Abstract
Results obtained from expression profilings of renal cell carcinoma using different “ome”-based approaches and comprehensive data analysis demonstrated that proteome-based technologies and cDNA microarray analyses complement each other during the discovery phase for disease-related candidate biomarkers. The integration of the respective data revealed the uniqueness and complementarities of the different technologies. While comparative cDNA microarray analyses though restricted to upregulated targets largely revealed genes involved in controlling gene/protein expression (19%) and signal transduction processes (13%), proteomics/PROTEOMEX-defined candidate biomarkers include enzymes of the cellular metabolism (36%), transport proteins (12%) and cell motility/structural molecules (10%). Candidate biomarkers defined by proteomics and PROTEOMEX are frequently shared, whereas the sharing rate between cDNA microarray and proteome-based profilings is limited. Putative candidate biomarkers provide insights into their cellular (dys)function and their diagnostic/prognostic value but still warrant further validation in larger patient numbers. Based on the fact that merely 3 candidate biomarkers were shared by all applied technologies, namely annexin A4, tubulin alpha-1A chain and ubiquitin carboxyl-terminal hydrolase L1 the analysis at a single hierarchical level of biological regulation seems to provide only limited results thus emphasizing the importance and benefit of performing rather combinatorial screenings which can complement the standard clinical predictors.
Keywords: RCC, transcriptomics, proteome-based technologies, biomarkers
Introduction
With the completion of the human genome project the emphasis is now moving towards the understanding of its functional products and the biological significance of approximately 25.000 human genes. This information has to be complemented by the determination of the protein expression pattern, which defines the basis to differentiate between normal and diseased stages. Overall this will lead to a better understanding of the molecular mechanisms associated with the initiation and progression of diseases and support the design of innovative therapeutic approaches. In this context a feasible goal is the identification of novel diagnostic and prognostic biomarkers as well as of therapeutic targets. Global gene expression profiling using cDNA microarrays allows the simultaneous analysis of the expression of thousands of genes in a high throughput setting and already has shed some light on cellular processes related to certain disease pathogenesis [1, 2] . Due to the existence of complex posttranscriptional processes such as transcript de/stabilization, translation, posttranslational modifications and protein degradation which determine and modulate the quality and quantity of expressed proteins, transcription is not coordinately associated with translation resulting in a limited correlation between mRNA levels and protein abundance [3] . The differences in protein concentrations are only to 20 – 40% attributable to altered mRNA levels [4, 5] . Therefore, proteomics represents a complementary strategy for biomarker detection in malignancies [6, 7] . So far, this approach mainly used 2-DE analysis followed by mass spectrometry. In addition PROTEOMEX/SERPA/SPEAR, a combination of classical proteomics with serology, has been implemented for the detection of putative immunogenic biomarkers in malignancies [8–11]. Using both transcriptome and proteome-based technologies a number of candidate biomarkers have been defined in different diseases [8–11]. Such candidate biomarkers could be employed in the diagnosis, prediction and prognosis of disease or even as therapeutic targets and are still urgently needed for the discovery, treatment and monitoring of some malignancies including renal cell carcinoma (RCC).
RCC represents the most prevalent cancer of the kidney, accounts for approximately 3% of adult malignancies in the Western world [12] and is the 6th leading cause of cancer-related deaths, whereas localized disease can be cured by nephrectomy. This is not the common option since symptoms arise late and approximately 55% of the patients exhibit locally advanced or metastatic RCC at the time of diagnosis. Unfortunately, these patients have a poor prognosis with a 5-years survival rate < 20%. This is based on the fact that RCC is moreover resistant to chemo- and radiation therapy. However, immunotherapy as well as a group of small molecules targeting growth factor receptors, their ligands or tyrosine kinases showed promising anti-tumor activity [13–15] .
Developments in cDNA microarrays and mass spectrometry (MS)-based functional proteomics have improved the understanding of the molecular mechanisms involved in neoplastic transformation and tumor progression at both the mRNA and protein level. Some transcriptome- and proteome-based studies using RCC lesions or RCC cell lines and corresponding normal renal epithelium or cell lines derived thereof led to the identification of RCC-specific changes associated with an altered expression pattern for a number of gene/protein families [11, 16–26]. These include prognostic factors associated with the growth rate and metastatic potential of RCC and predictive factors related to the sensitivity and/or resistance to therapeutic agents [16, 18, 20, 26]. However, further investigations are still required to receive a more comprehensive knowledge about genes/proteins that are selectively up- or down-regulated in RCC lesions and during disease progression. This information in association with clinicopatholigical parameters might lead to a more accurate stratification of RCC patients based on the risk and thus also influence the intensity of treatment and surveillance. Recently, a combination of selected tissue markers with standard clinical predictors was explored by integrating informative markers into a multivariate prognostic system [27].
The aim of the current study was to address in particular the complementation capacity of transcriptome- and proteome-based methods. In the context of discovery of candidate biomarkers in RCC. The integration of the heterogeneous data generated from these diverse global technology platforms in combination with the validation procedure using RCC lesions, autologous normal kidney epithelium as well as RCC and normal renal epithelium representing cell lines represent the first challenge to develop a database of putative RCC-specific candidate biomarkers. The low level of concordance (10%) in the mRNA and protein expression profiling of RCC systems investigated suggested that the analysis at a single hierarchical level of biological regulation provides only partial results since none of them can display the given complexity. The candidate biomarkers identified by these technologies belong to different gene/protein families. Some of them were novel and unexpected in the context of RCC, while others still await the further description of their physiological function and regulation. Thus, the implementation of cDNA microarrays in combination with proteome-based technologies and the subsequent validation of identified targets is a mandatory approach, which allows to assess the complexity of biological processes and minimizes the constriction of each technology in the discovery phase of diagnostic and prognostic candidate biomarkers for RCC.
Materials and methods
Tissue samples, sera and clinical data
Biopsies from RCC of distinct subtypes and normal kidney samples (n = 38) were obtained from the Department of Urology of the Johannes Gutenberg University (Mainz, Germany) (Table 1A) and have been previously described in detail [21, 23]. Specimens were collected following routine nephrectomy and tissue samples were directly cryo-conserved for subsequent “ome”-based analysis, whereas the remaining tissues were used for pathological confirmation. In addition, serum was obtained from RCC patients (n = 8) prior to and post surgery as well as from healthy controls (n = 7). Patients sample procurement followed standard ethical procedures according to the institutional policy and informed consent was obtained from each individual.
Table 1.
Characteristics of RCC samples analyzed by transcriptome- and proteome-based approaches.
| A: Details of the patients and samples. | ||||
|---|---|---|---|---|
| Group | Sex (m/f) | Mean age (y) | Clinical details | |
|
| ||||
| subtype | number | |||
|
|
||||
| oncocytoma | 1/1 | 49 (±11) | - | 2 |
|
| ||||
| leiomyosarcoma | 1/0 | 78 | - | 1 |
|
| ||||
| RCC | 20/12 | 62 (±24) | clear cell | 32 |
| 2/1 | 64 (±9) | chromophilic | 3 | |
| 0/2 | 58 (±16) | chromophobic | 2 | |
| B: Number of samples analyzed using different techniques | |||
|---|---|---|---|
| cDNA microarrays | classical proteomics | PROTEOMEX | |
| number of cases or cell lines (NN/RC) | 17/17 | 21/21 | 1/3 |
| number of serum samples (NN/RC) | - | - | 7/8 |
| number of targets | 119 | 334 | 50 |
RCC cell lines and tissue culture
The RCC cell lines established from primary RCC of various subtypes as well as SV40 large T antigen immortalized RCC and normal kidney epithelium representing cell lines have been described in detail elsewhere [28-30]. The cells were grown in DMEM supplemented with 10% fetal calf serum, 2 mM glutamine, 100 U/ml penicillin/100 μg/ml streptomycin, 1% MEM essential amino acids and 1% sodium pyruvate (all components purchased from Gibco/BRL, Life Technologies, Karlsruhe, Germany) Immortalized cells were grown in the presence of up to 600 μg/ml Hygromycin B (Roche Diagnostics, Mannheim, Germany). 5 x 106 to 1 x 107 cells were harvested, washed three times in phosphate buffered saline (PBS), shock frozen as dry pellets and stored in liquid nitrogen until further use.
cDNA microarrays
Total RNA was extracted either from frozen tissue specimens or from cell pellets using the Trizol reagent (Invitrogen, Carlsbad, CA, USA) and amplified into antisense RNA (aRNA) [31]. High quality aRNA was subsequently subjected to analysis. Similarly, total RNA from peripheral blood mononuclear cells pooled from six normal donors was extracted and amplified to serve as constant reference. Test and reference RNAs were labeled with Cy5 (red) and Cy3 (green) and co-hybridized to a custom-made 17.5K cDNA microarray. The 17.500 spots included 12.072 uniquely named genes, 875 duplicated genes, and about 4.000 expression sequence tags [16, 31].
Quality validation of transcriptome data
All statistical analyses were performed using the log2-based ratios normalizing the medial log2 ratio value across the array equal to zero. Validation and reproducibility were performed using our internal reference concordance system. The level of concordance was determined by periodically rehybridizing the same arbitrarily selected test sample with the reference sample as described [31]. The concordance analysis demonstrated a > 95% concordance level and non-concordant genes due to lack of reproducibility or random biases were excluded from subsequent analyses. However, the comparison is as yet limited to the set of upregulated transcripts.
