Summary
Biotransformation of arsenic includes oxidation, reduction, methylation and conversion to more complex organic arsenicals. Members of the class of arsenite [As(III)] S-adenosylmethyltransferase enzymes catalyze As(III) methylation to a variety of mono-, di- and trimethylated species, some of which are less toxic than As(III) itself. However, no methyltransferase gene has been identified in plants.
Here, an arsM gene from the soil bacterium Rhodopseudomonas palustris was expressed in Japonica rice (Oryza sativa L.) cultivar Nipponbare, and the transgenic rice produced methylated arsenic species, which were measured by inductively coupled plasma mass spectrometry (ICP-MS) and high performance liquid chromatography-inductively coupled plasma-mass spectrometry (HPLC-ICP-MS).
Both monomethylarsenate [MAs(V)] and dimethylarsenate [DMAs(V)] were detected in the root and shoot of transgenic rice. After 12-d exposure to As(III), the transgenic rice gave off 10-fold more volatile arsenicals.
The present study demonstrates that expression of an arsM gene in rice induces arsenic methylation and volatilization, providing a potential stratagem for phytoremediation theoretically.
Keywords: arsenic methylation, As(III) S-adenosylmethyltransferase, transgenic rice volatile arsenicals
Introduction
Arsenic is a ubiquitous toxic metalloid that is introduced into the environment from both anthropogenic and geochemical sources (Smith et al., 1998). Classified as a Group A human carcinogen by the US Environmental Protection Agency (EPA) (http://www.epa.gov/ttn/atw/hlthef/arsenic.html), arsenic poses a dietary risk for human health (Abernathy et al., 1999; Tchounwou et al., 2003). The irrigation of arsenic-contaminated water for rice (Meharg & Rahman, 2003), which is the primary source of nutrition for more than half of the world's population, leads to arsenic accumulation in soil and rice grain (Williams et al., 2006; Rahman et al., 2009), increasing arsenic exposure in the indigenous populations (Kile et al., 2007; Zhu et al., 2008; Pal et al., 2009).
Enzymatic reduction is an initial and crucial step of arsenic metabolism. As the predominant form of arsenic in aerobic soils, As(V) is thought to enter rice through phosphate transporters (Zhao et al., 2009), while As(III) is taken up by NIP superfamily of aquaporins in rice (Ma et al., 2008). Heretofore, there have been numerous studies on As(V) reductases in a variety of organisms, including microbes, plants and animals (Mukhopadhyay & Rosen, 2002). As(V) reductases has been identified and characterized from Arabidopsis (Dhankher et al., 2006; Ellis et al., 2006) and Pteris vitatta (Bleeker et al., 2006). Duan et al. (2007) cloned two ACR2-like genes (OsACR2.1 and OsACR2.2) in rice, which exhibited As(V) reductase activity in vivo and the purified gene products were demonstrated to reduce As(V) to As(III) in vitro. In some organisms, after reduction, As(III) is converted into various methylated metabolites by As(III) S-adenosylmethyltransferase enzymes, termed Cyt19 or AS3MT in mammals or ArsM in microbes (Thomas et al., 2004). The arsM gene from Rhodopseudomonas palustris was cloned and demonstrated to confer As(III) resistance in vivo when expressed in the arsenic sensitive E. coli strain AW3110, in which the arsRBC operon was deleted (Qin et al., 2006). ArsM catalyzes the formation of several methylated species, including dimethylarsenate [DMAs(V)], trimethylarsine oxide [(TMAs(V)O] and trimethylarsine [TMAs(III)] gas, which is nearly completely nontoxic (Cullen, 2005). Thus methylation and volatilization are clearly steps in a detoxification pathway in bacteria. Two other arsenic methyltransferase genes (termed cmarsM7 and cmarsM8) were cloned and characterized from the unicellular eukaryotic red alga Cyanidioschyzon from Yellowstone National Park (Qin et al., 2009).
Transgenic technology is being widely applied for improving metal resistance in plants (Dai et al., 2008; Wang et al., 2008). Indian mustard (Brassica juncea) expressing an E. coli gene encoding γ-glutamylcysteine synthetase (γ-ECS) was found to be more tolerant to arsenic (Doucleff & Terry, 2002); and Dhankher et al. (2002) reported that Arabidopsis thaliana plants transformed with two E. coli genes encoding ArsC and γ-ECS, respectively, exhibited enhanced arsenic tolerance as well as the ability to hyperaccumulate arsenic. To our knowledge, no methyltransferase gene has been identified in the rice genome or other higher plants. In this study, we constructed transgenic rice with the arsM gene from R. palustris and demonstrated that the resulting transgenic plant acquired the capability for methylating inorganic arsenic to a variety of organic species including volatile arsenicals..
