilation and the integration of carbon and nitrogen metabolism. Plant Physiol. 139, 64–78 10.1104/pp.105.066399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist M., Drincovich M. F., Fluegge U.-I., Maurino V. G. (2009). Two D-2-hydroxy-acid dehydrogenases in Arabidopsis thaliana with catalytic capacities to participate in the last reactions of the methylglyoxal and beta-oxidation pathways. J. Biol. Chem. 284, 25026–25037 10.1074/jbc.M109.021253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist M. K. M., Kuhn A., Wienstroer J., Weber K., Jansen E. E. W., Jakobs C., Weber A. P. M., Maurino V. G. (2011). Plant D-2-hydroxyglutarate dehydrogenase participates in the catabolism of lysine especially during senescence. J. Biol. Chem. 286, 11382–11390 10.1074/jbc.M110.194175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fait A., Fromm H., Walter D., Galili G., Fernie A. R. (2008). Highway or byway: the metabolic role of the GABA shunt in plants. Trends Plant Sci. 13, 14–19 10.1016/j.tplants.2007.10.005 [DOI] [PubMed] [Google Scholar]
- Feria Bourrellier A. B., Ferrario-Méry S., Vidal J., Hodges M. (2009). Metabolite regulation of the interaction between Arabidopsis thaliana PII and N-acetyl-l-glutamate kinase. Biochem. Biophys. Res. Commun. 387, 700–704 10.1016/j.bbrc.2009.07.088 [DOI] [PubMed] [Google Scholar]
- Fernie A. R., Aharoni A., Willmitzer L., Stitt M., Tohge T., Kopka J., Carroll A. J., Saito K., Fraser P. D., Deluca V. (2011). Recommendations for reporting metabolite data. Plant Cell 23, 2477–2482 10.1105/tpc.111.086272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie A. R., Roscher A., Ratcliffe R. G., Kruger N. J. (2001). Fructose 2,6-bisphosphate activates pyrophosphate: fructose-6-phosphate 1-phosphotransferase and increases triose phosphate to hexose phosphate cycling in heterotrophic cells. Planta 212, 250–263 10.1007/s004250000386 [DOI] [PubMed] [Google Scholar]
- Fernie A. R., Trethewey R. N., Krotzky A. J., Willmitzer L. (2004). Metabolite profiling: from diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 5, 763–769 10.1038/nrm1451 [DOI] [PubMed] [Google Scholar]
- Ferrario-Mery S., Bouvet M., Leleu O., Savino G., Hodges M., Meyer C. (2005). Physiological characterisation of Arabidopsis mutants affected in the expression of the putative regulatory protein PII. Planta 223, 28–39 10.1007/s00425-005-0063-5 [DOI] [PubMed] [Google Scholar]
- Forchhammer K. (2004). Global carbon/nitrogen control by P-II signal transduction in cyanobacteria: from signals to targets. FEMS Microbiol. Rev. 28, 319–333 10.1016/j.femsre.2003.11.001 [DOI] [PubMed] [Google Scholar]
- Foyer C. H., Noctor G., Hodges M. (2011). Respiration and nitrogen assimilation: targeting mitochondria-associated metabolism as a means to enhance nitrogen use efficiency. J. Exp. Bot. 62, 1467–1482 10.1093/jxb/erq453 [DOI] [PubMed] [Google Scholar]
- Gauthier P. P. G., Bligny R., Gout E., Mahé A., Nogués S., Hodges M., Tcherkez G. G. B. (2010). In folio isotopic tracing demonstrates that nitrogen assimilation into glutamate is mostly independent from current CO2 assimilation in illuminated leaves of Brassica napus. New Phytol. 185, 988–999 10.1111/j.1469-8137.2009.03130.x [DOI] [PubMed] [Google Scholar]
- Geigenberger P., Fernie A. R., Gibon Y., Christ M., Stitt M. (2000). Metabolic activity decreases as an adaptive response to low internal oxygen in growing potato tubers. Biol. Chem. 381, 723–740 10.1515/BC.2000.093 [DOI] [PubMed] [Google Scholar]
