SUMMARY
Piwi Argonautes and Piwi-interacting RNAs (piRNAs) mediate genome defense by targeting transposons. However, many piRNA species lack obvious sequence complementarity to transposons or other loci; only one C. elegans transposon is a known piRNA target. Here we show that, in mutants lacking the Piwi Argonaute PRG-1 (and consequently its associated piRNAs/21U-RNAs), many silent loci in the germline exhibit increased levels of mRNA expression and depletion of an amplified RNA-dependent RNA polymerase (RdRP)-derived species of small secondary RNA termed 22G-RNAs. Sequences depleted of 22G-RNAs are enriched at nearby potential target sites that base pair imperfectly but extensively to 21U-RNAs. We show that PRG-1 is required to initiate, but not to maintain, silencing of transgenes engineered to contain complementarity to endogenous 21U-RNAs. Our findings support a model in which C. elegans piRNAs utilize their enormous repertoire of targeting capacity to scan the germline transcriptome for foreign sequences, while endogenous germline-expressed genes are actively protected from piRNA-induced silencing.
INTRODUCTION
RNAi-related pathways play important roles in genome surveillance and fertility (Aravin et al., 2007; Ghildiyal and Zamore, 2009; Thomson and Lin, 2009). Argonaute (AGO) proteins bound to small RNA cofactors, 20-30 nucleotides (nt) in length, are the key effectors of RNAi pathways (Carmell et al., 2002). Structurally related to RNase H, AGOs are thought to present nts 2-8 (the seed region) of the guide RNA with a preformed helical pitch that is likely to reduce the free energy of initial base pairing with potential targets (Parker et al., 2005; Song et al., 2004). Base pairing between the seed region and target is thought to drive further pairing that positions the backbone of the target RNA within the DDH catalytic triad of AGO, which mediates target-strand cleavage and leaves the guide strand intact for additional rounds of targeting.
Not all AGOs encode a functional RNase H (or Slicer) domain. Moreover, catalytically competent AGOs do not always cleave their targets to silence gene expression (Liu et al., 2004; Yigit et al., 2006). In animals, mismatched nucleotides and G:U wobble base pairs are a common feature of micro (mi) RNA-mediated silencing (Bartel, 2009). During miRNA-mediated silencing, AGOs are thought to recruit accessory factors that block translation and/or increase mRNA turnover (Bartel, 2009).
AGOs related to Drosophila Piwi are key regulators of germline development (Lin and Spradling, 1997) and genome integrity in animals (Siomi et al., 2011). Piwi proteins engage small RNAs (piRNAs) complementary to transposons and silence transposons in the germline (Aravin et al., 2007; Thomson and Lin, 2009). In C. elegans an expanded group of Worm-specific AGOs (WAGOs) interact with small RNAs (22G-RNAs) synthesized by RNA-dependent RNA polymerase (RdRP) and silence transposons, pseudogenes and other loci in the germline (Gu et al., 2009). WAGOs also function downstream of the Argonaute RDE-1 in the response to exogenous double-stranded RNA (exo-RNAi). However, RDE-1 is not required for transposon silencing, nor is it required for endogenous silencing of most WAGO loci, so the upstream triggers (if any) that initiate 22G-RNA biogenesis are unknown in the majority of cases.
The C. elegans piwi-related gene, prg-1, regulates germline development and fertility, but it seems to have a limited role in transposon silencing (Batista et al., 2008; Cox et al., 1998; Das et al., 2008). Tc3 appears to be the only transposon family silenced by PRG-1 (Das et al., 2008), and PRG-1 interacts with piRNAs (or 21U-RNAs) complementary to Tc3 but not other transposons (Batista et al., 2008). Importantly, desilencing of Tc3 in a prg-1 mutant correlates with a reduction in WAGO-associated 22G-RNAs targeting Tc3. Because silencing of Tc3 is also dependent on the WAGO pathway, these findings suggest that piRNAs can recruit RdRP to generate 22G-RNAs and initiate the WAGO silencing pathway (Batista et al., 2008; Gu et al., 2009).
In C. elegans, over 15,000 21U-RNAs are expressed from two large clusters on chromosome IV (Ruby et al., 2006; Batista et al., 2008). The 21U-RNAs resemble mammalian “pachytene” piRNAs that are also expressed from large genomic clusters (Aravin et al., 2006; Batista et al., 2008; Das et al., 2008; Lau et al., 2006; Ruby et al., 2006). These worm and mammalian piRNA species are remarkable, in that most lack an obvious target; except for Tc3 piRNAs, nearly all of the C. elegans piRNAs lack extensive sequence complementarity to transposon or other endogenous gene targets. Nevertheless, the sequence diversity of 21U-RNAs and mammalian meiotic piRNAs is such that, if relaxed (miRNA-like) base pairing was allowed, then these piRNAs could target all mRNAs expressed in the germline.
Here we show that prg-1 mutants, which lack piRNAs, exhibit a striking depletion of a subset of RdRP-derived 22G-RNA species. Regions depleted of 22G-RNAs are enriched for sequences with potential for energetically-favored base-pairing to 21U-RNA species. By engineering transgenes containing complementarity to endogenous 21U-RNAs, we show that 21U-RNAs can induce silencing and do so through the WAGO-22G pathway. Interestingly, PRG-1 initiates but is not required to maintain trans-generational silencing on a transgene target. Together, our data support a model in which C. elegans piRNAs collaborate with the secondary WAGO-amplification system to initiate and reinforce trans-generational silencing on foreign and certain endogenous sequences. Furthermore, our findings suggest that other endogenous germline-expressed genes, targeted by CSR-1-22G-RNAs, are resistant to piRNA-induced silencing.