Materials and Methods
Bacterial strains and plasmids
Agrobacterium tumefaciens (A. tumefaciens) strain AGL1 was used for infection of rice. The transformation vector in this experiment is pCAM2300-35S-OCS (p35S), a binary vector carrying cauliflower mosaic virus (CaMV) 35S promoter, octopine synthase (OCS) terminator, and the neomycin phosphotransferase II (nptII) gene which serves as a bacterial selection marker (Wang et al., 2008). The detailed process of the construction of recombinant plasmid is presented in Supporting Information, Methods S1.
Generation of transgenic rice
Embryogenic calli were obtained from the mature seeds of Japonica rice (Oryza sativa L.) cultivar Nipponbare. The A. tumefaciens-mediated production of transgenic rice was carried out according to the modified method of Hiei et al. (1994), which is described in detail in Methods S1. Briefly, NB medium containing cephalosporins and G-418 was used for selection. After differentiation in MS medium containing 6-benzylaminopurine (6-BA), α-naphthalene acetic acid (NAA), and G-418, the transgenic rice plants were transplanted into soil and grown to maturity. Subsequently, T1 generation rice seeds were grown in Hainan Province of China for propagation and then T3 generation rice seeds were used for further experiments.
DNA isolation and PCR analysis
Total genomic DNA was extracted with a cetyl triethyl ammonium bromide (CTAB) method (Murray & Thompson, 1980) from the leaf tissues (0.1–0.5 g) of T0 generation rice in seedling medium before transplanting. The PCR program for the amplification of the antibiotic marker nptII was performed as follows: 5 min at 94°C, 30 cycles of 30 s at 94°C, 30 s at 56°C, and 30 s at 72°C, followed by 10 min at 72°C. The forward prime was 5'-TCGGCTATGACTGGGCACAACAGA-3', and the reverse prime was 5'-AAGAAGGCGATAGAAGGCGATGCG-3'.
RNA isolation and Northern blot analysis
Total RNA was isolated from the leaf tissues of T0 generation rice using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). DNase I (Takara, China) was used to remove contaminating genomic DNA. For Northern blot analysis, the loading buffer was prepared by mixing 7.5 μl of 10× MOPS buffer [10× MOPS is 0.2 M 3-(N-morpholino)propanesulfonic acid, 0.05 M sodium acetate (Amersco, USA), and 0.01 M EDTA], 9 μl of formaldehyde, and 30 μl of formamide together. Following incubation for 15 min at 70°C, the buffer was cooled in an ice bath, and 10–15 μg of total RNA with 0.5 μl of ethidium bromide was added. The sample was electrophoresed in 1.5% agarose gels containing 18% (v/v) formaldehyde in 1× MOPS buffer and blotted onto nitrocellulose membranes. After hybridized to 32P-labeled random-primed cDNA probes for 14–16 d at 65°C, the blots were stripped by washing once in 2× SSC (l× SSC is 0.15 M NaCl and 0.015 M sodium citrate)/0.1% (w/v) sodium dodecyl sulfate (SDS) for 15 min at room temperature and then twice in 0.2× SSC/0.1% (w/v) SDS for 15 min at 65°C. The membranes were analyzed by phosphoimaging.
Semi-quantitative PCR analysis
For semi-quantitative PCR, cDNA was synthesized in 20 μl reactions from total RNA isolated from the leaf tissues of T3 generation rice, using 200 U of MMLV reverse transcriptase (Invitrogen, Carlsbad, CA, USA) and oligo-(dT) as a primer. A rice actin1 gene was used as a constitutive internal standard to evaluate cDNA content. The PCR program for the amplification of actin1 gene was run with cDNA as follows: 5 min at 94°C, 35 cycles of 30 s at 94°C, and 1 min at 70°C, followed by 10 min at 72°C. The forward primer was 5'-GACTCTGGTGATGGTGTCAGC-3', and the reverse primer was 5'-GGCTGGAAGAGGACCTCAGG-3'. The PCR program for the amplification of arsM was carried out under the following conditions: 5 min at 94°C, 30 cycles of 30 s at 94°C, 45 s at 65°C, and 45 s at 72°C, followed by 10 min at 72°C, using a forward primer (5'-ATGCCCACTGACATGCAAGAC-3') and a reverse prime (5'-TCACCCGCAGCAGCGCGCCG-3'). Positive transgenic rice plants were used for further experiments.
Plant culture and treatment
After sterilization in 10% (v/v) H2O2 solution for 15 min, rice seeds were thoroughly washed with deionized water and then germinated in moist perlite. After 2 wk of germination, uniform rice seedlings were selected and transplanted to polyvinylchloride (PVC) pots (12 cm diameter and 15 cm height, six plants per pot) containing 1000 ml of one-ninth-strength nutrient solution. After cultivation in the one-ninth-strength nutrient solution for the first week, one-third-strength nutrient solution was changed. The composition of the full-strength nutrient solution was as follows: the macronutrients contain 5 mM NH4NO3, 2 mM K2SO4, 4 mM CaCl2, 1.5 mM MgSO4·7H2O, and 1.3 mM KH2PO4, the micronutrients contain 50 μM Fe(II)-EDTA, 10 μM H3BO4, 1 μM ZnSO4·7H2O, 1 μM CuSO4·5H2O, 5 μM MnSO4·H2O, 0.5 μM Na2MoO4·2H2O, and 0.2 μM CoSO4·7H2O, and the pH of the solution was adjusted to 5.5 using dilute HCl or KOH (Hewitt, 1966). The nutrient solution was changed twice weekly. The rice plants were cultivated in a growth room at a 14-h light period (260–350 μmol m−2·s−1; 25°C : 20°C day : night; 60% relative humidity (RH)). A hydroponic system was used to minimize the impact of soil bacteria on arsenic methylation and volatilization.
