Summary
Complexin prevents SNAREs from releasing neurotransmitters until an action potential arrives at the synapse. To understand the mechanism for this inhibition, we determined the structure of complexin bound to a mimetic of a pre-fusion SNAREpin lacking the portion of the v-SNARE which zippers last to trigger fusion. The “central helix” of complexin is anchored to one SNARE complex while its “accessory helix” extends away at ~45° and bridges to a second complex, occupying the vacant v-SNARE binding site to inhibit fusion. That the accessory helix competes with the v-SNARE for t-SNARE binding was expected, but surprisingly, the interaction occurs inter-molecularly. Thus complexin organizes the SNAREs into a zig-zag topology which, when interposed between the vesicle and plasma membranes, is incompatible with fusion.
Introduction
Release of neurotransmitter at the synapse must be timed precisely, to follow immediately the arrival of a nervous impulse. The physiological and anatomical mechanisms for this have long been known1,2. Synaptic vesicles containing neurotransmitter are already docked at the “active zones” of the pre-synaptic membrane, ready to respond to the elevated calcium levels that accompany an action potential by releasing neurotransmitter.
In recent years, much has also been learned about the molecular mechanisms underlying this physiology. The central players in neurotransmitter release are the SNARE proteins3. These are the engines that drive membrane fusion between cargo-carrying vesicles and the plasma membrane4,5 as v-SNAREs (anchored in the vesicle membrane) zipper into a coiled-coil four helix bundle with cognate t-SNAREs (anchored in the plasma membrane)3–6. In synapses, a major v-SNARE is VAMP2, and the t-SNARE proteins are SNAP25 and syntaxin1, where VAMP2 and syntaxin1 each contribute one helix to the coiled-coil and SNAP25 contributes two7. Another vital component is synaptotagmin, a synaptic vesicle protein8 that binds calcium ions9 and is the immediate sensor and trigger for vesicle fusion10–12. How precisely synaptotagmin couples to SNAREs to trigger fusion remains unknown.
But whatever the mechanism, rapid and synchronous release of neurotransmitter requires that the fusion process by SNARE proteins be frozen in place, or “clamped”3, when it is well advanced. This is because fusion by SNARE proteins is spontaneous4,5 and must therefore be inhibited to prevent continuous release of neurotransmitters. This is also because neurotransmitter release takes place on a much shorter time scale than the entire process of vesicle docking and fusion complex assembly. For example, fusion of artificial vesicles bearing v-SNAREs to planar lipid bilayers containing t-SNAREs requires 10–100 msec following docking13–15, whereas neurotransmitter release can take place in one millisecond or less after calcium entry. Thus, fusion must be clamped at a very late stage in synapses.
A combination of biochemical, genetic, and physiological results have clearly pinpointed complexin (CPX)16,17 as the central component of this clamp18–20. Since CPX both facilitates and inhibits synaptic fusion21–26, it has been proposed to act by catalyzing the initial stages of SNARE assembly, but then clamping further assembly until the arrival of an action potential (reviewed in27).
Structures of CPX bound to a post-fusion fully assembled SNAREpin28,29 yielded first insights regarding the facilitatory mechanism, but did not resolve how CPX inhibits fusion. In the post-fusion CPX–SNARE structures, CPX forms a continuous helix parallel to the SNAREpin coiled-coil, with a “central helix” portion of CPX (CPXcen, residues 48–70 in hCPX1) contacting both the v-SNARE and t-SNARE in the membrane-distal portion of the SNAREpin. This is the portion of the SNAREpin that zippers first, and it is thus possible that CPX facilitates initial assembly29. The remainder of the CPX helix, termed its “accessory helix” (CPXacc, residues 26–47 in hCPX1), parallels the C-terminal membrane-proximal portion of the fully zippered SNARE complex, but does not interact with it.