- Gibon Y., Blaesing O. E., Hannemann J., Carillo P., Hohne M., Hendriks J. H. M., Palacios N., Cross J., Selbig J., Stitt M. (2004). A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16, 3304–3325 10.1105/tpc.104.025973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf A., Kabysheva M., Klimuk E., Trofimova L., Dunaeva T., Zündorf G., Kahlert S., Reiser G., Storozhevykh T., Pinelis V., Sokolova N., Bunik V. (2009). Role of 2-oxoglutarate dehydrogenase in brain pathologies involving glutamate neurotoxicity. J. Mol. Catal. B Enzym. 61, 80–87 10.1016/j.molcatb.2009.02.016 [DOI] [Google Scholar]
- Gu L., Jones A. D., Last R. L. (2010). Broad connections in the Arabidopsis seed metabolic network revealed by metabolite profiling of an amino acid catabolism mutant. Plant J. 61, 579–590 10.1111/j.1365-313X.2009.04083.x [DOI] [PubMed] [Google Scholar]
- Guo S., Schinner K., Sattelmacher B., Hansen U. P. (2005). Different apparent CO2 compensation points in nitrate- and ammonium-grown Phaseolus vulgaris and the relationship to non-photorespiratory CO2 evolution. Physiol. Plant. 123, 288–301 10.1111/j.1399-3054.2005.00467.x [DOI] [Google Scholar]
- Hanning I., Heldt H. W. (1993). On the function of mitochondrial metabolism during photosynthesis in spinach (Spinacia oleracea L.) leaves (partitioning between respiration and export of redox equivalents and precursors for nitrate assimilation products). Plant Physiol. 103, 1147–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges M. (2002). Enzyme redundancy and the importance of 2-oxoglutarate in plant ammonium assimilation. J. Exp. Bot. 53, 905–916 10.1093/jexbot/53.370.905 [DOI] [PubMed] [Google Scholar]
- Hodges M., Flesch V., Galvez S., Bismuth E. (2003). Higher plant NADP(+)-dependent isocitrate dehydrogenases, ammonium assimilation and NADPH production. Plant Physiol. Biochem. 41, 577–585 10.1016/S0981-9428(03)00062-7 [DOI] [Google Scholar]
- Hsieh M.-H., Lam H.-M., Van De Loo F. J., Coruzzi G. (1998). A PII-like protein in Arabidopsis: putative role in nitrogen sensing. Proc. Natl. Acad. Sci. U.S.A. 95, 13965–13970 10.1073/pnas.95.23.13965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K., Larson T. R., Schauer N., Fernie A. R., Graham I. A., Leaver C. J. (2005). The critical role of Arabidopsis electron-transfer flavoprotein: ubiquinone oxidoreductase during dark-induced starvation. Plant Cell 17, 2587–2600 10.1105/tpc.105.035162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K., Schauer N., Larson T. R., Graham I. A., Fernie A. R., Leaver C. J. (2006). The mitochondrial electron transfer flavoprotein complex is essential for survival of Arabidopsis in extended darkness. Plant J. 47, 751–760 10.1111/j.1365-313X.2006.02826.x [DOI] [PubMed] [Google Scholar]
- Jenner H. L., Winning B. M., Millar A. H., Tomlinson K. L., Leaver C. J., Hill S. A. (2001). NAD malic enzyme and the control of carbohydrate metabolism in potato tubers. Plant Physiol. 126, 1139–1149 10.1104/pp.126.3.1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P., Ninfa A. J. (2009). Sensation and signaling of alpha-ketoglutarate and adenylylate energy charge by the Escherichia coli PII signal transduction protein require cooperation of the three ligand-binding sites within the PII trimer. Biochemistry 48, 11522–11531 10.1021/bi901158h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabysheva M. S., Storozhevykh T. P., Pinelis V. G., Bunik V. I. (2009). Synthetic regulators of the 2-oxoglutarate oxidative decarboxylation alleviate the glutamate excitotoxicity in cerebellar granule neurons. Biochem. Pharmacol. 77, 1531–1540 10.1016/j.bcp.2009.02.001 [DOI] [PubMed] [Google Scholar]
- Kochevenko A., Araújo W. L., Maloney G. S., Tieman D. M., Do P. T., Taylor M. G., Klee H. J., Fernie A. R. (2012). Catabolism of branched chain amino acids supports respiration but not volatile synthesis in tomato fruits. Mol. Plant 5, 366–375 10.1093/mp/ssr108 [DOI] [PubMed] [Google Scholar]