Uniform rice seedlings of 7-wk-old were selected for the experiment. Two of four independent transgenic rice lines of T3 generation were used, namely line 1 and line 5. In experiment 1, rice plants of line 5 were exposed to either 10 μM As(III) or 50 μM As(V) for 24 h, respectively. Six replicates were used for this experiment. In experiment 2, rice plants of line 1 and line 5 were exposed to either 10 μM As(III) for 4 wk or 100 μM As(III) for 72 h, respectively. Three replicates were used for this experiment. Long period of exposure to low levels of arsenic and short period of exposure to high levels of arsenic were applied to determine methylated arsenic species in transgenic rice of independengt lines in different conditions. For experiment 3 of trapping volatile arsenicals, rice plants of line 1 were selected to expose to 10 μM As(III) for 12 d, with nutrient solution changed every 3 d. Four replicates were used for the nontransgenic control, and ten replicates were used for the transgenic rice. The longer period of exposure to arsenic (12 d) in this experiment allowed collection of measurable amounts of volatile arsenicals. The harvested plants were thoroughly washed with tap water followed by deionized water. Adhering water was then removed with filter paper, and each plant was separated into roots and shoots, shock frozen with liquid nitrogen and homogenized to powder, which was stored at −80°C until further analysis.
Chemo-trapping system
Volatile arsenicals released from rice plants were trapped according to the method of Mestrot et al. (2009). Preparation of silica gel tubes was as follows: silica gel was immersed in 5% (v/v) HNO3 overnight, washed with deionized water and then impregnated with 10% AgNO3 solution (w/v) in an aluminum foil wrapped glass jar, placed in an oven overnight or longer at 70°C to remove H2O. Next, a clean 3 ml volume burette was filled with silver nitrate-impregnated silica gel held in with a small quantity of quartz wool at each end of the tube. The tube was covered with aluminum foil to avoid photodecomposition of silver nitrate. The tube was connected to a vessel (15 cm diameter and 60 cm height) made of transparent organic plastic with two tubules (0.5 cm diameter and 4.5 cm height) on the top, and a circular lid that could be removed from the bottom (Fig. S1). The silica gel tube was connected to one of the two tubules, and an air compressor (HAILEA ACO-318, 45 W power, 70 l min−1 output, China) was connected to the other to supply oxygen for growth of the rice plants in a PVC pot, which was placed inside the water-sealed vessel. Two chemo-trap controls were performed with no rice but only the nutrient solution in the PVC pot to determine the background level of arsenic in these assays. The trapping period lasted for 12 d, during which the nutrient solution with 10 μM As(III) was changed every 3 d. At the 6th and 12th day, the silica gel tubes were removed for analysis.
Analysis of total arsenic
For all experiments, total arsenic concentrations and speciation were measured. For the determination of total arsenic in rice roots and shoots, homogenized samples (0.1–0.2 g) were weighed into a 50 ml volume centrifuge tube and soaked in 2–3 ml of high purity HNO3. After standing overnight at room temperature, the samples were randomized and subjected to microwave digestion (Mars, Matthews Inc., USA). The digestion program was run as follows: 55°C for 10 min, 75°C for 10 min, 95°C for 30 min with 5 min ramp time between each stage. The digested samples were diluted to 40 ml with ultrapure water and then paper-filtered to remove impurities. Arsenic in the dilute solution was measured by inductively coupled plasma mass spectrometry (ICP-MS) 7500 (Agilent Technologies, China).
For determination of volatile arsenicals evolved from rice plants, the silica gel tubes were eluted as follows: 1.2 ml of 5% (v/v) hot boiling HNO3 was injected into one end of the trap and left for 5 min, and then 4 ml of 1% (v/v) hot boiling HNO3 was used to elute the tube (Mestrot et al., 2009). The eluates were paper-filtered and stored until quantification of total arsenic by inductively coupled plasma mass spectrometry (ICP-MS) 7500 (Agilent Technologies, China).