Nonetheless, the accessory helix is needed to create the clamped, pre-fusion state21,30 in which the membrane-distal N-terminal portions of the SNARE coiled-coil have zippered, but the membrane-proximal VAMP2 C-terminus has not yet associated with the corresponding regions of SNAP25 and syntaxin118,21,31–33. Biochemical and spectroscopic experiments strongly support a mechanism whereby CPXacc directly competes with the VAMP2 C-terminus for binding to the t-SNARE19,34 - but how this happens has been unclear in the absence of structural studies with pre-fusion SNARE complexes.
We have therefore designed a half-zippered soluble mimetic of the pre-fusion synaptic SNAREpin, and we have solved its structure when bound to complexin. Remarkably, we find that the CPX accessory helix extends away from the SNAREpin, and binds a second SNAREpin to inhibit its assembly. Solution and functional studies confirm both the CPX conformation and the interaction between the accessory helix and pre-fusion SNAREpin observed in the structure. Our studies thus suggest that complexin cross-links pre-fusion SNARE complexes into a zig-zag array. This array, when interposed between the vesicle and plasma membrane, provides a further barrier to fusion. Although cross-linking the CPX–SNARE array may block fusion, it also orients the SNAREs appropriately for fusion to proceed quickly upon clamp release.
Results
Structures of a “pre-fusion” SNAREpin and its complex with CPX
The pre-fusion form of the SNARE complex is a transient intermediate stabilized in part by the simultaneous insertion of SNAREpins into two membrane bilayers, and hence is not readily accessible for structural studies. Zippering up begins as the pre-folded N-terminal portions of VAMP2 associate with the pre-assembled t-SNARE complex35–37. In designing a soluble pre-fusion SNARE mimetic suitable for structural studies, we therefore prevented the completion of zippering by C-terminally truncating the VAMP2 SNARE motif. This SNARE complex (SNAREΔ60) also contains residues 190–253 of rSyntaxin1A and residues 10–82 and 141–203 of hSNAP25A.
We determined the structure of this truncated SNARE complex at 2.2 Å resolution (Table 1). Except for the absent VAMP2 C-terminus, the truncated SNARE complex in our studies superimposes well with fully assembled SNARE complexes studied previously (rmsd 0.77–0.97 Å)7,29. A notable finding is that the syntaxin1 and SNAP25 helices are almost fully formed even in the absence of the VAMP2 C-terminus (Figure 1a), suggesting that the t-SNAREs may be almost fully folded when the v-SNARE is only half zippered.
Table 1.
Data processing and refinement statistics for the structures presented in this study.
| SNARE | ScCPX– SNARE |
ScCPX F34M– SNARE |
|
|---|---|---|---|
| Data collection | |||
| Space group | P1 | C 21 | P1 |
| Cell dimensions | |||
| a, b, c (Å) | 27.6, 39,8, 102.3 | 75.9, 52.7, 128.7 | 53.7, 127.4, 142.7 |
| α, β, γ (°) | 83.4, 89.9, 89.9 | 90, 95.2, 90 | 107.5, 90.0, 90.1 |
| Resolution (Å) | 50-2.2 (2.28-2.2) | 50-3.5 (3.63-3.5) | 30-3.8 (3.94-3.8) |
| Rsym or Rmerge | 0.052 (0.204) | 0.062 (0.234) | 0.08 (0.253) |
| I / σI | 19.7 (5.7) | 15.7 (6.2) | 9.1 (2.6) |
| Completeness (%) | 91.9 (76.4) | 94.8 (91.3) | 83.5 (80.7) |
| Redundancy | 3.7 (3.3) | 3.4 (3.5) | 1.9 (1.8) |
| Refinement | |||
| Resolution (Å) | 40-2.2 | 25-3.5 | 30-3.8 |
| No. reflections | 20117 | 6172 | 32506 |
| Rwork / Rfree | 0.227 / 0.268 | 0.270 / 0.316 | 0.306 / 0.346 |
| No. atoms | |||
| Protein | 3760 | 2215 | 17672 |
| Ligand/ion | 4 | - | - |
| Water | 37 | - | - |
| B-factors | |||
| Protein | 56.9 | 99.8 | 116.8 |
| Ligand/ion | 68.7 | - | - |
| Water | 29.1 | - | - |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.017 | 0.054 | 0.035 |
| Bond angles (°) | 1.58 | 1.28 | 1.01 |
Values in parentheses are for highest-resolution shell.