RESULTS
prg-1 mutants primarily affect the WAGO 22G-RNA pathway
Previous studies indicated that the Tc3 transposon is expressed and mobile in the germline of a prg-1 mutant (Batista et al., 2008; Das et al., 2008). Importantly, 22G-RNAs targeting Tc3 were reduced proximal to, and on the same strand as, a 21U-RNA residing within the inverted repeat of Tc3. These findings raised the possibility that targeting of Tc3 by a PRG-1-21U-RNA complex can recruit RdRP to initiate WAGO-22G-dependent silencing (Batista et al., 2008; Das et al., 2008; Gu et al., 2009).
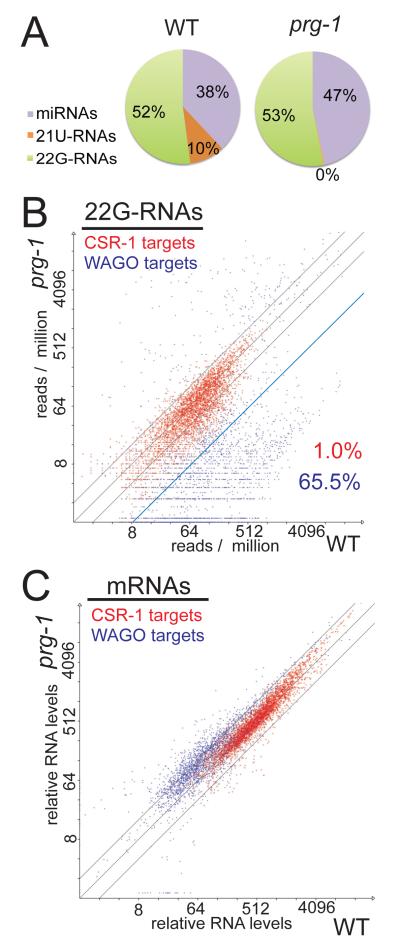
To further explore a link between 21U-RNAs and 22G-RNA accumulation, we first asked whether 22G-RNA levels were altered in prg-1 mutants (where 21U-RNAs are absent). When all 22G-targeted loci were considered together, we failed to observe a clear correlation between prg-1 activity and 22G-RNA levels (Figure 1A; Batista et al., 2008). However, recent reports have shown that 22G-RNA loci can be sorted into at least 4 distinct AGO pathways; the CSR-1 pathway, the WAGO pathway, the Eri (ERGO-1-WAGO) pathway, and the Eri (ALG-3/4-WAGO) pathway (Claycomb et al., 2009; Conine et al., 2010; Gu et al., 2009; Han et al., 2009; Maniar and Fire, 2011; Vasale et al., 2010). We therefore examined the fraction of loci in each pathway with increased, unchanged or decreased 22G-RNA levels in prg-1 mutants vs wild type (Figures 1B and S1). Strikingly, we found that non-Eri WAGO targets exhibited a strong tendency toward depletion of 22G-RNAs (Figure 1B). The other pathways (CSR-1, ERGO-1 and ALG-3/4) exhibited no clear trend, with most loci unchanged.
Figure 1. prg-1 primarily affects the WAGO-22G-RNA pathway.
(A) Pie charts showing the abundance of small RNA species in wild type (WT) and prg-1(n4357) mutants. (B,C) Scatter plots showing the abundance of 22G-RNAs (B) and target mRNAs (C) associated with WAGO (blue) or CSR-1 (red) targets in wild type (WT) compared to the prg-1 mutant. The two dashed diagonal lines indicate two-fold enrichment or depletion. In (B) the solid blue diagonal line indicates eight-fold depletion in the prg-1 mutant, and the percentage of WAGO and CSR-1 targets that exhibit 8-fold, or greater, depletion in 22G-RNA levels is indicated. See also Figure S1.
Consistent with the idea that WAGO 22G-RNAs negatively regulate their targets, we observed an overall trend toward increased mRNA expression of WAGO mRNA targets in prg-1 mutant worms (Figure 1C). By contrast, the expression of CSR-1 targets was not changed in prg-1 mutants (Figure 1C). Taken together, these findings indicate that PRG-1 is required to silence a subset of WAGO targets in the germline.
Production of 22G RNAs around predicted 21U-RNA targets
We noticed that 22G-RNAs are not distributed randomly along many endogenous target mRNAs, but instead exhibit significant hotspots where specific 22G-RNA species are tens or hundreds of times more abundant than elsewhere in the same target (Figures 2 and S2). We wondered if these hotspots might represent regions where RdRP is recruited by PRG-1/21U-RNA targeting. Only 29 WAGO targets exhibit perfect complementarity to annotated 21U-RNAs (Table S1). Therefore, we reasoned that if 21U-RNAs are directly responsible for the accumulation of the WAGO 22G-RNA species, they must do so through imperfect base-pairing interactions. We therefore decided to ask if 22G-RNA levels exhibit enrichment over mRNA coding regions with the potential for imperfect but energetically favorable base pairing with 21U-RNAs. Studies on miRNA target interactions indicate the importance of strong base pairing in the seed region, residues 2-8, and also suggest that a limited amount of G:U pairing is tolerated (Bartel, 2009). We chose parameters for pairing that were higher in stringency than that observed for most miRNA/target interactions, but sufficiently relaxed to allow an average of two piRNA sites in an average sized gene (Table 1). This involved allowing a total of, at most, two mismatched pairs and one G:U pair outside the seed region, with no mismatches (and at most one G:U pair) within the seed region. We then determined the level of 22G-RNAs within a 100 nt window of sequence centered around each potential 21U-RNA complementary site in wild type and prg-1 mutants. In addition, we separately considered 21U-RNAs matching WAGO targets and CSR-1 targets.