Analysis of arsenic speciation
For arsenic speciation in rice roots and shoots, digestion of samples was performed as for total arsenic analysis, except that 5–10 ml of 1% HNO3 was added after weighing, and after digestion, an appropriate volume of 1% HNO3 was added to the samples to replace the nitric acid lost during digestion. The samples were centrifuged and the supernatant solution was removed and filtered through 0.45 μm filters for analysis. The samples were divided into two equal parts. To one 10% (v/v) H2O2 solution (final concentration) was added to oxidize As(III) to As(V). This allowed for the determination of MAs(V) and DMAs(V), which otherwise would elute near the As(III) peak during the analysis with high performance liquid chromatography-inductively coupled plasma-mass spectrometry (HPLC-ICP-MS, Agilent Technologies, China). A PRP-X100 10 μm anion-exchange column (150×4.1 mm) (Hamilton, UK) was used to separate As(III), As(V), MAs(V) and DMAs(V). The mobile phase consisted of 10 mM (NH4)2HPO4 and 10 mM NH4NO3, adjusted to pH 6.2 using ammonia.
Statistical analysis
All statistical analyses were performed with the use of SPSS 13.0 software. One-Way ANOVA was used to examine the significance of difference between the transgenic and nontransgenic rice. One-Sample T Test was used to examine the significance of difference between the concentration of methylated arsenic species in planta and LOD. P value < 0.05 was judged to be statistically significant.
Results
Identification of transgenic rice
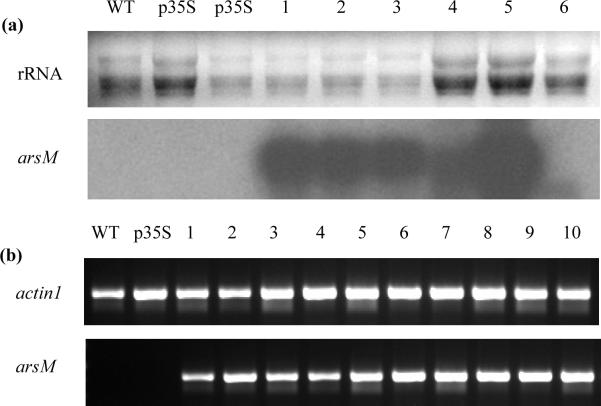
Integration of arsM into A. tumefaciens was verified by PCR before the transformation of rice. A fragment of 852 bp was shown to contain arsM by DNA sequencing. A. tumefaciens bearing the recombinant plasmid was then used to infect the rice calli. The antibiotic marker nptII in the p35S vector was used to detect the presence of arsM integrated into the rice genome. PCR was performed with DNA isolated from leaf tissues of 87 putative transgenic rice lines of T0 generation. 74 individual plants were determined to be nptII positive, of which 64 were confirmed to be transgenic rice bearing p35S-asrM. Northern blot analysis was carried out on six independent rice lines of T0 generation. Four independent transgenic rice lines expressing arsM at the transcriptional level were obtained, namely line 1, 2, 3 and 5 (Fig. 1a). Semi-quantitative PCR was performed to identify positive transgenic rice plants of line 1 and line 5 of T3 generation which were used in the experiments, with actin1 gene as a control for expression of arsM (Fig. 1b).
Fig. 1.
Identification of transgenic rice. (a) Northern blot analysis of T0 generation rice. WT, wild type, the nontransgenic control; p35S, transgenic rice with p35S vector; lanes 1, 2, 3 and 5, independent positive transgenic rice lines expressing arsM at the transcriptional level; lanes 4 and 6, negative transgenic rice lines expressing no arsM at the transcriptional level. (b) Semi-quantitative PCR analysis of T3 generation rice with actin1 gene as a constitutive internal standard. WT, wild type, the nontransgenic control; p35S, transgenic rice with p35S vector; lanes 1–10, individual positive transgenic rice plants with p35S-arsM.
Plant growth and total arsenic in roots and shoots
After 24-h exposure to 10 μM As(III) or 50 μM As(V), 4-wk exposure to 10 μM As(III), 72-h exposure to 100 μM As(III), or 12-d exposure to 10 μM As(III), rice plants were harvested, and total arsenic in roots and shoots was analyzed by ICP-MS. No obvious differences were observed between the transgenic rice and the nontransgenic control in appearance and growth. In none of the treatments were significant differences observed in biomass or total arsenic accumulation between the transgenic rice and the nontransgenic control (Table S1), except that the transgenic rice accumulated significantly less total arsenic in the root than the nontransgenic control after 12-d exposure to 10 μM As(III) (P < 0.05) (Table 1). Plants treated with As(III) had higher total arsenic concentration that those treated with As(V) (P < 0.05) (Table S1).
Table 1.
Arsenic in roots and shoots of rice in Experiment 3
| Total As (μg g−1 FW) | As(III) (μg g−1 FW) | As(V) (μg g−1 FW) | MAs(V) (μg g−1 FW) | DMAs(V) (μg g−1 FW) | As(III)% | |
|---|---|---|---|---|---|---|
| WT root | 50.5±5.1a | 29.6±3.8a | 12.2±0.5a | <LOD | <LOD | 70.7 |
| arsM 1 root | 41.3±1.5b | 22.9±1.1a | 9.32±0.36b | 0.13±0.00 | 0.10±0.01 | 70.6 |
| WT shoot | 4.94±2.22a | 4.37±1.58a | 0.49±0.25a | <LOD | <LOD | 90.0 |
| arsM 1 shoot | 6.53±0.92a | 5.19±1.15a | 0.32±0.05a | <LOD | <LOD | 94.2 |
Data are means ± SE (n = 4 for WT, n = 10 for arsM). Means followed by the same letter are not significantly different at P < 0.05 using One-Way ANOVA in SPSS. As, arsenic; WT, wild type, nontransgenic control; arsM 1, transgenic rice of line 1; FW, fresh weigh; SE, standard error; LOD, limit of detection.