- Kolbe A., Oliver S. N., Fernie A. R., Stitt M., Van Dongen J. T., Geigenberger P. (2006). Combined transcript and metabolite profiling of Arabidopsis leaves reveals fundamental effects of the thiol-disulfide status on plant metabolism. Plant Physiol. 141, 412–422 10.1104/pp.106.081208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka J., Schauer N., Krueger S., Birkemeyer C., Usadel B., Bergmuller E., Dormann P., Weckwerth W., Gibon Y., Stitt M., Willmitzer L., Fernie A. R., Steinhauser D. (2005). GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21, 1635–1638 10.1093/bioinformatics/bti236 [DOI] [PubMed] [Google Scholar]
- Lehmann M., Schwarzländer M., Obata T., Sirikantaramas S., Burow M., Olsen C. E., Tohge T., Fricker M. D., Møller B. L., Fernie A. R., Sweetlove L. J., Laxa M. (2009). The metabolic response of Arabidopsis roots to oxidative stress is distinct from that of heterotrophic cells in culture and highlights a complex relationship between the levels of transcripts, metabolites, and flux. Mol. Plant 2, 390–406 10.1093/mp/ssn080 [DOI] [PubMed] [Google Scholar]
- Lemaitre T., Urbanczyk-Wochniak E., Flesch V., Bismuth E., Fernie A. R., Hodges M. (2007). NAD-dependent isocitrate dehydrogenase mutants of Arabidopsis suggest the enzyme is not limiting for nitrogen assimilation. Plant Physiol. 144, 1546–1558 10.1104/pp.107.100677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Less H., Galili G. (2008). Principal transcriptional programs regulating plant amino acid metabolism in response to abiotic stresses. Plant Physiol. 147, 316–330 10.1104/pp.108.115733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J., Schauer N., Kopka J., Willmitzer L., Fernie A. R. (2006). Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 1, 387–396 10.1038/nprot.2006.59 [DOI] [PubMed] [Google Scholar]
- Little R., Reyes-Ramirez F., Zhang Y., Van Heeswijk W. C., Dixon R. (2000). Signal transduction to the Azotobacter vinelandii NIFL-NIFA regulatory system is influenced directly by interaction with 2-oxoglutarate and the PII regulatory protein. EMBO J. 19, 6041–6050 10.1093/emboj/19.22.6041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M., Nunes-Nesi A., Krueger P., Nagel A., Hannemann J., Giorgi F. M., Childs L., Osorio S., Walther D., Selbig J., Sreenivasulu N., Stitt M., Fernie A. R., Usadel B. (2010). Robin: an intuitive wizard application for R-based expression microarray quality assessment and analysis. Plant Physiol. 153, 642–651 10.1104/pp.109.152553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A. H., Hill S. A., Leaver C. J. (1999). Plant mitochondrial 2-oxoglutarate dehydrogenase complex: purification and characterization in potato. Biochem. J. 343, 327–334 10.1042/0264-6021:3430327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller I. M., Sweetlove L. J. (2010). ROS signalling – specificity is required. Trends Plant Sci. 15, 370–374 10.1016/j.tplants.2010.04.008 [DOI] [PubMed] [Google Scholar]
- Moorhead G. B. G., Smith C. S. (2003). Interpreting the plastid carbon, nitrogen, and energy status. A role for PII? Plant Physiol. 133, 492–498 10.1104/pp.103.025627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninfa A. J., Jiang P. (2005). PII signal transduction proteins: sensors of alpha-ketoglutarate that regulate nitrogen metabolism. Curr. Opin. Microbiol. 8, 168–173 10.1016/j.mib.2005.02.011 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A., Carrari F., Gibon Y., Sulpice R., Lytovchenko A., Fisahn J., Graham J., Ratcliffe R. G., Sweetlove L. J., Fernie A. R. (2007a). Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J. 50, 1093–1106 10.1111/j.1365-313X.2007.03115.x [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A., Sweetlove L. J., Fernie A. R. (2007b). Operation and function of the tricarboxylic acid cycle in the illuminated leaf. Physiol. Plant 129, 45–56 10.1111/j.1399-3054.2006.00778.x [DOI] [Google Scholar]