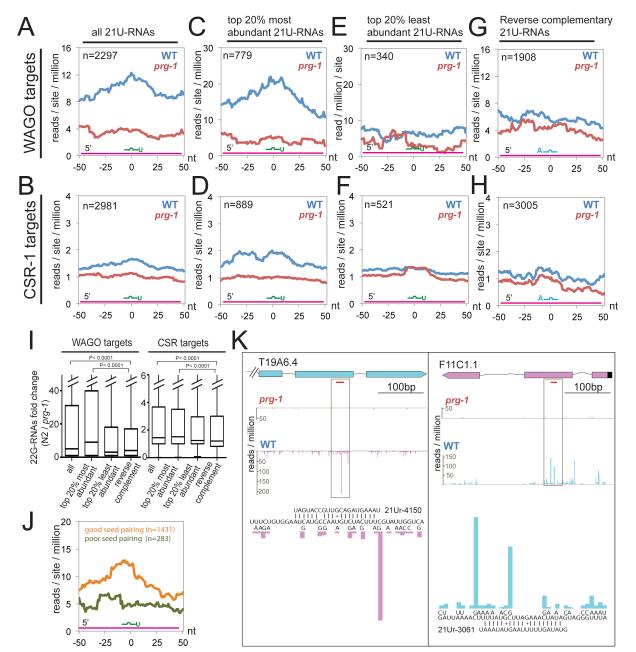
Figure 2. 22G-RNAs are enriched near predicted 21U-RNA target sites.
(A-H) Density of 22G-RNAs within a 100 nt window around predicted 21U-RNA (or control) target sites in the wild type (blue) or prg-1 mutant (red). The plots are centered on the 10th nucleotide of the small RNA shown schematically in each graph. WAGO targets (A,C,E,F) and CSR-1 targets (B,D,F,H) were analyzed separately. All 21U-RNAs (A,B), the top 20% most-(C,D) and least-abundant (E,F), as well as, binding sites of hypothetical reverse-complement 21U-RNAs (G,H) were analyzed separately (as indicated). (I) Box and whisker plots showing the fold change of 22G RNA levels for each category of target/small RNA interaction as indicated. The bottom and top of each box represents the value of the 25th and 75th percentile, the horizontal line inside the box represent the median value. The p-value was calculated using a one-sided t-test. (J) Density of 22G-RNAs at 21U-RNA predicted target sites with good or poor seed matches. (K) Distribution of 22G-RNAs at two predicted 21U-RNA targets in a prg-1 mutant and in wild type. The bars indicate the position of the first nucleotide, and the relative abundance of each 22G-RNA species. The positions of predicted 21U-RNA pairing sites are highlighted (red). Base-pairing alignments for the boxed regions are shown at single-nucleotide resolution below each diagram. See also Figure S2.
Table 1.
Target capacity of 21U-RNAs under different targeting parameters
| Number of mismatches | Number of genes targeted (%) |
Number of sites per gene |
||
|---|---|---|---|---|
| Maximum | Mismatch type | |||
| 0 | 0 | 289 (1.4%) | 1.6 | |
| 1 | 1 non-G:U | 355 (1.8%) | 1.5 | |
| 2 | 2 non-G:U | 1171 (5.8%) | 1.5 | |
| 3 | 3 non-G:U | 7670 (38.1%) | 2.1 | |
| 4 | 3 non-G:U + 1 G:U | 17934 (89.2%) | 6.7 | |
| Seed | Non-Seed | |||
| 4 | 1 G:U | 2 non-G:U + 1 G:U | 10969 (54.6%) | 2.0 |
| 6 | 2 G:U | 2 non-G:U + 2 G:U | 18131 (90.1%) | 6.7 |
This analysis revealed a significant enrichment of 22G-RNAs in wild type relative to prg-1 mutant animals for 22G-RNA production within +/− 50 nts of predicted 21U-binding sites at both WAGO targets and CSR-1 targets (Figure 2A-J). When all annotated 21U-RNAs were considered, we observed an approximately 3-fold enrichment of 22G-RNAs in wild type over prg-1 mutant animals, with a peak directly over the predicted 21U-binding sites in WAGO targets (Figure 2A,I). By contrast, for CSR-1 targets we observed a much more modest number of reads, and a median enrichment of approximately 1.4-fold (Figure 2B,I).
Not all predicted 21U-RNAs are expressed at equal levels, which led us to ask if local 22G-RNA levels were correlated with 21U-RNA expression levels. Strikingly, when only the most abundant (top 20%) 21U-RNA species were considered, we found that the number of reads per million in wild type increased dramatically, with a median enrichment of nearly 9-fold for 22G-RNA levels in wild type relative to the prg-1 mutant with a peak, once again, situated directly over the 21U-RNA complementary region (Figure 2C,I). By contrast, for CSR-1 targets there was, at most, a 2-fold increase in reads per million with a median increase of 1.5-fold (Figure 2D,I). No enrichment was observed when the least abundant 20% of 21U-RNAs were considered (Figure 2E,F) or when a conceptual library of reverse-complement 21U-RNA sequences was used to predict potential binding sites (Figure 2G-I).
In the above analysis we required nearly perfect seed pairing (at most one G:U pair). To examine the consequences of mismatches within the seed region, we identified a control set of hypothetical 21U-RNA/target interactions with similar overall base pairing potential but with very poor seed pairing. Consistent with the idea that seed pairing is important for targeting, we found little enrichment in 22G-RNA levels associated with poor seed-matched 21U-RNA/target pairs (Figure 2J).
As expected from the global analysis above, when specific potential-21U-RNA/target interactions were examined at single-nucleotide resolution, we observed peaks in 22G-RNA levels that are proximal to or overlap with predicted 21U-RNA target sites (Figures 2K and S2). Thus, high-affinity 21U-RNA-sites are correlated with the local production of 22G-RNAs on WAGO targets.
Nevertheless we were surprised to find that at least 89 WAGO targets failed to show prg-1 dependent accumulation of 22G-RNAs despite the presence of one or more high-affinity 21U-RNA binding sites (Table S1). This finding suggests that other unknown factors must influence 22G-RNA biogenesis on WAGO targets (See Discussion).