Arsenic speciation in roots and shoots
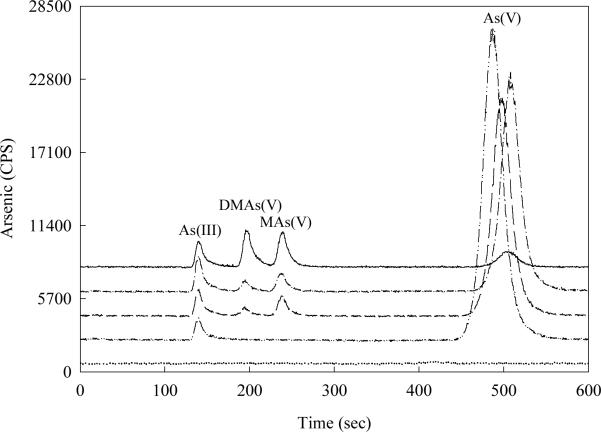
HPLC-ICP-MS was used for arsenic speciation in roots and shoots. All arsenic species are above the limit of detection (LOD) of HPLC-ICP-MS arsenic speciation analysis (LOD of As(III) and As(V) is 0.02 μg g−1 FW rice, LOD of MAs(V) and DMAs(V) is 0.008 μg g−1 FW rice). MAs(V) and DMAs(V) were detected in the root of transgenic rice of line 5 in experiment 1, with either As(III) exposure [0.44±0.11 and 0.25±0.06 μg g−1 fresh weight (FW), respectively] or As(V) exposure (0.52±0.13 and 0.25±0.05 μg g−1 FW, respectively) (Table S1, Fig. 2). No MAs(V) or DMAs(V) was detected in the shoot. As(III) was the primary arsenic species in both the transgenic rice and the nontransgenic control. The percentage of total arsenic as As(III) in the root was higher following As(III) exposure (c. 90%) than As(V) exposure (c. 70%) (P < 0.05) (Table S1).
Fig. 2.
HPLC-ICP-MS chromatograms of arsenic speciation in the root of transgenic rice, with the relative amounts of arsenic expressed as CPS. The standards (solid line) contain 10 μg l−1 of mixed As(III), DMAs(V), MAs(V) and As(V). The control (dotted line) contains 1% HNO3 only. With 24-h exposure to 10 μM As(III) (dashed line) or 50 μM As(V) (dash-dot line), DMAs(V) and MAs(V) were detected in the root of the transgenic rice. No DMAs(V) or MAs(V) was detected in the root of the nontransgenic control (dash-dot-dot line).
Moreover, MAs(V) and DMAs(V) were detected both in the root and shoot of transgenic rice of line 1 and line 5 in experiment 2 (Table S2). As(III) took up c. 70% of the total arsenic in the root and c. 90% in the shoot both in the transgenic rice and the nontransgenic control with exposure to 10 μM As(III) for 4 wk (Table S2a). However, following 72-h exposure to 100 μM As(III), As(V) rather than As(III) was the primary arsenic species in the root of both the transgenic rice and the nontransgenic control, in which the percentage of total arsenic as As(III) was only around 20% (Table S2b).
After 12-d exposure to 10 μM As(III), both MAs(V) (0.13±0.00 μg g−1 FW) and DMAs(V) (0.10±0.01 μg g−1 FW) were detected in the root of transgenic rice of line 1 (Table 1). The transgenic rice exhibited significantly lower concentration of As(V) in the root compared with the nontransgenic control (P < 0.05) (Table 1). Approximately 70% of total arsenic in the root and 90% in the shoot of both transgenic and nontransgenic rice were in the form of inorganic As(III) (Table 1).
Quantification of volatile arsenicals
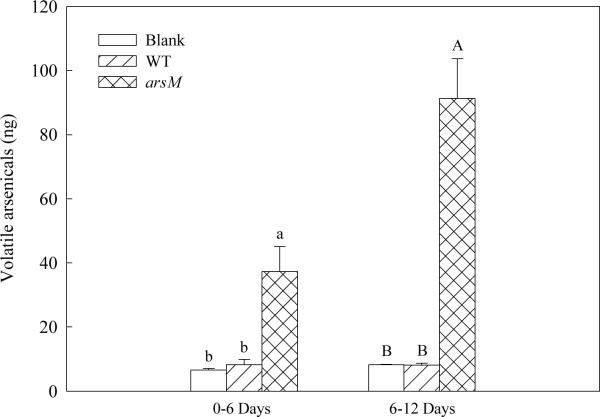
The HNO3 eluates from chemo-traps were analyzed by ICP-MS. For 0–6 day and 6–12 day of trapping, the amounts of volatile arsenicals trapped were 37.3±7.8 ng and 91.3±12.3 ng from the transgenic rice, respectively; only low background level of volatile products (around 8 ng) were found from the nontransgenic control or the chemo-trap control with no rice (P < 0.05) (Fig. 3). The amount of volatile arsenicals during the 12-d trapping period represented 0.06% of the total arsenic in rice plants.