Figure 1.
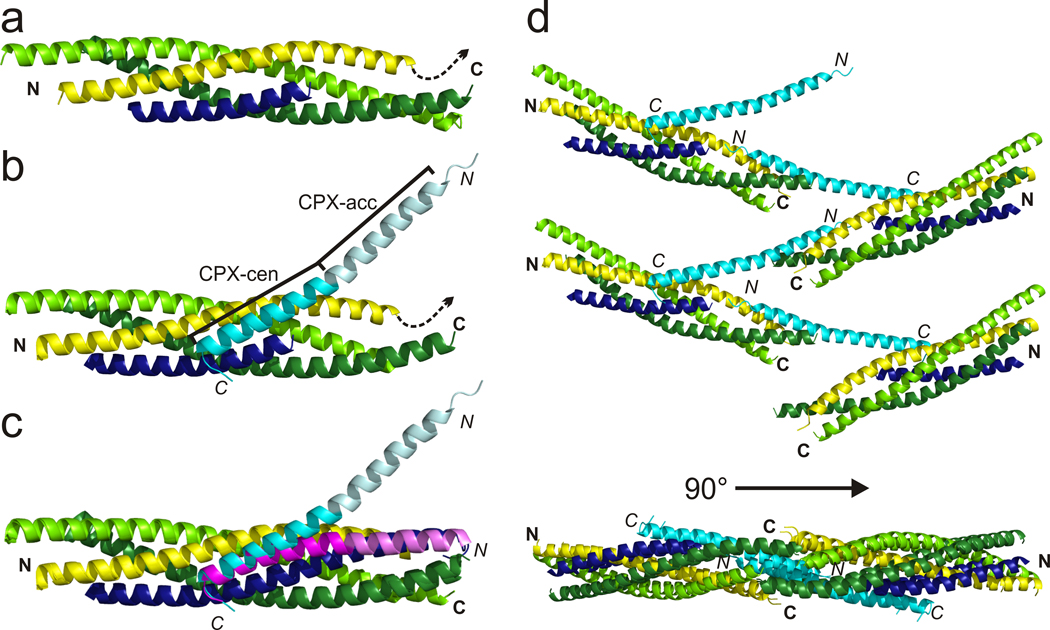
Structure of the pre-fusion CPX–SNARE complex. VAMP2 (residues 29–60) is blue, syntaxin yellow (residues 190–250), SNAP25 lime (N-terminal SNARE motif, residues 10–74) and green (C-terminal SNARE motif, residues 141–203) and CPX (residues 26–73) cyan. Model of (a) the truncated SNARE complex without and (b) with CPX bound. CPXcen is cyan, and CPXacc pale cyan. A dashed arrow indicates syntaxin membrane anchor. (c) Comparison of pre- and post-fusion CPX–SNARE complexes, with post-fusion CPX magenta (CPXacc is pale magenta, PDB ID 1KIL). The arrow indicates the conformational change of CPX during clamp release. (d) Top and side views of the zig-zag array of post-fusion CPX–SNARE complexes observed in crystals. SNAREpins are related by 180° rotation and translation along the zig-zag midline, so that on different sides of the mid-line the linkers that connect syntaxins and VAMPs to their trans-membrane helices are on opposite sides of the zig-zag plane.
