Abstract
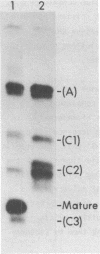
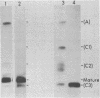
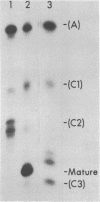
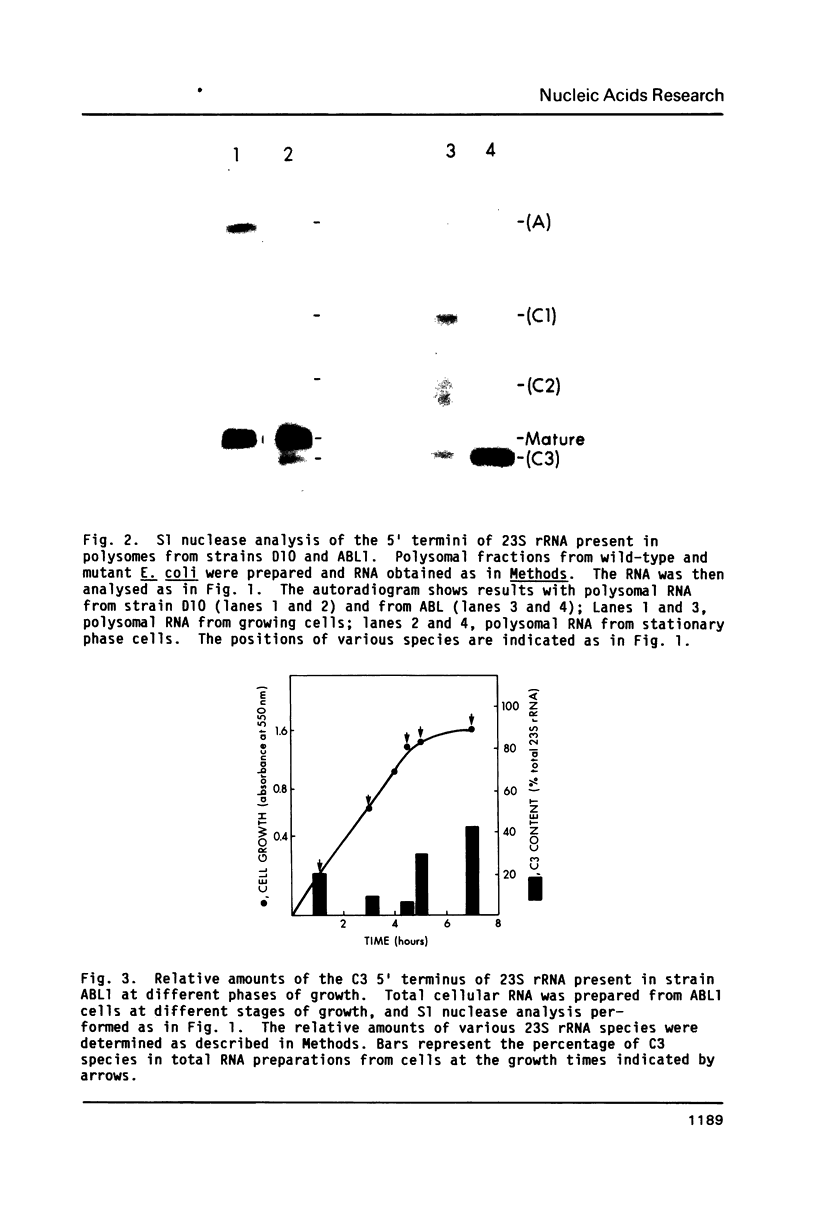
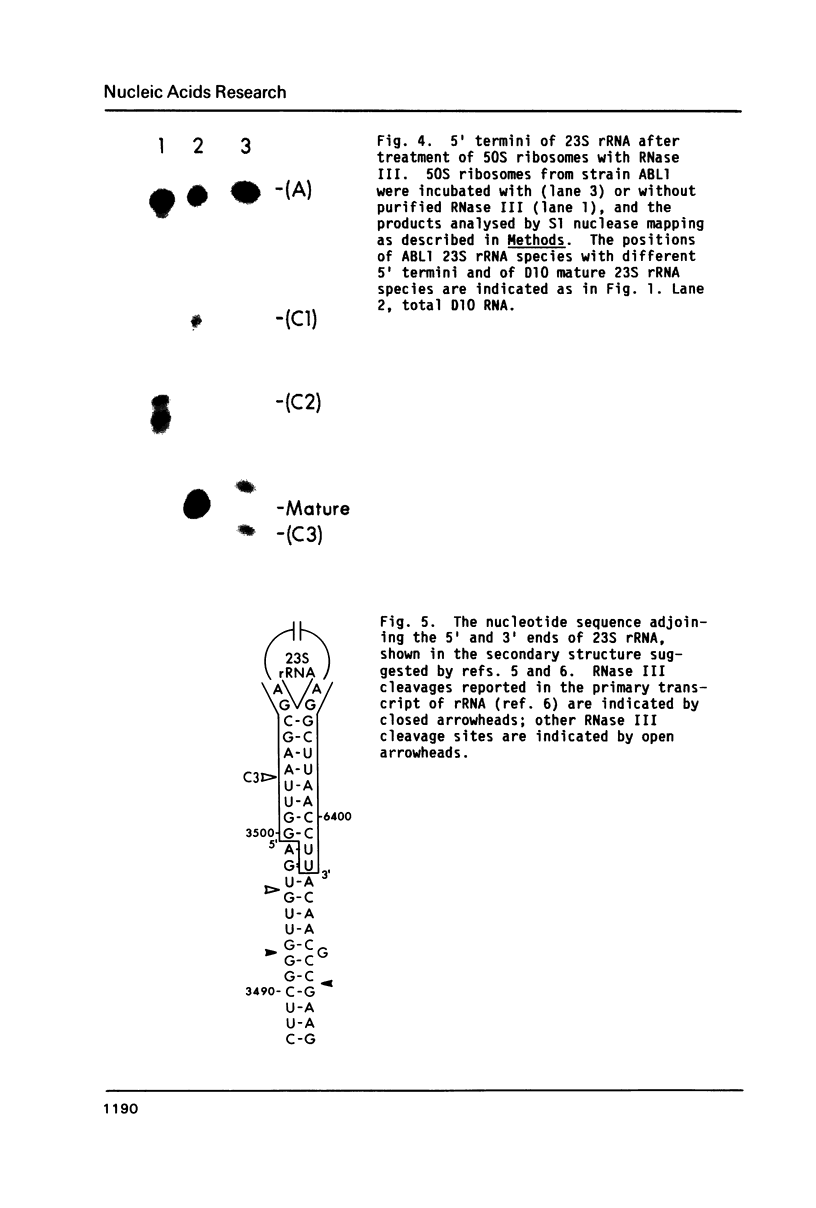
In a strain of E. coli deficient in RNase III (ABL1), 23S rRNA has been shown to be present in incompletely processed form with extra nucleotides at both the 5' and 3' ends (King et al., 1984, Proc. Natl. Acad. Sci. U.S. 81, 185-188). RNA molecules with four different termini at the 5' end are observed in vivo, and are all found in polysomes. The shortest of these ("C3") is four nucleotides shorter than the accepted mature terminus. In growing cells of both wild-type and mutant strains up to 10% of the 23S rRNA chains contain the 5' C3 terminus. In stationary phase cells, the proportion of C3 termini remains the same in the wild-type cells; but C3 becomes the dominant terminus in the mutant. Species C3 is also one of the 5' termini of 23S rRNA generated in vitro from larger precursors by the action of purified RNase III. We therefore suggest that some form of RNase III may still exist in the mutant; and since no cleavage is detectable at any other RNase III-specific site, the remaining enzyme would have a particular affinity for the C3 cleavage site, especially in stationary phase cells. We raise the question whether the C3 terminus has a special role in cellular metabolism.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bram R. J., Young R. A., Steitz J. A. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of E. coli. Cell. 1980 Feb;19(2):393–401. doi: 10.1016/0092-8674(80)90513-9. [DOI] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Gesteland R. F. Isolation and characterization of ribonuclease I mutants of Escherichia coli. J Mol Biol. 1966 Mar;16(1):67–84. doi: 10.1016/s0022-2836(66)80263-2. [DOI] [PubMed] [Google Scholar]
- Gitelman D. R., Apirion D. The synthesis of some proteins is affected in RNA processing mutants of Escherichia coli. Biochem Biophys Res Commun. 1980 Oct 16;96(3):1063–1070. doi: 10.1016/0006-291x(80)90060-1. [DOI] [PubMed] [Google Scholar]
- King T. C., Schlessinger D. S1 nuclease mapping analysis of ribosomal RNA processing in wild type and processing deficient Escherichia coli. J Biol Chem. 1983 Oct 10;258(19):12034–12042. [PubMed] [Google Scholar]
- King T. C., Sirdeshmukh R., Schlessinger D. RNase III cleavage is obligate for maturation but not for function of Escherichia coli pre-23S rRNA. Proc Natl Acad Sci U S A. 1984 Jan;81(1):185–188. doi: 10.1073/pnas.81.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer J. E., Schweiger M. RNase III is positively regulated by T7 protein kinase. J Biol Chem. 1983 May 10;258(9):5340–5343. [PubMed] [Google Scholar]
- Silengo L., Nikolaev N., Schlessinger D., Imamoto F. Stabilization of mRNA with polar effects in an Escherichia coli mutant. Mol Gen Genet. 1974;134(1):7–19. doi: 10.1007/BF00332808. [DOI] [PubMed] [Google Scholar]
- Talkad V., Achord D., Kennell D. Altered mRNA metabolism in ribonuclease III-deficient strains of Escherichia coli. J Bacteriol. 1978 Aug;135(2):528–541. doi: 10.1128/jb.135.2.528-541.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]