PRG-1 Slicer activity is not necessary for the accumulation of most 22G-RNAs
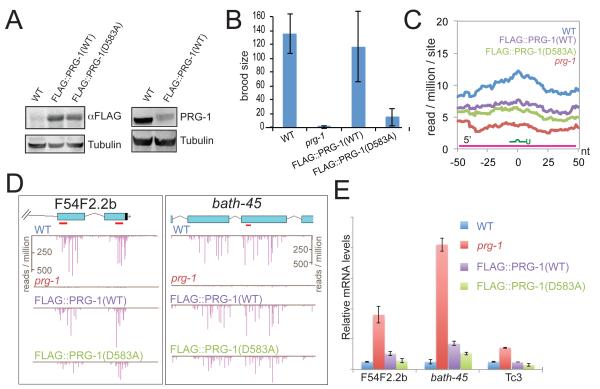
To ask if an intact Slicer domain is required for prg-1 activity, we mutated the first aspartic acid of the DDH catalytic triad of PRG-1 (D583A). Mutations in the corresponding aspartic acid have been shown to abolish Slicer activity of other AGO proteins (Liu et al., 2004; Maiti et al., 2007; Steiner et al., 2009). We generated flag::prg-1(WT) and flag::prg-1(D583A) transgene constructs and introduced them into a defined chromosomal locus using homologous recombination (Frokjaer-Jensen et al., 2008). Both transgenes were expressed at approximately 15% of wild-type PRG-1 level (Figure 3A), and both transgenes rescued 21U-RNA levels to a similar degree in a prg-1 mutant background (Figure S3). However, when we examined fertility in the rescued strains, we found that the Slicer defective transgenic lines were consistently less fertile than the wild-type rescued lines (Figure 3B).
Figure 3. Slicer-independent regulation of WAGO targets by PRG-1.
(A) Western blots of protein isolated from WT and prg-1 mutant transgenic strains (as indicated), probed with anti-FLAG antibody (left panel), anti-PRG-1 polyclonal antibody (right panel), and anti-tubulin antibody as a loading control. (B) Graphic representation of the brood size (at 25°C) observed for the wild type (WT), prg-1 mutant or prg-1 mutant rescued with WT or Slicer-deficient flag::prg-1 transgenes (as indicated). Error bars represent the standard deviation of the mean. (C) Graphic representation of 22G-RNA levels within a 100 nt window near predicted 21U-RNA target sites in the indicated strains. The base-paring parameters were as in Figure 2A-H). (D) Two examples of predicted 21U-RNA targets with the positions of 21U-RNAs highlighted (red) below the gene diagrams. The graphs show 22G-RNA levels in the WT, prg-1 mutant and rescued strains as indicated. (E) Quantitative RT-PCR analysis of three different predicted targets in the indicated strains. Error bars represent the standard deviation of the mean. See also Figure S3.
We next repeated the analysis described in Figure 2 to determine if the rescued strains restore the level of 22G-RNAs near predicted 21U-RNA target sites. Interestingly, both WT and D583A transgenes partially rescued the 22G-RNA accumulation (Figure 3C,D). Using quantitative PCR to analyze the mRNA levels of three target genes, we found that both the WT and D583A transgenes rescued the silencing defect of prg-1 to a similar degree (Figure 3E). These findings suggest that Slicer activity is not necessary for the rescue of most prg-1-dependent 22G-RNAs. However, we did find specific targets where 22G-RNA levels were either up or down in the D583A mutant relative to WT (Table S2), and it remains possible that one or more of these targets are responsible for the failure of the D583A mutant transgenes to rescue fertility.
PRG-1 is required for initiation but not maintenance of WAGO-22G-RNA-dependent trans-generational silencing
The above results suggest that some, but not all, WAGO targets are subject to prg-1-dependent 22G-induction and silencing. To further explore the consequences of 21U-RNA targeting, we engineered pie-1-promoter driven GFP::Histone H2B reporters with or without two 21U-RNA complementary sites embedded in the 3′UTR of the muscle myosin gene, unc-54 (See Experimental Procedures). We chose this reporter cassette because previous work had shown that transgenes driven with this reporter backbone have a low frequency of spontaneous silencing (data not shown). We used the MosSCI method to ensure that the transgenes are single-copy and are inserted at the same chromosomal site (Frokjaer-Jensen et al., 2008). We found that control reporters, with no 21U-RNA target sequences or with the reverse-complement sequences were always expressed (Figure 4A-C). However, reporters containing 21U-RNA target sites were always silenced when introduced into the wild-type background, but were expressed when introduced directly into the prg-1 mutant background (Figure 4A,C). We also tested reporters that contain a mismatch at nucleotide 10 relative to the 21U-RNA sequence, which has been shown to dramatically reduce the ability of Argonaute to slice a target (Martinez and Tuschl, 2004; Shin et al., 2010). Again, we found that all lines (4 of 4) containing the 21U-RNA mismatch reporters were completely silenced when introduced into wild-type animals (Figure 4C). Together these findings suggest that endogenous 21U-RNAs can silence a reporter by base pairing with complementary sequences in the 3′ UTR. Moreover, a mismatch at position 10 of the 21U-RNA does not prevent silencing, consistent with our finding that a Slicer-deficient PRG-1 can still silence (Figure 3E; Bagijn et al., 2012).
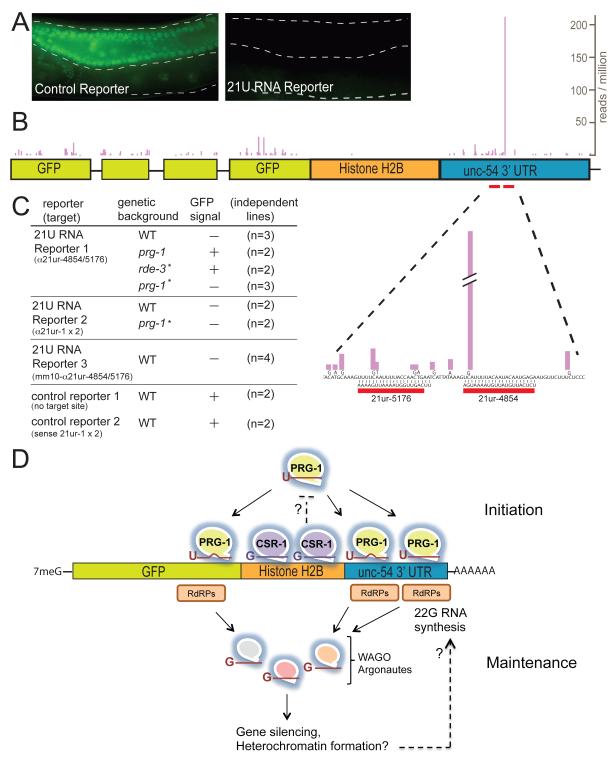
Figure 4. PRG-1 initiates trans-generational gene silencing.