Two-dimensional gel electrophoresis, immunoblotting and protein identification
Biopsy specimen or cell pellets (5 x106 to 1 x 107 cells sample) were extracted with lysis buffer in 7 M urea (Applichem, Darmstadt, Germany), 2 M thiourea (Sigma-Aldrich Chemie, Deisenhofen, Germany), 0.2 M dimethylbenzylammonium propane sulfonate (NDSB, ICN Biomedicals, Eschwege, Germany), 1% dithiothreitol (DTT; Applichem, Darmstadt, Germany), 4% 3-[(3-cholamidopropyl)dimethylamino]-1-propanesulfonate (CHAPS; Applichem), 0.5% pharmalytes (Amersham Biosciences, Freiburg, Germany) and a trace of the dye bromophenol blue (Serva Electrophoresis, Heidelberg, Germany) and the respective lysates were generated as recently described [21, 29]. Briefly, 300 – 500 μg total protein/sample were loaded on either linear pH 4 – 7 and pH 6 - 11 or non-linear pH 3 – 10 immobilized pH gradient strips (Immobiline DryStrips, Amersham Biosciences). All samples were run at least in triplicates. Isoelectric focusing, second dimension SDS-PAGE separation, gel staining with colloidal Coomassie blue, silver or Ruthenium II bathophenanthroline disulfonate chelate (RuBPS) was performed as recently published [29, 32].
Quantitative image analysis was performed using the Proteomeweaver™ software (Versions 1.1 – 4.0; Definiens AG/Bio-Rad GmbH, Munich, Germany). Spot patterns were first normalized for pattern recognition with a pre-match normalization algorithm and subsequently numerically normalized by a pair-matched normalization algorithm which computes a normalization factor for every gel in the analysis set resulting in a residual error between 1.01 and 1.1. Only proteins with a 2-fold altered expression level in the given match set (tumor/kidney epithelium) or displaying a restricted protein expression pattern to either tumor or normal kidney cells were considered as differentially expressed. The selected protein spots were statistically analyzed by the t-test (p < 0.05).
For immunoblotting, proteins from 2DE gels were transferred onto PVDF membranes (Roche Biochemicals, Mannheim, Germany), blocked in Tris-buffered saline (TBS, 140 mM NaCl, 10 mM Tris–HCl, pH 7.4) supplemented with 0.4% Tween 20, 5% skim milk (Difco Laboratories, USA) and 10% horse serum (PAA Laboratories GmbH, Coelbe, Germany) and then incubated over night at 4 °C with serum (1:50 diluted in TBS, 0.1% Tween 20, 2% skim milk). Sera were either obtained from RCC patients prior/post radical nephrectomy or from healthy donors. Specific binding of serum IgG was visualized by using the peroxidase-conjugated rabbit anti-human IgG antibody (Ab) PO214 (DAKO, Hamburg, Germany) in combination with a chemiluminescence substrate (Lumi-Light Western Blotting Substrate, Roche Molecular Biochemicals, Mannheim, Germany) according to the manufacturer's instructions. Immune reactive spots were subsequently matched with the 2DE spot gel pattern of the corresponding gel by superimposing the given pattern.
Microanalytical identification of proteins
Protein identification was performed as previously described [29]. Peptide mass fingerprinting analyses and post-source decay (PSD) fragmentation were performed by MALDI-TOF-MS using a Voyager STR Biospectrometry Workstation (Applied Biosystems, Foster City, CA, USA). Edman degradation was performed using a Procise 494 system (Applied Biosystems). Database searches employed the Mascot software package (Matrix Science, Durham, US) [32].
Bioinformatics
The raw data analysis was performed with an in house made web-based application which provides several tools such as calculating the overlap between different approaches, functional and subcellular distribution of the identified genes/proteins, generation of 2DE reference maps. The relevant “ome”-based data, such as mode of regulation, MS-based identification strategy or annotation of the genes/proteins from the three different approaches were collected and stored in an in house database. The database was automatically fed with the latest information from several public domain databases, such as SwissProt/TrEMBL (http://expasy.org/sprot/) or EBI (http://www.ebi.ac.uk/ego/GSearch) and used to classify the set of differentially expressed genes/proteins according to their biological functions and cellular compartments based on the retrieved gene ontology annotation for each candidate biomarker. Genes not described in the database or for which the biological process/function or cellular localization were unspecified were categorized as “unknown”.
Validation of differentially expressed genes/proteins
For RT-PCR analysis total cellular RNA was extracted, transcribed into cDNA and subjected to semi-quantitative RT-PCR using target-specific primers as recently described [30]. For Western blot analysis 20 μg protein/lane were size fractionated on SDS-PAGE prior to blotting on PVDF (Immobilon-P, Roche Biochemicals, Mannheim, Germany) or on nitrocellulose membranes (Protran, Schleicher & Schuell, Dassel, Germany), blocked, incubated with target-specific antibodies and subsequently developed with suitable secondary antibodies as previously published [23].
Immunohistochemistry
Representative paraffin blocks of tumor and normal tissues were selected and cut into 5 μm sections using a cryostat. For antigen retrieval, consecutive sections were incubated for 2–8 minutes in citrate buffer in a microwave oven followed by a washing procedure with Tris-buffered saline and an additional incubation with normal swine serum (dilution 1:5, DAKO, Hamburg, Germany) for 10 minutes. All sections were stained with HE to identify tumor tissue. Slides were incubated with the primary antibody for one hour at room temperature. Immunohistochemical stainings were performed as previously described [21, 33] with the polyclonal antibodies (pAb’s) directed against annexin (ANX) A3 (ab33068, Abcam, Cambridge, UK) or alpha-enolase (ab35075, Abcam, Cambridge, UK). Immunoreactivity was detected using the commercially available streptavidin biotin (LSAB)-peroxidase kit and amino-9-ethylcarbazol (AEC) (DAKO, Hamburg, Germany). Negative controls were performed by omitting the primary antibody. The extent of immunostaining was scored according to the following criteria: negative: < 5% positive cells; weak positive: 5 – 25% positive tumor cells; intermediate positive: 26 – 50% positive tumor cells; strong positive > 50% positive tumor cells.
Results
The profiling of RCC systems at the mRNA and protein level has become a widely used tool to identify and characterize molecules and/or pathways important for the development of RCC. Using three distinct experimental strategies, cDNA microarrays, proteomics and PROTEOMEX 423 unique differentially expressed/immunoreactive genes/proteins have been identified in the different RCC systems analyzed. Since the interpretation of the results requires the presentation of the data in the context of their functional processes and cellular localization, the sets of genes/proteins identified as differentially expressed/immunoreactive in RCC versus normal kidney (biopsies/cell lines) were sub-grouped into respective functional categories and according to their cellular localization based on the latest currently available information stored in public domain databases. This approach allows the discrimination between co- or discordant regulated biological pathways and/or processes disturbed in RCC when compared to normal kidney. Based on the collected experimental data, selected candidate biomarkers were further validated concerning their differential expression pattern in a series of RCC systems consisting of either tissue samples and/or cell lines.
Classical 2DE-based proteome analysis of RCC lesions and normal kidney epithelium
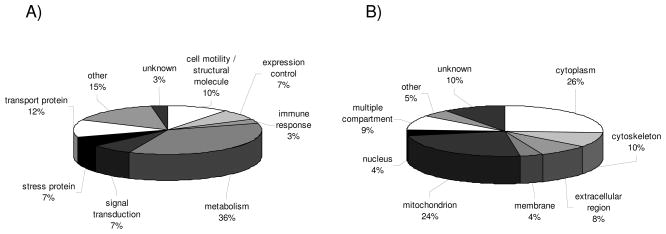
In this report 21 pairs of RCC samples and corresponding normal kidney epithelium have been analyzed by classical 2DE-based proteome analysis (Table 1B). This strategy allowed the detection of 334 proteins exhibiting a distinct/differential expression pattern between RCC lesions and normal kidney tissues. Protein ontology was employed to group the set of differentially expressed proteins into distinct protein families based on the closest affinity for their major biological function and according to their cellular localization. As shown in Fig. 1A the frequencies within the given functional categories significantly varied: in particular metabolic enzymes (36%), transport proteins (12%) and proteins associated with cell motility and cellular structure (10%) were found to be differentially expressed in RCC lesions when compared to normal kidney epithelium, whereas the frequency within the other functional protein subclasses in the respective biopsy pairs was < 10% (Fig. 1A). It is noteworthy that the group “other” (15%) is comprised of functional categories expressed at < 5% in the sample collection analyzed.
Figure 1.
Classification of unique proteins identified by classical proteomics. The pie charts display the classification of differentially expressed proteins identified by classical proteomics of 21 RCC lesions and corresponding normal kidney epithelium according to (A) functional protein classes and (B) cellular compartments. Each segment in the pie charts represents either a functional gene family (A) or a cellular compartment (B) as indicated by the respective labels. In addition, the relative distribution is indicated as % of the total target number analyzed. Functional protein classes and distribution into cellular compartments < 5% were combined and categorized as “other”.