Fig. 3.
Quantification of volatile arsenicals in eluates of chemo-traps from transgenic rice with 12-d exposure to 10 μM As(III). Data are means ± SE (n = 2 for blank, n = 4 for WT, n = 10 for arsM). Means followed by the same letter are not significantly different at P < 0.05 using One-Way ANOVA in SPSS. Open bars, blank (chemo-trap control with no rice); hatched bars, WT (wild type, the nontransgenic control); cross-hatched bars, arsM (transgenic rice); SE, standard error.
These results clearly demonstrate that the transgenic rice volatilizes arsenic by expression of the bacterial arsM gene. Since TMAs(III) is the nontoxic final product of the ArsM methylation pathway (Qin et al., 2006) and is the volatile species produced by the purified enzyme (Yuan et al., 2008), the main species of the trapped volatile arsenicals is presumed to be TMAs(III).
Discussion
In this study we constructed transgenic rice in which a bacterial As(III) S-adenosylmethyltransferas gene (arsM) (Qin et al., 2006) was stably integrated into the rice genome and expressed a functional ArsM enzyme capable of methylating arsenic in rice seedlings. When exposed to As(III), or As(V), the transgenic rice produced both MAs(V) and DMAs(V) in the root and shoot (Table S1, S2, Table 1, Fig. 2) and evolved volatile arsenicals (Fig. 3), thus demonstrating that the bacterial ArsM catalyzes methylation of arsenic in rice. The overall production of organic arsenic species produced by expression of arsM after a short term exposure (24 h) was small, and transgenic and nontransgenic rice plants had similar total arsenic concentrations in plant tissues (Table S1). With a longer term exposure (12 d) to As(III), total arsenic concentration in the root of the transgenic rice was significantly lower than the nontransgenic control (P < 0.05) (Table 1). Besides, long period (4 wk) or high levels (100 μM) of exposure to As(III) resulted in accumulation of MAs(V) and DMAs(V) in the shoot in addition to root (Table S2).
To date there has been no direct demonstration of in planta methylation and volatilization of arsenic by higher plants. Results from the present study are a proof of concept, demonstrating that expression of arsM can affect the speciation of arsenic in rice plants and lead to the volatilization of arsenic (Fig. 3). Only low amounts of methylated and volatile arsenicals were detected in this study. The volatile arsenicals represented 0.06% of the total arsenic in rice plants, which can be neglectable in practical phytoremediation. It is possible that the trapping system used in the present study is not sufficient to fully recover the volatile arsenicals. Firstly, although the silica gel tube is quantitative for trapping volatile arsenicals with high recovery of arsenic species, ranging from 80% to 96% (Mestrot et al., 2009), the arsenic recovery for the whole chemo-trap system is hard to be evaluated as the volatile arsenicals from rice plants could not be quantified. Secondly, during the 12-d trapping period, the nutrient solution with 10 μM As(III) was changed every 3 d, resulting in the unavoidable linkage of volatile arsenicals from the vessel, Alternatively, expression of arsM itself may be too low to produce abundant volatile arsenicals. Therefore, for such a system to be developed, optimization of gene expression and its regulation in higher plants will be necessary. Other arsM genes which exhibit higher activity in rice plants may be selected to construct transgenic rice in order to acquire more efficient removal of arsenic by volatilization, which may make this technology applicable and practical in phtoremediation of arsenic-contaminated soils and waters.
In our previous study, T0 generation rice was grown in an agricultural soil in Changping, Beijing, with arsenic concentration 9 mg/kg and pH 7.82 (Chen et al., 2002), and the arsenic accumulation in T1 generation rice grains was analyzed. The total arsenic concentration in the seeds of transgenic rice tended to be significantly lower than in the nontransgenic control (P < 0.05) (Table S3). Moreover, expressing arsM significantly decreased the concentrations of As(III) and As(V) both in the seeds and husks (P < 0.05), as well as the concentration of DMAs(V) in the husks (P < 0.05) (Table S3). Since arsenic is ubiquitous in paddy soils and rice, leading to accumulation of arsenic in the rice grain, this is a good indication of, representing a potential approach in improving the safety of this staple food. However, as this is the preliminary result of arsenic speciation in rice grains, more detailed work is to be carried out to reveal how expression of bacterial arsM may help alleviate health risks associated with arsenic accumulation in rice.