- Nunes-Nesi A., Carrari F., Lytovchenko A., Smith A. M. O., Loureiro M. E., Ratcliffe R. G., Sweetlove L. J., Fernie A. R. (2005). Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol. 137, 611–622 10.1104/pp.104.055566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A., Fernie A. R., Stitt M. (2010). Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol. Plant 3, 973–996 10.1093/mp/ssq049 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A., Sulpice R., Gibon Y., Fernie A. R. (2008). The enigmatic contribution of mitochondrial function in photosynthesis. J. Exp. Bot. 59, 1675–1684 10.1093/jxb/ern002 [DOI] [PubMed] [Google Scholar]
- Purnell M. P., Skopelitis D. S., Roubelakis-Angelakis K. A., Botella J. R. (2005). Modulation of higher-plant NAD(H)-dependent glutamate dehydrogenase activity in transgenic tobacco via alteration of beta subunit levels. Planta 222, 167–180 10.1007/s00425-005-1510-z [DOI] [PubMed] [Google Scholar]
- Randall D. D., Miernyk J. A. (1990). “The mitochondrial pyruvate dehydrogenase complex,” in Methods in Plant Biochemistry: Enzymes of Primary Metabolism, ed. Lea P. J. (London: Academic Press; ), 175–199 [Google Scholar]
- Redman J. C., Haas B. J., Tanimoto G., Town C. D. (2004). Development and evaluation of an Arabidopsis whole genome Affymetrix probe array. Plant J. 38, 545–561 10.1111/j.1365-313X.2004.02061.x [DOI] [PubMed] [Google Scholar]
- Rhoads D. M., Subbaiah C. C. (2007). Mitochondrial retrograde regulation in plants. Mitochondrion 7, 177–194 10.1016/j.mito.2007.01.002 [DOI] [PubMed] [Google Scholar]
- Rocha M., Licausi F., Araújo W. L., Nunes-Nesi A., Sodek L., Fernie A. R., Van Dongen J. T. (2010). Glycolysis and the tricarboxylic acid cycle are linked by alanine aminotransferase during hypoxia induced by waterlogging of Lotus japonicus. Plant Physiol. 152, 1501–1513 10.1104/pp.109.150045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali U., Liu J. L., Leisse A., Balbo I., Perez-Melis A., Willmitzer L., Fernie A. R. (2004). Kinetics of labelling of organic and amino acids in potato tubers by gas chromatography-mass spectrometry following incubation in (13)C labelled isotopes. Plant J. 39, 668–679 10.1111/j.1365-313X.2004.02157.x [DOI] [PubMed] [Google Scholar]
- Sá Santos S., Gibson G. E., Cooper A. J. L., Denton T. T., Thompson C. M., Bunik V. I., Alves P. M., Sonnewald U. (2006). Inhibitors of the α-ketoglutarate dehydrogenase complex alter [1-13C]glucose and [U-13C]glutamate metabolism in cerebellar granule neurons. J. Neurosci. Res. 83, 450–458 10.1002/jnr.20749 [DOI] [PubMed] [Google Scholar]
- Schauer N., Steinhauser D., Strelkov S., Schomburg D., Allison G., Moritz T., Lundgren K., Roessner-Tunali U., Forbes M. G., Willmitzer L., Fernie A. R., Kopka J. (2005). GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett. 579, 1332–1337 10.1016/j.febslet.2005.01.029 [DOI] [PubMed] [Google Scholar]
- Schippers J. H. M., Nunes-Nesi A., Apetrei R., Hille J., Fernie A. R., Dijkwel P. P. (2008). The Arabidopsis onset of leaf death5 mutation of quinolinate synthase affects nicotinamide adenine dinucleotide biosynthesis and causes early ageing. Plant Cell 20, 2909–2925 10.1105/tpc.107.056341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelp B. J., Mullen R. T., Waller J. C. (2012). Compartmentation of GABA metabolism raises intriguing questions. Trends Plant Sci. 17, 57–59 10.1016/j.tplants.2011.12.006 [DOI] [PubMed] [Google Scholar]