These differentially expressed proteins were mainly found in the cytoplasm (26%), mitochondria (24%) as well as in the cytoskeleton (10%), but rarely detected on the membrane (4%) or in the nucleus (4%, Fig. 1B). In addition, 10% of the proteins identified could not be localized to a specific compartment.
PROTEOMEX as a suitable tool for candidate biomarker detection in RCC
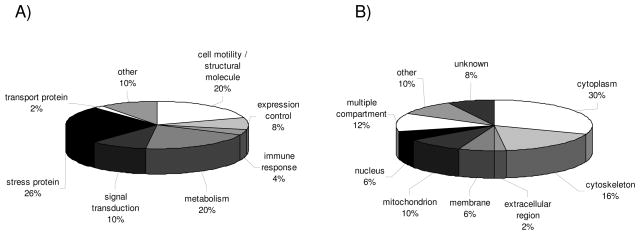
For PROTEOMEX analyses the three RCC cell lines MZ1257RC, MZ1940RC and MZ2733RC and the normal kidney epithelium representing cell line MZ2733NN were subjected to 2DE, blotted onto membranes, blocked and subsequently incubated with either serum obtained from 7 healthy volunteers or from 8 RCC patients prior to the detection of immunoreactive proteins with an anti-human lgG-specific pAb. Each sample set consisting of the 4 cell lines was tested with the given serum sample individually (Table 1B). Employing this approach still provides a challenge which is underlined by the fact that more than 50% of the immunoreactive proteins detected by this methodology have not yet been identified. So far, only 50 immunogenic proteins have been identified successfully, which is partially linked to the enormous complexity at the protein level, their low expression level and/or the limited sensitivity of mass spectrometry. The highest percentage of putative biomarkers identified by PROTEOMEX represent stress proteins (26%), proteins essential for cell motility and restructuring or maintenance of the cytoskeleton (20%) and metabolic enzymes (20%; Fig. 2A). The classification of the immunoreactive targets into cellular compartments revealed the highest frequency within the cytoplasm (30%) and in the cytoskeleton (16%), whereas 8% of the immunoreactive proteins exhibit an unspecified cellular localization (Fig. 2B). In contrast to the classical proteomics approach, PROTEOMEX also allowed the identification of targets which were not necessarily differentially expressed but likely rather defined by posttranslational modifications, as previously demonstrated for isoforms of the heat shock protein (HSP) 27 [21].
Figure 2.
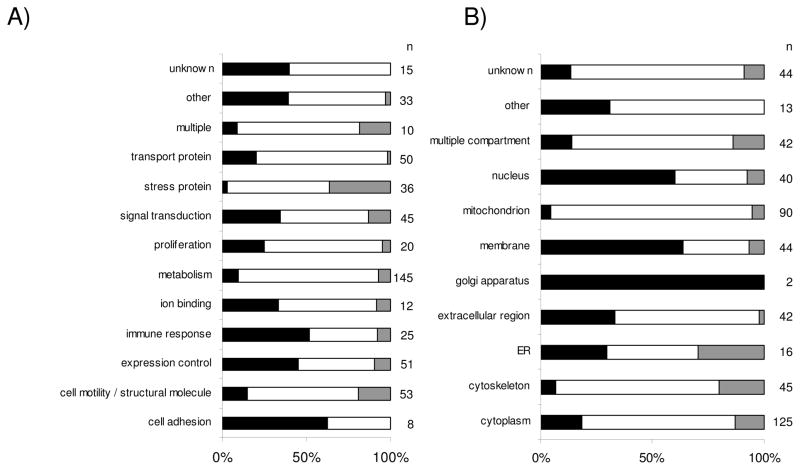
Classification of unique proteins identified by PROTEOMEX. The pie charts display the classification of differentially expressed protein spots defined by PROTEOMEX using three RCC cell lines and one normal renal epithelium representing cell line and based on immunostainings obtained with serum samples from 7 healthy individuals and 8 RCC patients, respectively according to (A) functional protein classes and (B) the subcellular distribution of the identified targets. The layout is in analogy to Fig. 1
Putative candidate biomarkers of RCC identified by transcriptomics
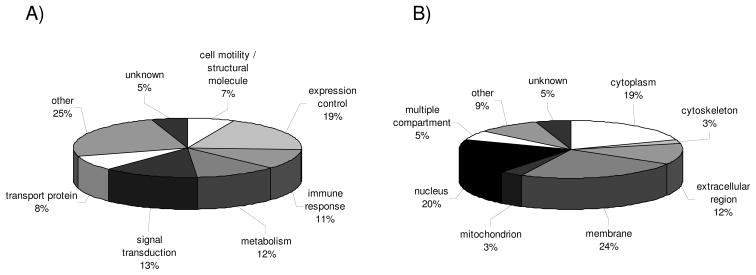
Using cDNA microarrays 17 RCC lesions versus corresponding normal kidney epithelium were analyzed for their distinct mRNA expression pattern (Table 1B) [16] leading to the detection of 119 genes with significant upregulated transcript levels. Gene ontology was used to classify the set of upregulated genes according to their biological functions and cellular compartments. The comparative profiling at the transcriptional level revealed a increased mRNA expression pattern for genes involved in the control of gene/protein expression (19%), in signal transduction processes (13%), in cellular metabolism (12%), in the immune response (11%), transport proteins (8%) as well as molecules involved in cell motility and cell structure maintenance (7%). The 25% of genes classified as “others” by this approach were involved in cellular stress (1%), proliferation (4%), ion binding (3%), cell adhesion (4%), multiple function (1%) next to a group of differentially expressed genes which despite having ontology annotations could not be assigned to any of the given functional classes (12%, Fig. 3A). Aside from their physiological function the subset of upregulated candidate biomarkers were also categorized according to their distinct cellular localization (Fig. 3B). The leading compartments here are the membrane (24%), the nucleus (20%), as well as the cytoplasm (19%). Moreover, it is noteworthy that the transcriptomic profiling revealed the highest frequency of genes assigned to the extracellular region (12%).
Figure 3.
Classification of differentially expressed genes identified by transcriptome analyses. The pie charts display the data sets obtained by transcriptomics of 13 RCC lesions of the clear cell type, 1 chromophobic and 2 chromophilic RCC lesion along with corresponding normal kidney epithelium and 1 leiomyosarcoma according to their function (A) and cellular localization (B). The layout is in analogy to Fig. 1.
Target sharing of candidate biomarkers
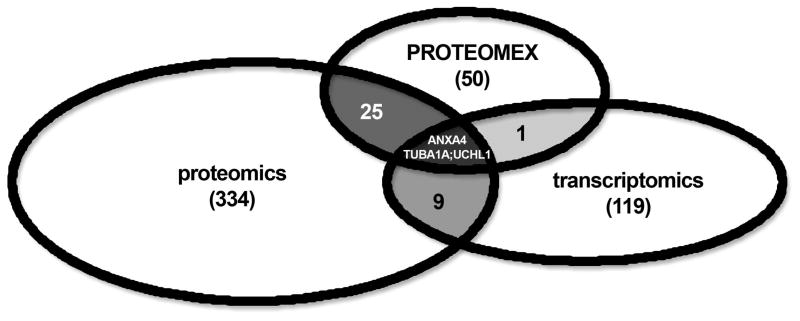
As shown in Fig. 4, the target sharing rates between these three technologies strongly differed. As expected there exists the broadest overlap of candidate biomarkers between the 2DE gel-based proteomics and PROTEOMEX approaches. 28 of the 50 proteins identified by PROTEOMEX were also found to be differentially expressed using classical proteomics (Table 2, Fig. 4, 5). In particular, proteins involved in the metabolism and stress responses as well as in cell proliferation and cytoskeletal formation, such as for example aldose reductase, HSP 27, endoplasmin, annexin (ANX) A2, moesin and vimentin were found in the overlapping set of candidate biomarkers/segment (Table 2, Fig. 4, 5).
Figure 4.
Combined target sharing between the different “ome”-based approaches. Each of the distinct “ome”-based technologies is represented by an oval. The total number of targets identified via the given approach is listed in brackets. The number of shared proteins between the different approaches is stated in the overlapping segments, whereas the three candidate biomarkers defined by all three approaches are stated in the core segment according to their gene names.
Table 2.