In conclusion, our study demonstrates that expression of an arsM gene in rice induces arsenic methylation and volatilization, as well as alters arsenic speciation in root and shoot,. This is a first step in the production of rice capable of methylating and volatilizing environmental arsenic. Since no gene encoding arsM has as yet been identified in the genome of any higher plants, including the rice genome, this study provides insight into the consequences of arsenic methylation in transgenic rice. Future research will focus on optimizing expression and activity of the bacterial ArsM or orthologues with the aim of reducing the accumulation of more toxic arsenic species in rice plant including rice grain, and on enhancing the production of volatile arsenicals
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health Grant GM55425 (to B.P.R.) and National Natural Science of China 20720102042 (to Y.G.Z.).
Abbreviations
- As(III)
arsenite
- As(V)
arsenate
- MAs(V)
monomethylarsenate
- DMAs(V)
dimethylarsenate
- TMAs(III)
trimethylarsine
- TMAs(V)O
trimethylarsine oxide
Footnotes
Supporting Information Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
References
- Abedin MJ, Cresser MS, Meharg AA, Feldmann J, Cotter-Howells J. Arsenic accumulation and metabolism in rice (Oryza sativa L.) Environmental Science and Technology. 2002;36:962–968. doi: 10.1021/es0101678. [DOI] [PubMed] [Google Scholar]
- Bleeker PM, Hakvoort HWJ, Bliek M, Souer E, Schat H. Enhanced arsenate reduction by a CDC25-like tyrosine phosphatase explains increased phytochelatin accumulation in arsenate-tolerant Holcus lanatus. The Plant Journal. 2006;45:917–929. doi: 10.1111/j.1365-313X.2005.02651.x. [DOI] [PubMed] [Google Scholar]
- Chen TB, Wei CY, Huang ZC, Huang QF, Lu QG, Fan ZL. Arsenic hyperaccumulator Pteris vittata L. and its arsenic accumulation. Chinese Science Bulletin. 2002;47:902–905. [Google Scholar]
- Cullen WR. The toxicity of trimethylarsine: An urban myth. Journal of Environmental Monitoring. 2005;7:11–15. doi: 10.1039/b413752n. [DOI] [PubMed] [Google Scholar]
- Cullen WR, Reimer KJ. Arsenic speciation in the environment. Chemical Reviews. 1989;89:713–764. [Google Scholar]
- Dai SH, Wei XP, Alfonso AA, Pei LP, Duque UG, Zhang ZH, Babb GM, Beachy RN. Transgenic rice plants that overexpress transcription factors RF2a and RF2b are tolerant to rice tungro virus replication and disease. Proceedings of the National Academy of Sciences. 2008;105:21012–21016. doi: 10.1073/pnas.0810303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhankher OP, Li YJ, Rosen BP, Shi J, Salt D, Senecoff JF, Sashti NA, Meagher RB. Engineering tolerance and hyperaccumulation of arsenic in plants by combining arsenate reductase and γ-glutamylcysteine synthetase expression. Nature Biotechnology. 2002;20:1140–1145. doi: 10.1038/nbt747. [DOI] [PubMed] [Google Scholar]
- Dhankher OP, Rosen BP, McKinney EC, Meagher RB. Hyperaccumulation of arsenic in the shoots of Arabidopsis silenced for arsenate reductase (ACR2) Proceedings of the National Academy of Sciences. 2006;103:5413–5418. doi: 10.1073/pnas.0509770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucleff M, Terry N. Pumping out the arsenic. Nature Biotechnology. 2002;20:1094–1095. doi: 10.1038/nbt1102-1094. [DOI] [PubMed] [Google Scholar]
- Duan GL, Zhou Y, Tong YP, Mukhopadhyay R, Rosen BP, Zhu YG. A CDC25 homologue from rice functions as an arsenate reductase. New Phytologist. 2007;174:311–321. doi: 10.1111/j.1469-8137.2007.02009.x. [DOI] [PubMed] [Google Scholar]
- Ellis DR, Gumaelius L, Indriolo E, Pickering IJ, Banks JA, Salt DE. A novel arsenate reductase from the arsenic hyperaccumulating fern Pteris vittata. Plant Physiology. 2006;141:1544–1554. doi: 10.1104/pp.106.084079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt EJ. Technical Communication No.22. 2ed. Commonwealth Agriculture Bureau; Farnham Royal, UK: 1966. Sand and water culture methods used in the study of plant nutrition; pp. 67–69. [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza satival L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. The Plant Journal. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Kile ML, Houseman EA, Breton CV, Smith T, Quamruzzaman Q, Rahman M, Mahiuddin G, Christiani DC. Dietary arsenic exposure in Bangladesh. Environmental Health Perspectives. 2007;115:889–893. doi: 10.1289/ehp.9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZJ, Shen J, Carbrey JM, Mukhopadhyay R, Agre P, Rosen BP. Arsenite transport by mammalian aquaglyceroporins AQP7 and AQP9. Proceedings of the National Academy of Sciences. 2002;99:6053–6058. doi: 10.1073/pnas.092131899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JF, Yamaji N, Mitani N, Xu XY, Su YH, McGrath SP, Zhao FJ. Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proceedings of the National Academy of Sciences. 2008;105:9931–9935. doi: 10.1073/pnas.0802361105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meharg AA, Hartley-Whitaker J. Arsenic uptake and metabolism in arsenic resistant and nonresistant plant species. New Phytologist. 2002;154:29–43. [Google Scholar]