(A) Fluorescence micrographs showing GFP::H2B expression in transgenic animals carrying a control reporter (left) or a 21U-RNA reporter (right). (B) Schematic of 21U-RNA reporter #1 showing the distribution of 22G-RNAs identified in a silent transgenic strain. The bars indicate reads in the antisense (pink) or sense (blue) orientation. (C) Genetics of reporter silencing. The reporters were either directly injected into the indicated strain or (as indicated by the star) were crossed into the strains after establishment in a wild-type background. “mm10” indicates mismatch mutations in both target sites at position 10 of the 21U-RNA. (D) Model showing the proposed role of PRG-1 in the initiation of silencing on a 21U-RNA target reporter. See also Figure S4.
The preceding analysis indicated that 22G-RNA production occurs locally within a 100 nt window near predicted 21U-RNA target sites. We therefore wished to ask if 22G-RNAs were produced locally around the synthetic 21U-RNA target sites in the reporter 3′ UTR. Deep sequencing revealed a dramatic increase of 22G-RNA levels originating mostly 5′ of the 21U-RNA target site (Figure 4B). The single most abundant 22G-RNA species in the silent reporter strains was found to overlap by 2 nucleotides with the 3′ end of the 21ur-4854 complementary sequence in the reporter 3′UTR. No such induction of 22G-RNAs was observed in a strain carrying the actively-expressed control reporter lacking the artificial 21U-RNA target sites (Figure S4). Interestingly, 22G-RNAs were also significantly enriched within the GFP region, but were not enriched within the transgene region corresponding to histone sequences (Figure 4B; see Discussion).
Surprisingly, we found that maintenance of silencing on this reporter did not depend on prg-1 (Figure 4C). When a prg-1 mutation was crossed into an already silent reporter strain, the transgene did not recover expression even after propagating for more than 6 generations in the prg-1 mutant background (Figure 4C). By contrast, we found that crossing a WAGO-pathway-specific RNAi deficient mutant, rde-3, into the strain resulted in full recovery of transgene expression, indicating that the maintenance of silencing requires the WAGO-22G pathway. Conversely, when an active version of this transgene, established by injecting the transgene directly into prg-1 mutants, was outcrossed to wild-type, we found that the transgene was silenced in 100% of the F1 cross progeny. Furthermore, the transgene remained silent in all F2 segregants from this cross, including prg-1 mutant homozygotes. Silencing was also observed when the active transgene generated in the prg-1 mutant background was crossed to either the flag::prg-1(WT) or flag::prg-1(D583A) rescued strains. However, consistent with the idea that the Slicer mutant partially compromises PRG-1 function, the reporter was silenced with a significant delay when crossed into the flag::prg-1(D583A) strain. For example, we found that only one of five F1 flag::prg-1(D583A) cross progeny were silenced, while ten of ten F1 flag::prg-1(WT) cross progeny were silenced. Nevertheless by the F3, 100% of reporter lines in both the flag::prg-1(WT) and (D583A) backgrounds were fully silenced. These results suggest that Slicer activity contributes to PRG-1 function but is not essential for silencing. Taken together our findings indicate that PRG-1 and its 21U-RNA guides can initiate a long-lasting trans-generational mode of silencing that, once established, is maintained by the WAGO 22G-RNA pathway.
DISCUSSION
Piwi/piRNA complexes recognize targets through imperfect pairing and trigger local production of WAGO 22G-RNAs
Although Piwi proteins and their associated piRNAs have been linked to transposon suppression in flies, mammals and C. elegans, there remain many piRNAs whose targets cannot readily be deduced from their sequences alone. For example, in both mammals and nematodes the majority of piRNAs lack obvious sequence complementarity to transposons or other expressed mRNA sequences in the cell. Here we have shown that C. elegans mutants lacking the Piwi Argonaute PRG-1 not only lack piRNA (21U-RNAs), but also exhibit a striking depletion of some, but not all, RdRP-derived 22G-RNAs. Interestingly, the prg-1-dependent 22G-RNAs were overwhelmingly associated with the WAGO pathway, which has been linked to the silencing of transposons, pseudogenes, and non-annotated loci, as well as 1021 annotated genes (Gu et al., 2009). We have also shown that WAGO targets exhibit a local enrichment of 22G-RNAs (of up to 9-fold) surrounding potential high-affinity 21U-RNA binding sites. This correlation was strengthened when the most abundant 21U-RNAs were considered separately, and was lost when the least abundant 21U-RNAs were considered. This enrichment for local 22G-RNA accumulation was much less pronounced for CSR-1 (approximately 1.5- to 2-fold).
Consistent with these global correlations between predicted 21U-RNA targeting and 22G-RNA production and mRNA silencing, we have shown that individual transgenes engineered to contain sites complementary to specific endogenous 21U-RNAs are subject to potent prg-1-dependent silencing. Furthermore consistent with the idea that PRG-1 initiates 22G-RNA biogenesis locally on its target RNAs, we observed a dramatic production of 22G-RNAs overlapping the synthetic 21U-RNA complementary site in the reporter construct. While initiation of reporter silencing depended on PRG-1 activity, maintenance of silencing did not, and was instead dependent on the WAGO/RdRP silencing pathway. Together with a parallel study (Shirayama et al., this issue), these findings are consistent with a model (Figure 4D) in which the WAGO, CSR-1 and PRG-1 Argonaute pathways converge to recognize “self” and “non-self” sequences. The induction of silencing on foreign sequences is initiated by PRG-1, while an epigenetic memory of self/active is maintained by the CSR-1 pathway, and of non-self/silent by the WAGO pathway.