- Shen W., Wei Y., Dauk M., Tan Y., Taylor D. C., Selvaraj G., Zou J. (2006). Involvement of a glycerol-3-phosphate dehydrogenase in modulating the NADH/NAD+ ratio provides evidence of a mitochondrial glycerol-3-phosphate shuttle in Arabidopsis. Plant Cell 18, 422–441 10.1105/tpc.105.039750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q., Risa Ø., Sonnewald U., Gibson G. E. (2009). Mild reduction in the activity of the alpha-ketoglutarate dehydrogenase complex elevates GABA shunt and glycolysis. J. Neurochem. 109, 214–221 10.1111/j.1471-4159.2009.05949.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siehl D. L., Subramanian M. V., Walters E. W., Lee S. F., Anderson R. J., Toschi A. G. (1996). Adenylosuccinate synthetase: site of action of hydantocidin, a microbial phytotoxin. Plant Physiol. 110, 753–758 10.1104/pp.110.3.753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienkiewicz-Porzucek A., Nunes-Nesi A., Sulpice R., Lisec J., Centeno D. C., Carillo P., Leisse A., Urbanczyk-Wochniak E., Fernie A. R. (2008). Mild reductions in mitochondrial citrate synthase activity result in a compromised nitrate assimilation and reduced leaf pigmentation but have no effect on photosynthetic performance or growth. Plant Physiol. 147, 115–127 10.1104/pp.108.117978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson J. P., Clark S. M., Portt A., Allan W. L., Makhmoudova A., Rochon A., Shelp B. J. (2010). γ-Aminobutyrate transaminase limits the catabolism of γ-aminobutyrate in cold-stressed Arabidopsis plants: insights from an overexpression mutant. Botany 88, 522–527 10.1139/B10-020 [DOI] [Google Scholar]
- Smith C. S., Morrice N. A., Moorhead G. B. G. (2004). Lack of evidence for phosphorylation of Arabidopsis thaliana PII: implications for plastid carbon and nitrogen signaling. Biochim. Biophys. Acta 1699, 145–154 [DOI] [PubMed] [Google Scholar]
- Smith C. S., Weljie A. M., Moorhead G. B. G. (2003). Molecular properties of the putative nitrogen sensor PII from Arabidopsis thaliana. Plant J. 33, 353–360 10.1046/j.1365-313X.2003.01634.x [DOI] [PubMed] [Google Scholar]
- Smith C. S., Zaplachinski S. T., Muench D. G., Moorhead G. B. G. (2002). Expression and purification of the chloroplast putative nitrogen sensor, PII, of Arabidopsis thaliana. Protein Expr. Purif. 25, 342–347 10.1016/S1046-5928(02)00018-9 [DOI] [PubMed] [Google Scholar]
- Studart-Guimarães C., Fait A., Nunes-Nesi A., Carrari F., Usadel B., Fernie A. R. (2007). Reduced expression of succinyl-coenzyme A ligase can be compensated for by up-regulation of the γ-aminobutyrate shunt in illuminated tomato leaves. Plant Physiol. 145, 626–639 10.1104/pp.107.103101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulpice R., Sienkiewicz-Porzucek A., Osorio S., Krahnert I., Stitt M., Fernie A. R., Nunes-Nesi A. (2010). Mild reductions in cytosolic NADP-dependent isocitrate dehydrogenase activity result in lower amino acid contents and pigmentation without impacting growth. Amino Acids 39, 1055–1066 10.1007/s00726-010-0617-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove L. J., Beard K. F. M., Nunes-Nesi A., Fernie A. R., Ratcliffe R. G. (2010). Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci. 15, 462–470 10.1016/j.tplants.2010.05.006 [DOI] [PubMed] [Google Scholar]
- Sweetlove L. J., Fait A., Nunes-Nesi A., Williams T., Fernie A. R. (2007). The mitochondrion: an integration point of cellular metabolism and signalling. CRC Crit. Rev. Plant Sci. 26, 17–43 10.1080/07352680601147919 [DOI] [Google Scholar]