- Meharg AA, Rahman MM. Arsenic contamination of Bangladesh paddy field soils: Implications for rice contribution to arsenic consumption. Environmental Science and Technology. 2003;37:229–234. doi: 10.1021/es0259842. [DOI] [PubMed] [Google Scholar]
- Mestrot A, Uroic MK, Plantevin T, Islam MR, Krupp EM, Feldmann J, Meharg AA. Quantitative and qualitative trapping of arsines deployed to assess loss of volatile arsenic from paddy soil. Environmental Science and Technology. 2009;43:8270–8275. doi: 10.1021/es9018755. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R, Rosen BP. Arsenate reductases in prokaryotes and eukaryotes. Environmental Health Perspectives. 2002;110:745–748. doi: 10.1289/ehp.02110s5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal A, Chowdhury UK, Mondal D, Das B, Nayak B, Ghosh A, Maity S, Chakraborti D. Arsenic burden from cooked rice in the populations of arsenic affected and nonaffected areas and Kolkata City in West-Bengal, India. Environmental Science and Technology. 2009;43:3349–3355. doi: 10.1021/es803414j. [DOI] [PubMed] [Google Scholar]
- Qin J, Lehr CR, Yuan CG, Le XC, McDermott TR, Rosen BP. Biotransformation of arsenic by a Yellowstone thermoacidophilic eukaryotic alga. Proceedings of the National Academy of Sciences. 2009;106:5213–5217. doi: 10.1073/pnas.0900238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Rosen BP, Zhang Y, Wang GJ, Franke S, Rensing C. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proceedings of the National Academy of Sciences. 2006;103:2075–2080. doi: 10.1073/pnas.0506836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MM, Owens G, Naidu R. Arsenic levels in rice grain and assessment of daily dietary intake of arsenic from rice in arsenic-contaminated regions of Bangladesh-implications to groundwater irrigation. Environmental Geochemistry and Health. 2009;31:179–187. doi: 10.1007/s10653-008-9238-x. [DOI] [PubMed] [Google Scholar]
- Smith E, Naidu R, Alston AM. Arsenic in the soil environment: A review. Advances in Agronomy. 1998;64:149–195. [Google Scholar]
- Tchounwou PB, Patlolla AK, Centeno JA. Carcinogenic and systemic health effects associated with arsenic exposure-a critical review. Toxicologic Pathology. 2003;31:575–588. doi: 10.1080/01926230390242007. [DOI] [PubMed] [Google Scholar]
- Thomas DJ, Waters SB, Stybloc M. Elucidating the pathway for arsenic methylation. Toxicology and Applied Pharmacology. 2004;198:319–326. doi: 10.1016/j.taap.2003.10.020. [DOI] [PubMed] [Google Scholar]
- Turpeinen R, Pantsar-Kallio M, Kairesalo T. Role of microbes in controlling the speciation of arsenic and production of arsines in contaminated soils. Science of the Total Environment. 2002;285:133–145. doi: 10.1016/s0048-9697(01)00903-2. [DOI] [PubMed] [Google Scholar]
- Wang QY, Guan YC, Wu YR, Chen HL, Chen F, Chu CC. Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Molecular Biology. 2008;67:589–602. doi: 10.1007/s11103-008-9340-6. [DOI] [PubMed] [Google Scholar]
- Williams PN, Islam MR, Adomako EE, Raab A, Hossain SA, Zhu YG, Feldmann J, Meharg AA. Increase in rice grain arsenic for regions of Bangladesh irrigating paddies with elevated arsenic in groundwaters. Environmental Science and Technology. 2006;40:4903–4908. doi: 10.1021/es060222i. [DOI] [PubMed] [Google Scholar]
- Yuan CG, Lu XF, Qin J, Rosen BP, Le XC. Volatile arsenic species released from Escherichia coli expressing the AsIII S-adenosylmethionine methyltransferase gene. Environmental Science and Technology. 2008;42:3201–3206. doi: 10.1021/es702910g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao FJ, Ma JF, Meharg AA, McGrath SP. Arsenic uptake and metabolism in plants. New Phytologist. 2009;181:777–794. doi: 10.1111/j.1469-8137.2008.02716.x. [DOI] [PubMed] [Google Scholar]
- Zhu YG, Williams PN, Meharg AA. Exposure to inorganic arsenic from rice: A global health issue? Environmental Pollution. 2008;154:169–171. doi: 10.1016/j.envpol.2008.03.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.