A recent study suggests that C. elegans piRNAs are depleted for self-complementary sequences and may avoid silencing endogenous protein encoding genes by selecting against self-recognition (Bagijn et al., 2012). We did observe what appears to be a slight (~2-fold) trend for the depletion of perfect matches to 21U-RNAs among CSR-1 targets. For example 22 of 3659 annotated CSR-1 targets have perfect complementarity to 21U-RNA species, while 29 of 2911 annotated WAGO targets have perfect pairing. When compared to WAGO target loci, there was also a slight bias against CSR-1 targets with 1 (1.18-fold), 2 (1.86-fold) or 3 (1.2-fold) mismatches. It is important to note, however, that despite these slight biases there are nevertheless at least 1380 of germline expressed CSR-1 targets with three or fewer mismatches to 21U-RNAs, whose mRNA expression was unaffected in prg-1 mutants. Thus if selection against 21U-RNA pairing exists within germline expressed mRNAs, it is very mild and cannot explain why CSR-1 targets do not undergo prg-1-dependent silencing. Furthermore, there are over one thousand annotated endogenous (non-transposon) germ-line expressed genes that are silenced by WAGO-pathway 22G-RNAs (Gu et al., 2009), and it was among these targets that we observed a strong (3-9 fold) depletion of 22G-RNAs near 21U-RNA complementary sites. So, our analysis indicates that WAGO targets are sensitive to prg-1 induced silencing, while many other genes (primarily CSR-1 targets) have similar levels of complementarity to 21U-RNAs but are resistant to silencing.
The Bagijn et al. study also reported that piRNAs are enriched for complementarity to transposon ends. We observed a similar trend, however the numbers are low. Out of a total of ten 21U-RNAs with perfect matches to transposons in our data sets, five matches were located at the first or last 50 nt of the transposon. This is an intriguing trend indicating 21U-loci can trap transposons perhaps in a manner similar to that observed for piRNA clusters in the fly. If a transposon inserts within the 40 nt window between a 21U promoter element and the 21U transcription start site, then it is possible that a new transposon-targeted 21U species could be generated. However it is not clear whether or how such 21U-RNAs could mediate transposon silencing, since the resulting end-associated piRNA is unlikely to be complementary to an expressed portion of the transposon.
Not all WAGO targets exhibited decreased 22G-RNA levels in prg-1 mutant animals even though many of these targets contained one or more high-affinity 21U-RNA complementary sites. There are several possible explanations for these findings. First, WAGO-22G-RNAs are known to function downstream of RDE-1 in the dsRNA pathway, and downstream of ERGO-1 and ALG-3/4 in the Eri pathway, and we found that RDE-1 and ERI-pathway 22G-RNAs were not depleted in prg-1 mutants. However, these alternative pathways do not account for all of the WAGO targets whose 22G-RNA levels remain high in prg-1 mutants. A second possibility is that, perhaps just as PRG-1 was required for the initiation but not the maintenance of WAGO-pathway silencing on our reporter construct, these endogenous loci were targeted by PRG-1 in a previous generation, but now the WAGO-pathway maintains their silencing independently.
Does CSR-1 protect its targets from PRG-1 and WAGO-mediated silencing?
Since 21U-RNAs have the potential to target essentially all germline transcripts, a mechanism must exist to prevent silencing of functional germline genes. CSR-1 and its associated 22G-RNAs target, but do not silence thousands of germline-expressed protein-encoding mRNAs (Claycomb et al., 2009), therefore CSR-1 22G-RNAs are ideal candidates for this protective function. Consistent with this idea, we have shown that despite an abundance of potential high affinity 21U-RNA binding sites, CSR-1 targets exhibit no change in mRNA expression and at most a slight (1.5- to 2-fold) change in 22G-RNA levels near predicted 21U-target sites. In our analysis of a 21U-RNA reporter strain, we observed 22G-RNAs produced from the 3′ UTR region, and also within the GFP region of the transgene. However, very few 22G-RNAs were observed within the transgenic region corresponding to the germline-expressed Histone H2B, which is a known target of CSR-1 22G-RNAs (Claycomb et al., 2009). Therefore CSR-1 may prevent PRG-1 recruitment to its targets, perhaps by selectively destroying PRG-1-bound template RNAs before RdRP can produce WAGO 22G-RNAs (see model in Figure 4D). Such a self-protection pathway could explain why high-affinity pairing is tolerated in many actively-expressed endogenous mRNAs (e.g. CSR-1 targets).
In a parallel study, the Miska group generated a 21U-RNA reporter for which PRG-1 was continuously required to maintain silencing. In their reporter, the 21U-RNA target site was flanked by the his-58 coding sequence and the tbb-2 3′ UTR (Bagijn et al., 2012); both are targets of CSR-1 22G-RNAs (Claycomb et al., 2009). By contrast, PRG-1 is only required to initiate (not to maintain) silencing of our reporter, in which the 21U-RNA target sites are embedded in the unc-54 3′ UTR, a somatic transcript that is not targeted by CSR-1 22G-RNAs. Thus one intriguing explanation for the difference in the behavior of these 21U-RNA reporters is that the presence of flanking sequences targeted by CSR-1 can help drive the recovery of the transgene from silencing once prg-1(+) is crossed out of the strain. Such a mechanism could explain the existence of endogenous genes that (like the Miska-study transgene) require PRG-1 for maintenance of silencing. These loci might represent genes that are targeted at least partially by both the WAGO and CSR-1 pathways.