- Tcherkez G., Bligny R., Gout E., Mahe A., Hodges M., Cornic G. (2008). Respiratory metabolism of illuminated leaves depends on CO2 and O2 conditions. Proc. Natl. Acad. Sci. U.S.A 105, 797–802 10.1073/pnas.0708947105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkez G., Cornic G., Bligny R., Gout E., Ghashghaie J. (2005). In vivo respiratory metabolism of illuminated leaves. Plant Physiol. 138, 1596–1606 10.1104/pp.105.062141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkez G., Mahe A., Boex-Fontvieille E., Gout E., Guerard F., Bligny R. (2011). Experimental evidence of phosphoenolpyruvate resynthesis from pyruvate in illuminated leaves. Plant Physiol. 157, 86–95 10.1104/pp.111.180711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkez G., Mahe A., Gauthier P., Mauve C., Gout E., Bligny R., Cornic G., Hodges M. (2009). In folio respiratory fluxomics revealed by (13)C isotopic labeling and H/D isotope effects highlight the noncyclic nature of the tricarboxylic acid “cycle” in illuminated leaves. Plant Physiol. 151, 620–630 10.1104/pp.109.142976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton G. W., Moorhead G. B. G. (2004). A renaissance of metabolite sensing and signaling: from modular domains to riboswitches. Plant Cell 16, 2252–2257 10.1105/tpc.104.160930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman D., Taylor M., Schauer N., Fernie A. R., Hanson A. D., Klee H. J. (2006). Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proc. Natl. Acad. Sci. U.S.A. 103, 8287–8292 10.1073/pnas.0602469103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trofimova L., Lovat M., Groznaya A., Efimova E., Dunaeva T., Maslova M., Graf A., Bunik V. (2010). Behavioral impact of the regulation of the brain 2-oxoglutarate dehydrogenase complex by synthetic phosphonate analog of 2-oxoglutarate: implications into the role of the complex in neurodegenerative diseases. Int. J. Alzheimers Dis. 10.4061/2010/749061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrig R. G., Ng K. K. S., Moorhead G. B. G. (2009). PII in higher plants: a modern role for an ancient protein. Trends Plant Sci. 14, 505–511 10.1016/j.tplants.2009.07.003 [DOI] [PubMed] [Google Scholar]
- Usadel B., Nagel A., Steinhauser D., Gibon Y., Blaesing O. E., Redestig H., Sreenivasulu N., Krall L., Hannah M. A., Poree F., Fernie A. R., Stitt M. (2006). PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7, 535. 10.1186/1471-2105-7-535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B., Nagel A., Thimm O., Redestig H., Blaesing O. E., Palacios-Rojas N., Selbig J., Hannemann J., Piques M. C., Steinhauser D., Scheible W. R., Gibon Y., Morcuende R., Weicht D., Meyer S., Stitt M. (2005). Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol. 138, 1195–1204 10.1104/pp.105.060459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Merwe M. J., Osorio S., Araújo W. L., Balbo I., Nunes-Nesi A., Maximova E., Carrari F., Bunik V. I., Persson S., Fernie A. R. (2010). Tricarboxylic acid cycle activity regulates tomato root growth via effects on secondary cell wall production. Plant Physiol. 153, 611–621 10.1104/pp.109.149047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal E. A., Gutierrez R. A. (2008). A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr. Opin. Plant Biol. 11, 521–529 10.1016/j.pbi.2008.07.003 [DOI] [PubMed] [Google Scholar]
- Zündorf G., Kahlert S., Bunik V. I., Reiser G. (2009). α-Ketoglutarate dehydrogenase contributes to production of reactive oxygen species in glutamate-stimulated hippocampal neurons in situ. Neuroscience 158, 610–616 10.1016/j.neuroscience.2008.10.015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full transcriptome data set.