Unique targets shared between different approaches.
| Swiss- entry name Prot ID | identification strategy | cov % | score | family | compartment | regulation at the protein level | sharing | |||
|---|---|---|---|---|---|---|---|---|---|---|
| UP | DN | |||||||||
| O00299 Chloride intracellular channel protein 1 | MS / MSMS | 51 | 98 | signal transduction | multiple compartment | 1 | 2 | p | t | |
| P12110 Collagen alpha-2(VI) chain | MS | NA | 41 | cell adhesion | extracellular region | 1 | p | t | ||
| P49411 Elongation factor Tu | MS | 35 | 97 | expression control | mitochondrion | 1 | p | t | ||
| P09104 Gamma-enolase | MS / MSMS | 33 | 155 | metabolism | cytoplasm | 1 | p | t | ||
| P04406 Glyceraldehyde-3-phosphate dehydrogenase | MS / MSMS | 41 | 55 | metabolism | cytoplasm | 6 | 1 | p | t | |
| P00338 L-lactate dehydrogenase A chain | MSMS | 24* | 1e506* | metabolism | cytoplasm | 2 | p | t | ||
| P07237 Protein disulfide-isomerase | MS / MSMS | 60 | 271 | multiple | multiple compartment | 4 | 1 | p | t | |
| Q14554 Protein disulfide-isomerase A5 | MS | 60 | 271 | stress protein | ER | 2 | p | t | ||
| P50991 T-complex protein 1 subunit delta | MSMS | 27* | 1e456* | proliferation | cytoplasm | 1 | p | t | ||
| P10809 60 kDa heat shock protein | MS / MSMS | 48 | 187 | stress protein | mitochondrion | 10 | 8 | p | px | |
| P11021 78 kDa glucose-regulated protein | MS / MSMS | 34 | 146 | stress protein | ER | 10 | 1 | p | px | |
| P63261 Actin, cytoplasmic 2 | MS / MSMS / Edman | 41 | 82 | cell motility | cytoskeleton | 12 | 10 | p | px | |
| P15121 Aldose reductase | MS / MSMS / Edman | NA | NA | stress protein | extracellular region | 1 | p | px | ||
| P06733 Alpha-enolase | MS / MSMS | 55 | 187 | expression control | multiple compartment | 4 | 2 | p | px | |
| P07355 Annexin A2 | MS / MSMS | 53 | 161 | proliferation | membrane | 2 | 2 | p | px | |
| P08758 Annexin A5 | MS | 54 | 127 | signal transduction | cytoplasm | 2 | 1 | p | px | |
| P14625 Endoplasmin | MS / MSMS | 19 | 134 | stress protein | ER | 7 | 4 | p | px | |
| P30084 Enoyl-CoA hydratase | MS / MSMS | 53 | 113 | metabolism | mitochondrion | 2 | 6 | p | px | |
| P08263 Glutathione S-transferase A1 | MS / MSMS | 34 | 103 | metabolism | cytoplasm | 4 | p | px | ||
| P09211 Glutathione S-transferase P | MS / MSMS | 52 | 85 | metabolism | cytoplasm | 2 | 3 | p | px | |
| P48637 Glutathione synthetase | MS / MSMS | 43 | 127 | metabolism | cytoplasm | 1 | 2 | p | px | |
| P11142 Heat shock cognate 70 kDa protein | MS | 21 | NA | stress protein | cytoplasm | 1 | 1 | p | px | |
| Q12931 Heat shock protein 75 kDa | MS | NA | 90 | stress protein | mitochondrion | 1 | p | px | ||
| P04792 Heat shock protein beta-1 | MS / MSMS | 55 | 133 | stress protein | multiple compartment | 11 | 6 | p | px | |
| P08238 Heat shock protein HSP 90-beta | MS | 16 | NA | stress protein | cytoplasm | p | px | |||
| P05787 Keratin, type II cytoskeletal 8 | MS / MSMS | 38 | 168 | structural molecule | cytoskeleton | 6 | 2 | p | px | |
| P26038 Moesin | MS / MSMS | 33 | 121 | cell motility | cytoskeleton | 2 | 1 | p | px | |
| P38646 Stress-70 protein | MS / MSMS | 50 | 203 | stress protein | multiple compartment | 7 | 5 | p | px | |
| P04179 Superoxide dismutase [Mn] | MS / MSMS | 34 | 64 | stress protein | mitochondrion | 10 | p | px | ||
| P10599 Thioredoxin | MS / MSMS | 32 | 35 | signal transduction | cytoplasm | 2 | p | px | ||
| P60174 Triosephosphate isomerase | MS / MSMS | 50 | 107 | metabolism | unknown | 8 | 2 | p | px | |
| P06753 Tropomyosin alpha-3 chain | MS | 34 | 141 | cell motility | cytoskeleton | 1 | 1 | p | px | |
| P68366 Tubulin alpha-4A chain | MS / MSMS | 35 | 127 | structural molecule | cytoskeleton | 4 | 2 | p | px | |
| P08670 Vimentin | MS / MSMS | 35 | 87 | cell motility | cytoskeleton | 11 | 6 | p | px | |
| P40261 Nicotinamide N-methyltransferase | MS / Edman | 54 | NA | other | cytoplasm | t | px | |||
| P09525 Annexin A4 | MS / MSMS | 56 | 158 | signal transduction | cytoplasm | 8 | 6 | p | t | px |
| Q71U36 Tubulin alpha-1A chain | MS / MSMS | 47 | 128 | structural molecule | mitochondrion | 1 | p | t | px | |
| P09936 Ubiquitin carboxyl-terminal hydrolase isozyme L1 | Edman | NA | NA | metabolism | cytoplasm | 3 | 12 | p | t | px |
Coverage (cov) and score values mostly represent data obtained with the Mascot search engine whereas values marked with an * were derived from the Profound search engine; upregulated (UP) or downregulated (DN) in independent RCC systems, p: proteomics, px: PROTEOMEX, t: transcriptomics.
Figure 5.
Relative distribution pattern of the distinct gene/protein families and compartments using the three ”ome”-based technologies. The bar plots display the relative distribution pattern of genes/proteins which are differentially expressed/immunoreactive applying the various techniques. Black bars represent the number of genes identified by transcriptomics, white bars the number of proteins identified by classical proteomics and grey bars represent the PROTEOMEX targets. (A) illustrates the relative distribution of the gene/protein families, (B) the subcellular distribution of the proteins. The total number of genes/proteins in each bar segment is listed under n.
In contrast, the number of shared candidate biomarkers between proteome-based methods and transcriptomics is much lower, in the range of 10%, respectively. Whereas the overlapping segment between proteomics and transcriptomics is defined by only twelve candidate biomarkers merely 4 candidate biomarkers are shared between PROTEOMEX and transcriptomics (Table 2, Fig. 4, 5). These data suggest that transcriptomic profiling detects indeed a distinct set of genes, which was further supported by differences in the targeting of membrane and nuclear proteins. The importance of posttranscriptional regulatory mechanisms in RCC was suggested since for example the candidate biomarker ubiquitin carboxyl-terminal hydrolase L1 (UCHL1), was detectable at the transcript level, but was not found at the protein level of some RCC cell lines [30].
Taken together each of these technologies differs in terms of their sensitivity, complexity and advantages/disadvantages. The total number of candidate biomarkers detected by all of the experimental strategies is currently limited to 3, namely annexin A4 (ANXA4), tubulin alpha-1A chain (TUBA1A) and ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) (Table 2, Fig. 4).
Validation of candidate biomarkers identified by “ome”-based technologies
In order to determine whether the results obtained by comparative transcriptomics and proteomics reflect indeed a common gene/protein expression pattern of RCC lesions, of distinct subtypes, of tumor gradings and stagings, target validation of differentially expressed genes and/or proteins was performed employing RT-PCR, Western blot analysis and/or immunohistochemistry.
For immunohistochemical stainings either single paraffin-embedded tissue samples or tissue micro arrays (TMA) consisting of a large series of tumor lesions and corresponding normal renal epithelium (>200) were employed as recently described [30], whereas for RT-PCR and Western blot analysis either sets of 7 to 21 established RCC and/or normal renal epithelium representing cell lines and/or series of respective biopsy specimens were used. Concerning our own data the initially selected proteins were prioritized concerning their biological significance, availability of antibodies and relative novelity in RCC. In addition to our own validation data also validation data obtained by other groups were considered (Table 3).
Table 3.
Currently available validation data on selected candidate biomarkers as defined by “ome”-based technologies in renal cell carcinoma.