Finally, Shirayama and coworkers (this issue) have shown that a stable trans-generational form of WAGO-dependent silencing, referred to as RNAe for RNA-induced epigenetic silencing, also requires prg-1(+) activity for initiation, but not for the maintenance of silencing. Single copy transgenes containing a GFP sequence fused to an endogenous gene can be permanently silenced or expressed. Interestingly, WAGO 22G-RNAs targeting these silent transgenes were found to accumulate only within the GFP sequences and did not spread into neighboring transgene sequences shared with endogenous germline-expressed mRNAs. These findings are also consistent with a CSR-1 protection model. Taken together these findings suggest that, a perfectly complementary 21U-RNA sequence can overcome local CSR-1 22G-RNA protection, while transgenes lacking perfectly complementary 21U-RNA sequences (like those analyzed in Shirayama et al) are prone to PRG-1-initiated silencing through imperfect 21U-RNA binding at one or more partially complementary 21U-RNA sites within the foreign sequences (Shirayama et al., this issue).
Argonautes as mediators of genome-wide surveillance of gene expression
Macronuclear development in the ciliated protozoan Tetrahymena provides a remarkable example of genome surveillance mediated by an Argonaute small RNA pathway (Mochizuki et al., 2002). In this system, an AGO pathway scans the entire contents of two nuclei that share a common cytoplasm. This process is thought to build an inventory of genes previously expressed in the parental macronucleus, in order to direct massive chromosomal rearrangements during assembly of the new macronuclear genome. Could other metazoan Piwi Argonautes provide a correlate for this type of genome-wide surveillance? Although, most metazoans do not undergo massive chromosomal rearrangements like those observed in Tetrahymena and other ciliates (Klobutcher and Jahn, 1991; Smith et al., 2009), animal genomes nevertheless exhibit extensive chromatin remodeling during gametogenesis, and especially during spermatogenesis and in zygotes shortly after fertilization (Sassone-Corsi, 2002). Perhaps in other animals DNA methylation and chromatin marks, as well as RNA-binding proteins that stably associate with RNAs produced within different chromatin environments could provide surrogates for the specificities associated with CSR-1 (memory of self/licensed) and WAGO (memory of non-self/silent). For example, piRNA complexes might only initiate or reinforce silencing on targets whose chromatin or RNA products lack marks indicative of “self/licensed” genes. In a context where “self” sequences are protected from silencing, Piwi Argonautes could use their vast array of piRNAs, along with relaxed base-pairing, to ensure that silencing remains focused on foreign (or unlicensed) genes.
EXPERIMENTAL PROCEDULES
Worm Strains
C. elegans culture and genetics were essentially as described (Brenner, 1974). The Bristol strain N2 was used as standard wild-type strain. Alleles used in this study: prg-1(n4357), and prg-1(tm873), and rde-3(ne3370). The MosSCI recipient strain EG4322 Mos1(ttTi5605) II; unc-119(ed3) III and the direct insertion method (Frokjaer-Jensen et al., 2008) was used to create single copy insertion lines listed in Supplemental Experimental Procedures.
Creation of MosSCI donor vectors
The flag::prg-1(WT) and flag::prg-1(D583A) constructs were made as follows: a 5.0 kb PCR fragment containing the genomic sequence of prg-1 with 1 kb upstream and downstream was cloned into pCR-Blunt-II-TOPO using the Zero-Blunt-TOPO cloning kit (Invitrogen). A unique NotI site was removed from the plasmid and reintroduced immediately after the ATG of prg-1, and the 3xflag sequence was inserted into the NotI site. The D583A mutation was introduces by site-directed mutagenesis using the QuickChange Mutagenesis Kit (Stratagene). The flag::prg-1(WT) and flag::prg-1(D583A) fragments were subcloned into the MosSCI vector pCFJ151 (Frokjaer-Jensen et al., 2008) using SpeI and XhoI sites.
The 21U-RNA reporter and control GFP reporter constructs were created by Multisite Gateway cloning (Invitrogen). The pie-1 promoter, GFP/Histone 2B, and unc-54 3′ UTR fragments were recombined using plasmids pCG142, pCM1.35 and pCM5.37 plasmid, respectively, in a Gateway reaction with the pCFJ150 destination vector to create final vector for insertion at Mos1(ttTi5605) on LGII. The 21U-RNA target sites or reverse complement were introduced into unc-54 3′ UTR in pCM5.37 between the sequences ttactcttcaacatccctacatgc and tctttctccctgtgctcccacc by plasmid PCR with primers containing the inserted sequences, followed by DpnII digestion, PNK and DNA ligation region.
Small RNA cloning and analysis
About 100,000 gravid adults were washed 3 times with M9 buffer and RNA was extracted using TRI Reagent (MRC, Inc.). Small RNA was enriched from 200 mg of total RNA using the MirVana Kit (Life Technologies). RNAs from 15-30 nt were gel purified on a 15% acrylamide/7M Urea gel. To efficiently clone 22G-RNAs, eluted small RNAs were treated with TAP (Tobacco Acid Pyrophosphatase, Epicentre) to convert 5′ triphosphate RNAs into monophosphate. The small RNAs were ligated to a 3′ linker (miRNA cloning linker 1, IDT) and a 5′ linker containing 4nt barcode using T4 RNA ligase with gel purifications following each ligation. Ligated RNA products were converted to cDNA using Superscipt III (Invitrogen). Libraries were amplified and sequenced using an Illumina GAII to obtain single-end 36 nt sequence at the UMass Medical School Deep Sequencing Core.
A custom Perl script was used to remove the 5′ barcode and the 3′ adapter sequences. If the 3′ adapter was not identified, then incomplete 3′ adapters CTGTA, CTGT, CTG, or CT were removed. Reads of at least 17 nt in length were mapped to the C. elegans genome (WormBase release WS215) and miRBase 16 using Bowtie 0.12.7 with the parameter ‘-v 3 -a --best --strata -m 400′. A custom Perl script was used to perform a post-match analysis, only allowing mis-matches with reads ≥19 nts: one mismatch for 19 – 21 nt, two for 22 – 24 nt and three for ≥25 nt. The Bowtie parameter ‘- a --best –strata’ was used to return only the best matches. The read count of each sequence was normalized to the number of matches in the genome. To account for differences in sequencing volume between samples, we normalized the total of matched non-structural RNAs to 5 million reads. A custom Perl script and Bioperl was used to draw scatter plots. The single nt histogram for the start site of matched RNA was obtained using a custom Perl script and the generic genome browser 1.70. All scripts are available upon request.