| Target validation | |||||
|---|---|---|---|---|---|
| Swiss- Prot ID | entry name | regulation pattern1) | approach | validation method | reference |
| P10809 | 60 kDa heat shock protein | (+/−) | p, px | WB | [21] |
| P11021 | 78 kDa glucose-regulated protein | (+/−) | p, px | WB | [21] |
| Q9BSE5 | Agmatinase | (−) | p | IHC, RT- PCR, WB | [57] |
| P02511 | Alpha-crystallin B chain | (+) | p | RT-PCR, WB | [18] |
| P06733 | Alpha-enolase | (+) | p, px | IHC | this report |
| P04083 | Annexin A1 | (+) | p | IHC | [36] |
| P07355 | Annexin A2 | (+) | p, px | IHC | [36] |
| P12429 | Annexin A3 | (+) | px | IHC | this report |
| P09525 | Annexin A4 | (+) | p, px, t | IHC, RT- PCR, WB | [18, 36, 48] |
| P23528 | Cofilin-1 | (+) | p | IHC | [36] |
| P31930 | Cytochrome b-c1 complex subunit 1 | (−) | p | WB | [24] |
| P22695 | Cytochrome b-c1 complex subunit 2 | (−) | p | WB | [24] |
| P14625 | Endoplasmin | (+) | p, px | WB | [21] |
| P30084 | Enoyl-CoA hydratase | (−) | p, px | RT-PCR | [37] |
| P15311 | Ezrin | (+/−) | p | WB | [24] |
| O15540 | Fatty acid-binding protein, brain | (+) | p | RT-PCR | [23] |
| P07148 | Fatty acid-binding protein, liver | (−) | p | RT-PCR, IHC | [23] |
| Q16658 | Fascin | (+) | p | WB | [36] |
| P04075 | Fructose-bisphosphate aldolase A | (+) | p | WB | [36] |
| P05062 | Fructose-bisphosphate aldolase B | (−) | p | WB | [24] [36] |
| P09211 | Glutathione S-transferase P | (−) | p, px | WB, IHC | [22] |
| P04406 | Glyceraldehyde-3-phosphate dehydrogenase | (+) | p, t | WB | [36] |
| P08108 | Heat shock cognate 70 kDa protein | (+/−) | p, px | WB | [21] |
| P04792 | Heat shock Protein 27 | (+/−) | p, px | WB, qRT- PCR, IHC | [20, 21, 25] |
| P08238 | Heat shock protein HSP 90-beta | (−) | p, px | WB | [21] |
| P05787 | Keratin, type II cytoskeletal 8 | (−) | p, px | WB, IHC | [19] |
| P26038 | Moesin | (+/−) | p, px | WB | [24] |
| Q96RQ3 | Methylcrotonoyl-CoA carboxylase subunit alpha | (+) | p | WB | [24] |
| Q9P0J0 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13 | (−) | p | WB, IHC | [58] |
| O75489 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 3 | (−) | p | WB | [24] |
| P05165 | Propionyl-CoA carboxylase alpha chain | (+) | p | WB | [24] |
| P11498 | Pyruvate carboxylase | (−) | p | WB | [24] |
| P11177 | Pyruvate dehydrogenase E1 component subunit beta | (−) | p | WB | [24] |
| P14618 | Pyruvate kinase isozymes M1/M2 | (+) | p | WB | [24, 36] |
| P35241 | Radixin | (+) | p | WB | [24] |
| Q15019 | Septin-2 | (+) | p | WB, IHC | [24] |
| P38646 | Stress-70 protein | (−) | p, px | WB | [21] |
| P04179 | Superoxide dismutase [Mn] | (+) | p, px | qRT-PCR , WB, IHC | [18, 22, 25] |
| P10599 | Thioredoxin | (+) | p, px | WB, | [22] |
| P19971 | Thymidine phosphorylase | (+) | p | IHC | [36] |
| P60174 | Triosephosphate isomerase | (+) | p, px | WB, IHC | [22] |
| P06753 | Tropomyosin alpha-3 chain | (+) | p, px | WB | [19] |
| Q71U36 | Tubulin alpha-3 chain | (+/−) | p, px | WB | [19] |
| P09936 | Ubiquitin carboxyl-terminal hydrolase L1 | (−) | p, px, t | WB, IHC, RT-PCR | [22, 30] |
| P08670 | Vimentin | (+) | p, px | IHC, WB | [19, 20] |
according to currently available RCC literature, (+) upregulated in RCC, (−) downregulated in RCC, p: proteomics, px: PROTEOMEX, t: transcriptomics, RT-/qRT-PCR: semi-quantitative/real time polymerase chain reaction, WB Western blot analysis, IHC: immunohistochemistry. Potential candidates for forthcoming biomarkers are highlighted in bold.
The simultaneous analysis of the expression pattern in different RCC systems demonstrated that various members of the HSP family known to be involved in the induction of stress responses, apoptosis and in the development of anti-tumor immunity were differentially expressed in RCC lesions when compared to normal kidney epithelium [21, 34]. This was confirmed by RT-PCR and Western blot analysis on a series of RCC cell lines and short-term cultures of normal kidney epithelium. In addition, a number of post-translational modifications for HSP 27 were identified, which were also differentially detected by the PROTEOMEX approach demonstrating indeed a distinct immune recognition pattern of HSP 27 variants [21].
Structural proteins like cytoskeletal components are mainly co-regulated at the transcriptomic and proteomic level in RCC. Using these approaches a significantly heterogeneous expression pattern of different members of the cytoskeleton including for example cytokeratin (CK) 8, vimentin and stathmin was found between distinct RCC subtypes and corresponding normal kidney epithelium. The validation of their expression patterns by Western blot analysis and immunohistochemistry in a series of RCC systems, comprised of tumor and corresponding normal epithelium, revealed similar results therefore confirming the “ome”-based data. Thus, the co-expression of CK8, vimentin and/or stathmin can be used as a tool to distinguish between the different RCC subtypes, since their combined expression is in particular associated with RCC of the clear cell type [35]. Moreover, antibody responses in RCC patients were detected against various cytokeratins, cytoskeletal tropomyosin, F-actin capping protein and vimentin. Thus, some of the cytoskeletal proteins identified by proteomics and/or PROTEOMEX characterized RCC subtype-specific changes, which had not been previously reported.
A discordance between mRNA and protein abundance in RCC systems was found for the differentially expressed family of metabolic enzymes [22, 36, 37], such as enzymes involved in the carbohydrate, lipid and phosphate metabolism, but also in the maintenance of the cellular redox potential as well as components of the antigen processing machinery. This suggests that posttranscriptional alterations in these different cellular processes might play an important role in RCC. Some of these markers were also detected by the PROTEOMEX approach and were further validated by Western blot and/or immunohistochemistry. As representatively shown for members of the fatty acid binding protein (FABP) family [23] an almost complete loss of L-FABP was found in RCC lesions of the clear cell, chromophilic and chromophobic subtype, whereas the loss of L-FABP was restricted to about 50% of benign oncocytoma. Thus, lack of L-FABP is independent of the RCC subtype, but associated with the malignant phenotype of this disease. In contrast, a RCC subtype-specific expression pattern was detected for B-FABP in RCC of the clear cell subtype and for H-FABP in tumors of the oncocytic subtype. Due to the small number of chromophobic and chromophilic RCC and oncocytomas analyzed thus far, which is based on the relative low frequencies of these RCC subtypes, the validation of candidate biomarkers both at the mRNA and protein level has yet to be performed on a significantly higher number of specimens in order to bolster their statistical relevance.
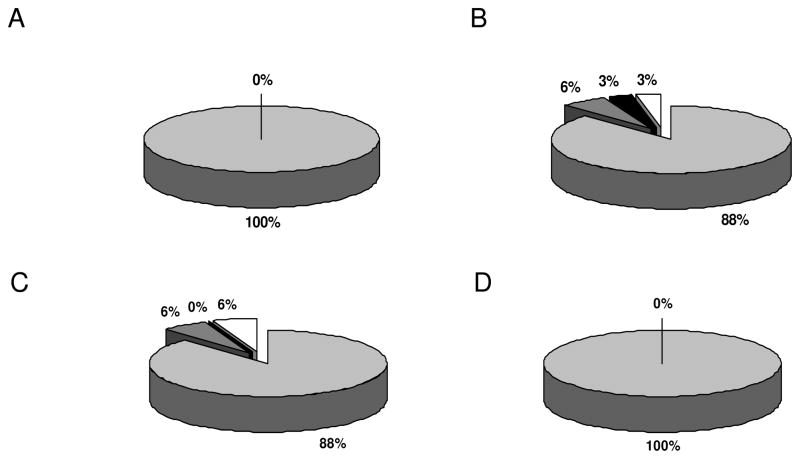
For one differentially expressed target, UCHL1, which represents one of the 3 candidate biomarkers defined by all three “ome”-based strategies employed, the validation data were further extended by performing functional assays. The expression of UCHL1 is down-regulated in the early phase of RCC tumorigenesis, whereas it is re-expressed at the metastatic stage [30] as determined by RT-PCR, Western blot analysis and immunohistochemistry on a large series of RCC specimens and RCC cell lines. In addition, UCHL1 gain of function variants not only exhibited increased cell cycle rates, but also an improved cell motility and migration capacity, which is in line with features required by metastatic cells. The down-regulation or lack of UCHL1 is in most cases due to an epigenetic silencing and can be reverted by demethylating agents (Seliger, personal communication). Enolase is a surface protein and a key enzyme of the glycolysis. It exerts multiple functions, serves as a plasminogen receptor on the surface of different cell types, including epithelial cells and appears to be involved in systemic autoimmune disorders [38, 39] . Alpha-enolase has been shown to be recognized by autoantibodies infections and autoimmune diseases [40, 41]. In addition, alpha-enolase has been recently identified as a tumor antigen in non-small-lung carcinoma [42]. We here extended the analysis of alpha-enolase to RCC, which is mainly expressed at high levels in all RCC subtypes. Only 6% of chromophilic and chromophobic RCC, respectively, exhibit intermediate and no more than 6% show low or even lack alpha-enolase expression (Fig. 6). Since alpha-enolase was detected by both proteomics and PROTEOMEX analyses it could be traced by autoantibodies in the serum of RCC patients. Furthermore, alpha-enolase has the capacity to function as a HSP and binds cytoskeletal and chromatin structures. Its coordinated expression together with various HSPs and cytoskeletal components suggests a role in gene transcription and the pathophysiology of RCC. In this context, it is moreover noteworthy that HSP 27 and alpha enolase expression in pancreatic cancer might represent candidate biomarkers for resistance to chemotherapy as well as response prediction [43].
Figure 6.
Immunohistochemical analysis for alpha-enolase expression by tissue micro arrays technology. RCC tissue micro arrays were stained with the alpha-enolase specific antibody as described in Material and methods section. Panel A shows the staining pattern of RCC of clear cell subtype, panel B that of chromophilic subtype, panel C that of chromophobic subtype and panel D renal cell adenomas of oncocytic type. Light grey segments indicate strong positive, dark grey segments intermediate, black segments weak staining and blank segments no staining. The relative staining frequencies are indicated by providing the respective percentage for each segment.
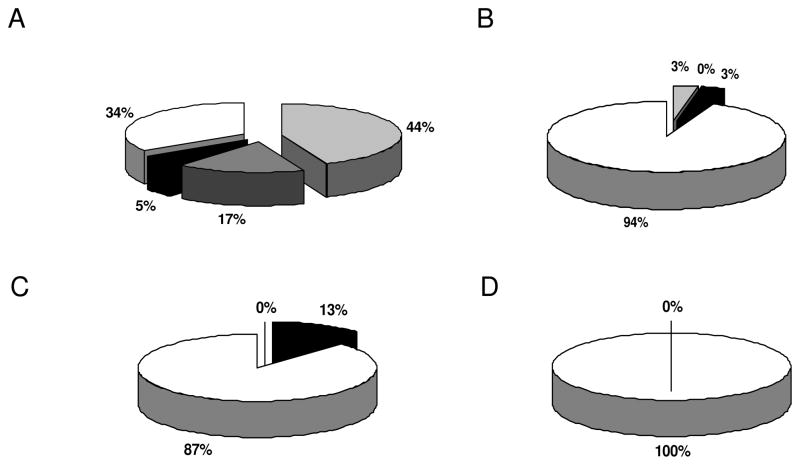
Annexin A3 (ANXA3) represents a rare member of a family of calcium-binding proteins, which are involved in membrane trafficking, lymphocyte migration and stimulation of specific immune responses [44]. Furthermore, ANXA3 plays an important role in angiogenesis [45]. Although ANXA3 has not been intensely analyzed in human tumors, it appears to represent a promising candidate biomarker for prostate carcinoma when combined with standard prognostic parameters. Negative staining of ANXA3 correlated with prognosis and progression of prostate cancer [46, 47]. We here extended the analysis to RCC demonstrating an extremely heterogeneous expression pattern in the different RCC subtypes (Fig. 7). In particular, in RCC of clear cell subtype 44% of the tumor lesions exhibit a strong, 17% an intermediate and 5% a weak expression of ANXA3, whereas 34% of these RCC lesions totally lack ANXA3 staining. In contrast, the loss of ANXA3 expression was detected in 94% of chromophilic, 87% of chromophobic RCC and in 100% of adenomas of the oncocytic type (Fig. 7), which is in line with the results obtained in prostate cancer. In addition, it is noteworthy that ANXA4 another member of the annexin family was also detected to be differentially expressed/immunoreactive in the RCC systems analysed, independent from the employed mRNA and/or protein profiling strategy. Furthermore, these findings are in line with the data provided by Zimmermann and co-authors [48] describing an increased expression of ANXA4 in RCC, which was even associated with its altered cellular localization. Thus, ANX family members may be involved in the development, progression and/or dissemination of RCC and thus way well serve as putative candidate biomarkers. Taken together the expression/recognition pattern of the selected putative biomarkers identified by either transcriptomics, classical proteomics or PROTEOMEX was largely confirmed for a number of putative RCC biomarkers by at least one, for the majority of targets even by two of the three independent validation methods proving that the alterations detected by the different “ome”-based approaches were reliable.
Figure 7.
Immunohistochemical analysis for annexin A3 expression by tissue micro arrays technology. RCC tissue micro arrays were stained with the annexin A3 specific antibody as described in Material and methods section. Panel A shows the staining pattern of RCC of clear cell subtype, panel B that of chromophilic subtype, panel C that of chromophobic subtype and panel D renal cell adenomas of oncocytic type. Light grey segments indicate strong positive, dark grey segments intermediate, black segments weak staining and blank segments indicate no staining. The relative staining frequencies are indicated by providing the respective percentage for each segment.
Discussion
Analyses integrating mRNA and protein profilings in mammalian cells suggest that there exists an approximately 60–80% discordance between mRNA and protein abundances, which is mainly due to posttranscriptional/posttranslational changes [3, 49]. Based on this knowledge, a combination of transcriptomics and proteome-based technologies seems to be a promising strategy to provide interesting candidate genes and proteins that can be developed as potential indicators of diagnostic and prognostic values for RCC or even used as novel therapeutic target structures. Although it is generally accepted that proteins represent key molecular entities that play a more direct role in dictating cellular activities than RNA. Proteome-based analysis are hampered by three major kinds of limitations: (i) the high dynamic range within the protein expression profiles [50], (ii) the vast complexity of the proteome to be analyzed and (iii) the technical limitations of the available analytical methods [51]. Therefore the implementation of both transcriptome and proteome defining strategies appear to complement each other and facilitate the identification of novel candidate biomarkers in RCC. However, it is noteworthy that the integration of data from different technology platforms is a challenge due to the heterogeneity of results. Statistical and bioinformatic approaches are required to develop a comprehensive data set, which includes annotations, functional correlations as well as the results of validation studies. The systematic analysis of the mRNA and/or protein expression pattern of RCC lesions versus normal renal epithelium provides the first clue into alterations involved in the development of this disease and further delineates the signaling networks and the regulation of cell functions during malignant transformation. In addition the identification of markers can be used for tumor class discovery, by developing algorithms based on transcriptome and proteome data and by determining the physiological and clinical meaning of the putative classes [27, 52]. So far, the present study describes the most extensive effort of data integration from RCC systems using such distinct technology platforms as transcriptome- and proteome-based analyses and demonstrates advantages and disadvantages of these various approaches in terms of searching for candidate biomarker detection. We have used 2DE-based classical proteomics and PROTEOMEX in combination with cDNA microarrays to screen paired RCC lesions and normal kidney. Whereas proteomics enables the identification of posttranscriptional as well as posttranslational modifications including degradation products, transcriptome analysis facilitates the identification of genes/candidate biomarkers coding for hydrophobic, extreme basic or weakly expressed proteins, which are still difficult to determine by 2DE gel-based analysis [50, 51]. Thus the data obtained with these different experimental approaches were first collected in an in house designed data base, converted into a common format by extracting all of the relevant public domain available information from various databases and subsequently classified into various functional families and according to their cellular compartments in order to allow their presentation in comparable charts (Fig. 1–3).
This sub-grouping allows the discrimination between co- and dysregulated gene/protein families involved in the neoplastic phenotype of RCC. If we compare the categories and frequencies of shared targets there exists a larger difference between candidate biomarkers identified by transcriptomics and proteomics than between the proteome-based technologies employed.
PROTEOMEX appears not only to represent a powerful tool for the detection of immune reactive proteins, but moreover helps to trim down the long lists of candidate biomarkers defined by proteomics through highlighting target structures which are recognized by the immune system [19–21]. Thus far in particular putative candidates shared between these approaches showed promising biomarker features. In addition, this technology allows the identification of putative targets which cannot be detected by the other two technologies demonstrating the necessity of its implementation to provide complementary information regarding RCC-specific alterations.
The observed discrepancy of mRNA and protein expression levels in RCC systems is in accordance with other publications analyzing hematopoietic cells of distinct differentiation status [53] and lung carcinoma systems [54]. Regarding the comparison between transcriptomics and PROTEOMEX only 4 targets were shared. These belong to different proteins families, such as metabolic enzymes (nicotinamide-N-methyltransferase, UCHL1), structural molecules, tubulin alpha1A chain (TUBA1A) and signal transduction molecules (ANXA4) (Table 2; Fig. 4, 5). While transcriptomics might identify in particular genes coding for membrane and/or low abundant proteins, such as genes involved in signal transduction processes and the regulation of gene/protein expression levels, these markers might not be easily detected by proteome-based methods due to the extreme complexity of the proteome and the obvious restrictions concerning both the staining and detection sensitivity of these methods. The rate of candidate biomarkers defined by all three methods is currently limited to just three proteins, ANXA4, TUBA1A and UCHL1 (Table 2; Fig. 4).
Based on the low frequency of shared targets between proteome-based methods and transcriptomics, there exists a capacity of falsification. Proteomics might falsify transcriptomics, but not vice versa due to the limited correlation of these approaches. Thus validations of selected markers by Western blot and/or immunohistochemistry and in some cases RT-PCR analyses were performed. The distinct protein expression patterns between RCC lesions and normal kidney epithelium obtained by proteomics and PROTEOMEX was confirmed employing different validation technologies. Since some candidate biomarkers were monitored on a broad series of tissue samples even RCC subtype- or grading-dependent target expression pattern were found [20, 23, 30]. Some of the differentially expressed candidate biomarkers like alpha-enolase and UCHL1 were defined in RCC by classical proteomics and by PROTEOMEX. The strong expression of alpha-enolase observed in all RCC subtypes indicates that this molecule might serve as a candidate biomarker for the discrimination between RCC lesions and normal kidney epithelium. The detection of autoantibodies directed against this molecule in different infectious and autoimmune disorders [38], but also in RCC might further suggest that alpha-enolase represents a potent antibody target. These data stress that (i) overlapping identifications can be defined by analyzing both the mRNA and protein expression profile, (ii) that all three technologies are able to identify independently a substantial number of biological targets with high reproducibility and robustness and (iii) that the discrepancy between the mRNA and protein abundance is mainly due to the biology of gene expression rather than measurement errors. In this context it is noteworthy that falsification was shown for UCHL1 [30].