Lists of CSR-1 and WAGO targets are provided in Table S3 and Table S4. CSR-1 targets were defined as genes with small RNAs enriched in three independent CSR-1 IP experiments (Claycomb et al., 2009). WAGO targets were defined as genes depleted of small RNAs in rde-3, mut-7 and drh-3 mutants (Gu et al., 2009).
Bioinformatic prediction and analysis of 21U-RNA targets
21U-RNA targets were identified using custom Perl scripts and Bowtie 0.12.7. We used Bowtie with parameter “-n 3 -e 300 -l 6 -a -m 4000000” to map annotated 21U-RNAs to annotated transcripts (WormBase release WS215) with up to 10 mismatches. Mismatches at position 1 of the 21U-RNA or that correspond to G:U wobble base pairs were then discounted, and alignments with up to 4 mismatches were analyzed further. A custom Perl script was used to filter subsets of 21U-RNA targets with desired pairing criteria and/or 21U-RNA abundance. Unless a specific criterion is defined, we required a perfect match, with no more than one G:U pair, in the seed region (nts 2-8), and up to 2 mismatches and an additional G:U pair outside the seed region. The abundance of each 21U-RNA was based on deep sequencing of small RNAs cloned from a PRG-1 IP (Batista et al., 2008). A library of reverse-complement 21U-RNAs was also generated for the control analysis.
The density of antisense 22G-RNAs within a 100 nt window of predicted 21U-RNA target sites centered around position 10 of the 21U-RNA was determined using a custom Perl script. 21U-RNA target sites on chromosome IV or within 100 nt of another target site were excluded. The density profile is a sum of reads with the 5′ nt mapping to a given position within the 100 nt window around predicted 21U-RNA target sites.
qRT-PCR analysis and Tiling microarray analysis
qRT-PCR analysis for mRNA and small RNA was performed as described (Batista et al., 2008; Das et al., 2008). Primer sequences are provided in the Supplemental Experimental Procedures. Tiling array data were published previously (Batista et al., 2008). A Custom Perl script was used to convert Affymetrix probe coordinates (release WS170) to release WS215 coordinate. We identified probe signals present in both data sets with a p value < 0.1. Gene expression values were calculated as the geometric mean of probe signals within a gene and normalized to total signal of all histone transcripts prior to comparison.
Protein analysis
Western blotting was performed as previously described (Batista et al., 2008). Antibodies used for Western blotting were anti-FLAG (M2, Sigma), anti-PRG-1 (Batista et al., 2008), and anti-alpha-Tubulin (MCA78A, Serotec) antibodies.
Supplementary Material
HIGHLIGHTS.
Transcriptome-wide surveillance of germline transcripts by C. elegans piRNAs
C. elegans piRNAs use imperfect base pairing to initiate silencing
Maintenance of silencing depends on the WAGO/RdRP pathway
mRNAs targeted by the CSR-1 Argonaute appear to be protected from silencing
ACKNOWLEDGEMENTS
We thank E. Miska for sharing unpublished results and G. Seydoux for providing the multisite gateway system. H.C.L. is supported by a Ruth L. Kirschstein National Research Service Awards (GM099372). C.C.M. is a Howard Hughes Medical Institute Investigator. This work is supported by an R01 grant (GM058800) to C.C.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers Illumina data is available from GEO under the accession number GSE38723.
REFERENCES
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, E.-M. W, Bouasker S, Lehrbach NJ, Simard MJ, Miska EA. Function, Targets, and Evolution of Caenorhabditis elegans piRNAs. Science. 2012 doi: 10.1126/science.1220952. DOI: 10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine CC, Batista PJ, Gu W, Claycomb JM, Chaves DA, Shirayama M, Mello CC. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2010;107:3588–3593. doi: 10.1073/pnas.0911685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell. 2008;31:79–90. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D, Jr., Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T, Manoharan AP, Harkins TT, Bouffard P, Fitzpatrick C, Chu DS, Thierry-Mieg D, Thierry-Mieg J, Kim JK. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2009;106:18674–18679. doi: 10.1073/pnas.0906378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobutcher LA, Jahn CL. Developmentally controlled genomic rearrangements in ciliated protozoa. Curr Opin Genet Dev. 1991;1:397–403. doi: 10.1016/s0959-437x(05)80306-5. [DOI] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Maiti M, Lee HC, Liu Y. QIP, a putative exonuclease, interacts with the Neurospora Argonaute protein and facilitates conversion of duplex siRNA into single strands. Genes Dev. 2007;21:590–600. doi: 10.1101/gad.1497607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniar JM, Fire AZ. EGO-1, a C. elegans RdRP, modulates gene expression via production of mRNA-templated short antisense RNAs. Curr Biol. 2011;21:449–459. doi: 10.1016/j.cub.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Tuschl T. RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev. 2004;18:975–980. doi: 10.1101/gad.1187904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 2002;296:2176–2178. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Smith JJ, Antonacci F, Eichler EE, Amemiya CT. Programmed loss of millions of base pairs from a vertebrate genome. Proc Natl Acad Sci U S A. 2009;106:11212–11217. doi: 10.1073/pnas.0902358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Steiner FA, Okihara KL, Hoogstrate SW, Sijen T, Ketting RF. RDE-1 slicer activity is required only for passenger-strand cleavage during RNAi in Caenorhabditis elegans. Nat Struct Mol Biol. 2009;16:207–211. doi: 10.1038/nsmb.1541. [DOI] [PubMed] [Google Scholar]
- Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, Duchaine TF, Mello CC, Conte D., Jr. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc Natl Acad Sci U S A. 2010;107:3582–3587. doi: 10.1073/pnas.0911908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.