Despite the major advances in recent RCC studies obtained by functional transcriptome and proteome analysis, the discovery of suitable biomarkers for early diagnosis and prognosis of RCC is still urgently needed. This might in fact be due to an enormous heterogeneity of this tumor entity. Clearly subsequent studies have to be designed with larger numbers of cell lines and tissues. However, also on a more proper quantitative comparison of transcriptome versus proteome data by implementation of quantitative rather than semi-quantitative proteome techniques such as ICAT and ICPL labeling or 2D-DIGE in combination with mass spectrometry and a more extensive characterization of the proteins identified so that a priorization could occur at an early step of the analysis [54]. In order to achieve this goal it will be important in the future to combine the complementary quantitative ”ome”-based technologies and to focus the analysis on different subtypes of RCC as well as different tumor grading and staging profiles and to perform multivariable analysis using clinicopathological parameters including the survival of patients. Furthermore, a detailed view on molecular changes in pathways known to be altered in RCC, like the Von-Hippel-Lindau (VHL) protein pathway, will present an innovative approach to identify novel markers [55, 56]. Although a new group of targeting kinase inhibitors recently showed promising results in the treatment of RCC patients [14, 15], the identification of molecular biomarkers suitable as clinical tools still remains a priority of research in this disease.
Acknowledgments
We would like to thank C. Stoerr and A. Wasilewski for excellent secretarial help. This work represents part of the PhD thesis of Sven Dressler. This work was funded by the BMBF (BMBF project 031U101H (B.S.), NGFN II EP grant 0313376 (B.S) and the Roux program of the Martin-Luther-University Halle-Wittenberg (FKZ 16/30 (M.B.).
Abbreviations
- Ab
antibody
- AEC
amino-β-ethylcarbazol
- ANX
annexin
- aRNA
anti-sense RNA
- CK
cytokeratin
- FABP
fatty acid binding protein
- HSP
heat shock protein
- pAb
polyclonal antibody
- PROTEOMEX
combination of classical proteomics and SEREX
- RCC
renal cell carcinoma
- TUBA1A
tubulin alpha 1A chain
- UCHL1
ubiquitin carboxyl-terminal hydrolase L1
References
- 1.Ding C, Cantor CR. Quantitative analysis of nucleic acids--the last few years of progress. J Biochem Mol Biol. 2004;37:1–10. doi: 10.5483/bmbrep.2004.37.1.001. [DOI] [PubMed] [Google Scholar]
- 2.Guo QM. DNA microarray and cancer. Curr Opin Oncol. 2003;15:36–43. doi: 10.1097/00001622-200301000-00005. [DOI] [PubMed] [Google Scholar]
- 3.Waters KM, Pounds JG, Thrall BD. Data merging for integrated microarray and proteomic analysis. Brief Funct Genomic Proteomic. 2006;5:261–272. doi: 10.1093/bfgp/ell019. [DOI] [PubMed] [Google Scholar]
- 4.Tian Q, Stepaniants SB, Mao M, Weng L, et al. Integrated genomic and proteomic analyses of gene expression in Mammalian cells. Mol Cell Proteomics. 2004;3:960–969. doi: 10.1074/mcp.M400055-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Brockmann R, Beyer A, Heinisch JJ, Wilhelm T. Posttranscriptional expression regulation: what determines translation rates? PLoS Comput Biol. 2007;3:e57. doi: 10.1371/journal.pcbi.0030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ideker T, Thorsson V, Ranish JA, Christmas R, et al. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 7.Graham DR, Garnham CP, Fu Q, Robbins J, Van Eyk JE. Improvements in two-dimensional gel electrophoresis by utilizing a low cost "in-house" neutral pH sodium dodecyl sulfate-polyacrylamide gel electrophoresis system. Proteomics. 2005;5:2309–2314. doi: 10.1002/pmic.200401249. [DOI] [PubMed] [Google Scholar]
- 8.Miles AK, Matharoo-Ball B, Li G, Ahmad M, Rees RC. The identification of human tumour antigens: Current status and future developments. Cancer Immunol Immunother. 2006;55:996–1003. doi: 10.1007/s00262-005-0115-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merrick BA, Bruno ME. Genomic and proteomic profiling for biomarkers and signature profiles of toxicity. Curr Opin Mol Ther. 2004;6:600–607. [PubMed] [Google Scholar]
- 10.Hu YF, Kaplow J, He Y. From traditional biomarkers to transcriptome analysis in drug development. Curr Mol Med. 2005;5:29–38. doi: 10.2174/1566524053152915. [DOI] [PubMed] [Google Scholar]
- 11.Seliger B, Lichtenfels R, Kellner R. Detection of renal cell carcinoma-associated markers via proteome- and other 'ome'-based analyses. Brief Funct Genomic Proteomic. 2003;2:194–212. doi: 10.1093/bfgp/2.3.194. [DOI] [PubMed] [Google Scholar]
- 12.Mostofi FKaD, CJ . Histological typing of kidney tumors. 1998. [Google Scholar]
- 13.Gouttefangeas C, Stenzl A, Stevanovic S, Rammensee HG. Immunotherapy of renal cell carcinoma. Cancer Immunol Immunother. 2007;56:117–128. doi: 10.1007/s00262-006-0172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmad T, Eisen T. Kinase inhibition with BAY 43-9006 in renal cell carcinoma. Clin Cancer Res. 2004;10:6388S–6392S. doi: 10.1158/1078-0432.CCR-040028. [DOI] [PubMed] [Google Scholar]
- 15.Marx J. Cancer. Encouraging results for second-generation antiangiogenesis drugs. Science. 2005;308:1248–1249. doi: 10.1126/science.308.5726.1248. [DOI] [PubMed] [Google Scholar]
- 16.Wang E, Lichtenfels R, Bukur J, Ngalame Y, et al. Ontogeny and oncogenesis balance the transcriptional profile of renal cell cancer. Cancer Res. 2004;64:7279–7287. doi: 10.1158/0008-5472.CAN-04-1597. [DOI] [PubMed] [Google Scholar]
- 17.Scherer A, Krause A, Walker JR, Sutton SE, et al. Optimized protocol for linear RNA amplification and application to gene expression profiling of human renal biopsies. Biotechniques. 2003;34:546–550. 552–544, 556. doi: 10.2144/03343rr01. [DOI] [PubMed] [Google Scholar]
- 18.Shi T, Dong F, Liou LS, Duan ZH, et al. Differential protein profiling in renal-cell carcinoma. Mol Carcinog. 2004;40:47–61. doi: 10.1002/mc.20015. [DOI] [PubMed] [Google Scholar]
- 19.Kellner R, Lichtenfels R, Atkins D, Bukur J, et al. Targeting of tumor associated antigens in renal cell carcinoma using proteome-based analysis and their clinical significance. Proteomics. 2002;2:1743–1751. doi: 10.1002/1615-9861(200212)2:12<1743::AID-PROT1743>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 20.Seliger B, Menig M, Lichtenfels R, Atkins D, et al. Identification of markers for the selection of patients undergoing renal cell carcinoma-specific immunotherapy. Proteomics. 2003;3:979–990. doi: 10.1002/pmic.200300404. [DOI] [PubMed] [Google Scholar]
- 21.Lichtenfels R, Kellner R, Bukur J, Beck J, et al. Heat shock protein expression and anti-heat shock protein reactivity in renal cell carcinoma. Proteomics. 2002;2:561–570. doi: 10.1002/1615-9861(200205)2:5<561::AID-PROT561>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 22.Lichtenfels R, Kellner R, Atkins D, Bukur J, et al. Identification of metabolic enzymes in renal cell carcinoma utilizing PROTEOMEX analyses. Biochim Biophys Acta. 2003;1646:21–31. doi: 10.1016/s1570-9639(02)00547-2. [DOI] [PubMed] [Google Scholar]
- 23.Seliger B, Lichtenfels R, Atkins D, Bukur J, et al. Identification of fatty acid binding proteins as markers associated with the initiation and/or progression of renal cell carcinoma. Proteomics. 2005;5:2631–2640. doi: 10.1002/pmic.200401264. [DOI] [PubMed] [Google Scholar]
- 24.Craven RA, Stanley AJ, Hanrahan S, Dods J, et al. Proteomic analysis of primary cell lines identifies protein changes present in renal cell carcinoma. Proteomics. 2006;6:2853–2864. doi: 10.1002/pmic.200500549. [DOI] [PubMed] [Google Scholar]
- 25.Perego RA, Bianchi C, Corizzato M, Eroini B, et al. Primary cell cultures arising from normal kidney and renal cell carcinoma retain the proteomic profile of corresponding tissues. J Proteome Res. 2005;4:1503–1510. doi: 10.1021/pr050002o. [DOI] [PubMed] [Google Scholar]
- 26.Banks RE, Craven RA, Harnden P, Madaan S, et al. Key clinical issues in renal cancer: a challenge for proteomics. World J Urol. 2007;25:537–556. doi: 10.1007/s00345-007-0199-y. [DOI] [PubMed] [Google Scholar]
- 27.Kim HL, Seligson D, Liu X, Janzen N, et al. Using protein expressions to predict survival in clear cell renal carcinoma. Clin Cancer Res. 2004;10:5464–5471. doi: 10.1158/1078-0432.CCR-04-0488. [DOI] [PubMed] [Google Scholar]
- 28.Seliger B, Hohne A, Knuth A, Bernhard H, et al. Analysis of the major histocompatibility complex class I antigen presentation machinery in normal and malignant renal cells: evidence for deficiencies associated with transformation and progression. Cancer Res. 1996;56:1756–1760. [PubMed] [Google Scholar]
- 29.Lichtenfels R, Ackermann A, Kellner R, Seliger B. Mapping and expression pattern analysis of key components of the major histocompatibility complex class I antigen processing and presentation pathway in a representative human renal cell carcinoma cell line. Electrophoresis. 2001;22:1801–1809. doi: 10.1002/1522-2683(200105)22:9<1801::AID-ELPS1801>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 30.Seliger B, Fedorushchenko A, Brenner W, Ackermann A, et al. Ubiquitin COOH-terminal hydrolase 1: a biomarker of renal cell carcinoma associated with enhanced tumor cell proliferation and migration. Clin Cancer Res. 2007;13:27–37. doi: 10.1158/1078-0432.CCR-06-0824. [DOI] [PubMed] [Google Scholar]
- 31.Wang E, Miller LD, Ohnmacht GA, Liu ET, Marincola FM. High-fidelity mRNA amplification for gene profiling. Nat Biotechnol. 2000;18:457–459. doi: 10.1038/74546. [DOI] [PubMed] [Google Scholar]
- 32.Recktenwald CV, Mendler S, Lichtenfels R, Kellner R, Seliger B. Influence of Ki-ras-driven oncogenic transformation on the protein network of murine fibroblasts. Proteomics. 2007;7:385–398. doi: 10.1002/pmic.200600506. [DOI] [PubMed] [Google Scholar]
- 33.Atkins D, Ferrone S, Schmahl GE, Storkel S, Seliger B. Down-regulation of HLA class I antigen processing molecules: an immune escape mechanism of renal cell carcinoma? J Urol. 2004;171:885–889. doi: 10.1097/01.ju.0000094807.95420.fe. [DOI] [PubMed] [Google Scholar]
- 34.Atkins D, Lichtenfels R, Seliger B. Heat shock proteins in renal cell carcinomas. Contrib Nephrol. 2005;148:35–56. doi: 10.1159/000086042. [DOI] [PubMed] [Google Scholar]
- 35.Young AN, Amin MB, Moreno CS, Lim SD, et al. Expression profiling of renal epithelial neoplasms: a method for tumor classification and discovery of diagnostic molecular markers. Am J Pathol. 2001;158:1639–1651. doi: 10.1016/S0002-9440(10)64120-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Unwin RD, Craven RA, Harnden P, Hanrahan S, et al. Proteomic changes in renal cancer and co-ordinate demonstration of both the glycolytic and mitochondrial aspects of the Warburg effect. Proteomics. 2003;3:1620–1632. doi: 10.1002/pmic.200300464. [DOI] [PubMed] [Google Scholar]
- 37.Balabanov S, Zimmermann U, Protzel C, Scharf C, et al. Tumour-related enzyme alterations in the clear cell type of human renal cell carcinoma identified by two-dimensional gel electrophoresis. Eur J Biochem. 2001;268:5977–5980. doi: 10.1046/j.0014-2956.2001.02546.x. [DOI] [PubMed] [Google Scholar]
- 38.Pancholi V. Multifunctional alpha-enolase: its role in diseases. Cell Mol Life Sci. 2001;58:902–920. doi: 10.1007/PL00000910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoneda M, Fujii A, Ito A, Yokoyama H, et al. High prevalence of serum autoantibodies against the amino terminal of alpha-enolase in Hashimoto's encephalopathy. J Neuroimmunol. 2007;185:195–200. doi: 10.1016/j.jneuroim.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 40.Yavlovich A, Rechnitzer H, Rottem S. Alpha-enolase resides on the cell surface of Mycoplasma fermentans and binds plasminogen. Infect Immun. 2007;75:5716–5719. doi: 10.1128/IAI.01049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Magrys A, Anekonda T, Ren G, Adamus G. The role of anti-alpha-enolase autoantibodies in pathogenicity of autoimmune-mediated retinopathy. J Clin Immunol. 2007;27:181–192. doi: 10.1007/s10875-006-9065-8. [DOI] [PubMed] [Google Scholar]
- 42.He P, Naka T, Serada S, Fujimoto M, et al. Proteomics-based identification of alpha-enolase as a tumor antigen in non-small lung cancer. Cancer Sci. 2007;98:1234–1240. doi: 10.1111/j.1349-7006.2007.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mori-Iwamoto S, Kuramitsu Y, Ryozawa S, Mikuria K, et al. Proteomics finding heat shock protein 27 as a biomarker for resistance of pancreatic cancer cells to gemcitabine. Int J Oncol. 2007;31:1345–1350. [PubMed] [Google Scholar]
- 44.Gerke V, Creutz CE, Moss SE. Annexins: linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol. 2005;6:449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- 45.Park JE, Lee DH, Lee JA, Park SG, et al. Annexin A3 is a potential angiogenic mediator. Biochem Biophys Res Commun. 2005;337:1283–1287. doi: 10.1016/j.bbrc.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 46.Wozny W, Schroer K, Schwall GP, Poznanovic S, et al. Differential radioactive quantification of protein abundance ratios between benign and malignant prostate tissues: cancer association of annexin A3. Proteomics. 2007;7:313–322. doi: 10.1002/pmic.200600646. [DOI] [PubMed] [Google Scholar]
- 47.Kollermann J, Schlomm T, Bang H, Schwall GP, et al. Expression and Prognostic Relevance of Annexin A3 in Prostate Cancer. Eur Urol. 2008 doi: 10.1016/j.eururo.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 48.Zimmermann U, Balabanov S, Giebel J, Teller S, et al. Increased expression and altered location of annexin IV in renal clear cell carcinoma: a possible role in tumour dissemination. Cancer Lett. 2004;209:111–118. doi: 10.1016/j.canlet.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 49.Chen G, Gharib TG, Huang CC, Taylor JM, et al. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics. 2002;1:304–313. doi: 10.1074/mcp.m200008-mcp200. [DOI] [PubMed] [Google Scholar]
- 50.Corthals GL, Wasinger VC, Hochstrasser DF, Sanchez JC. The dynamic range of protein expression: a challenge for proteomic research. Electrophoresis. 2000;21:1104–1115. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1104::AID-ELPS1104>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 51.Rabilloud T. Two-dimensional gel electrophoresis in proteomics: old, old fashioned, but it still climbs up the mountains. Proteomics. 2002;2:3–10. [PubMed] [Google Scholar]
- 52.Shi T, Seligson D, Belldegrun AS, Palotie A, Horvath S. Tumor classification by tissue microarray profiling: random forest clustering applied to renal cell carcinoma. Mod Pathol. 2005;18:547–557. doi: 10.1038/modpathol.3800322. [DOI] [PubMed] [Google Scholar]
- 53.Lian Z, Kluger Y, Greenbaum DS, Tuck D, et al. Genomic and proteomic analysis of the myeloid differentiation program: global analysis of gene expression during induced differentiation in the MPRO cell line. Blood. 2002;100:3209–3220. doi: 10.1182/blood-2002-03-0850. [DOI] [PubMed] [Google Scholar]
- 54.Gygi SP, Han DK, Gingras AC, Sonenberg N, Aebersold R. Protein analysis by mass spectrometry and sequence database searching: tools for cancer research in the post-genomic era. Electrophoresis. 1999;20:310–319. doi: 10.1002/(SICI)1522-2683(19990201)20:2<310::AID-ELPS310>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 55.Abdulrahman M, Maina EN, Morris MR, Zatyka M, et al. Identification of novel VHL targets that are associated with the development of renal cell carcinoma. Oncogene. 2007;26:1661–1672. doi: 10.1038/sj.onc.1209932. [DOI] [PubMed] [Google Scholar]
- 56.Wykoff CC, Pugh CW, Maxwell PH, Harris AL, Ratcliffe PJ. Identification of novel hypoxia dependent and independent target genes of the von Hippel-Lindau (VHL) tumour suppressor by mRNA differential expression profiling. Oncogene. 2000;19:6297–6305. doi: 10.1038/sj.onc.1204012. [DOI] [PubMed] [Google Scholar]
- 57.Dallmann K, Junker H, Balabanov S, Zimmermann U, et al. Human agmatinase is diminished in the clear cell type of renal cell carcinoma. Int J Cancer. 2004;108:342–347. doi: 10.1002/ijc.11459. [DOI] [PubMed] [Google Scholar]
- 58.Alchanati I, Nallar SC, Sun P, Gao L, et al. A proteomic analysis reveals the loss of expression of the cell death regulatory gene GRIM-19 in human renal cell carcinomas. Oncogene. 2006;25:7138–7147. doi: 10.1038/sj.onc.1209708. [DOI] [PubMed] [Google Scholar]