We next co-crystallized SNAREΔ60 with a CPX fragment (scCPX) consisting of its central and accessory helices (residues 26–83) and containing three “superclamp” mutations (D27L, E34F, R37A) that increase its clamping efficiency both in vitro19 and in vivo38. The structure was determined at 3.5 Å resolution using the truncated SNARE complex as a search model in the molecular replacement method (Table 1), and CPX was modeled into difference electron density. The final model includes residues 190–250 of syntaxin1, 10–74 and 141–203 of SNAP25, 29–60 of VAMP2, and 26–73 of CPX (Figure 1b).
To confirm the sequence alignment along CPX, we used selenomethionine substituted forms of scCPX, where residues Leu27 and Phe34 in the accessory helix were mutated to methionine (scCPX-L27M, scCPX-F34M). The selenomethionine-substituted forms of CPX were co-crystallized with the truncated SNARE complex, and anomalous data (Table 1) were used to calculate difference maps that unambiguously locate the position of residues 27 and 34 as well as 55, a methionine in the wild-type sequence (Supplementary Figure 1). We also determined the structure of the selenomethionine-substituted scCPX-F34M bound to the truncated SNARE complex (Table 1). While the crystals of this complex belong to a different spacegroup (P1) from the scCPX–SNARE crystals we initially obtained (C2), our findings regarding CPX-SNARE interactions are similar.
The CPX–SNAREΔ60 structures resemble the fully-zippered, post-fusion structures observed previously in several aspects (Figure 1c). The conformations of the SNARE proteins are essentially unaltered (rmsd 0.83 Å). Further, as in the post-fusion forms of the CPX–SNARE complex28,29 as well as alone in solution39, CPX forms a continuous helix. The interactions between CPXcen and the SNARE complex observed in the post-fusion structure are also largely unperturbed (small positional shifts in CPXcen are detailed in Supplementary Table 1; see also Figure 1c). CPXcen binds in the groove between syntaxin1 and VAMP2, with key residues Arg59, Arg63, Ile66, Tyr70, and Ile72 inserted into two high affinity binding pockets observed in earlier studies28,29. A third previously identified binding interface involves residues Asp64, Asp65 and Asp68 in the VAMP2 C-terminus, which are missing in our construct. This interaction is not necessary for clamping40, and so its absence in our structure likely will not affect conclusions regarding the CPX clamping mechanism.
Despite these similarities, the arrangement of CPX relative to the SNAREs in our structure differs markedly from that in post-fusion forms of the CPX–SNARE complex. The accessory helix undergoes a dramatic reorientation (rmsd of CPX as compared to PDB ID 1KIL29 is 2.2 Å). Rather than running alongside the SNARE complex, CPXacc now bends away at a ~ 45° angle (Figure 1c). The reorientation likely results from small differences in CPXcen docking (Supplementary Table 1) as well as small changes in phi and psi torsion angles in the transition region between CPXcen and CPXacc (Supplementary Table 2). This result was unexpected given biochemical data indicating that CPXacc should occupy the binding site for the VAMP2 C-terminus, since CPX and the VAMP2 C-terminus compete for binding to the t-SNAREs19,34. While CPXacc does not interact with the same SNARE complex bound by CPXcen, however, it does interact with a second, symmetry-related complex.
Overall, the crystal packing is such that CPX–SNARE complexes are arranged in a continuous zig-zag (Figure 1d), leaving the middle of the accessory helix entirely solvent exposed. This region has high thermal motion, as evidenced by high B-factors (Supplementary Figure 1). SNAREpins on opposite sides of the zig-zag mid-line are related by a 180 degree rotation about (and a translation along) the mid-line. This means that on different sides of the mid-line, the linkers that connect the syntaxins and the VAMPs to their trans-membrane helices in the plasma membrane and the synaptic vesicle, respectively, are on opposite sides of the zig-zag plane. Although the CPX-F34M mutant crystallized in a different space group, it cross-links different SNARE complexes the same way, and the complexes are arranged in a zig-zag (Supplementary Figure 2).
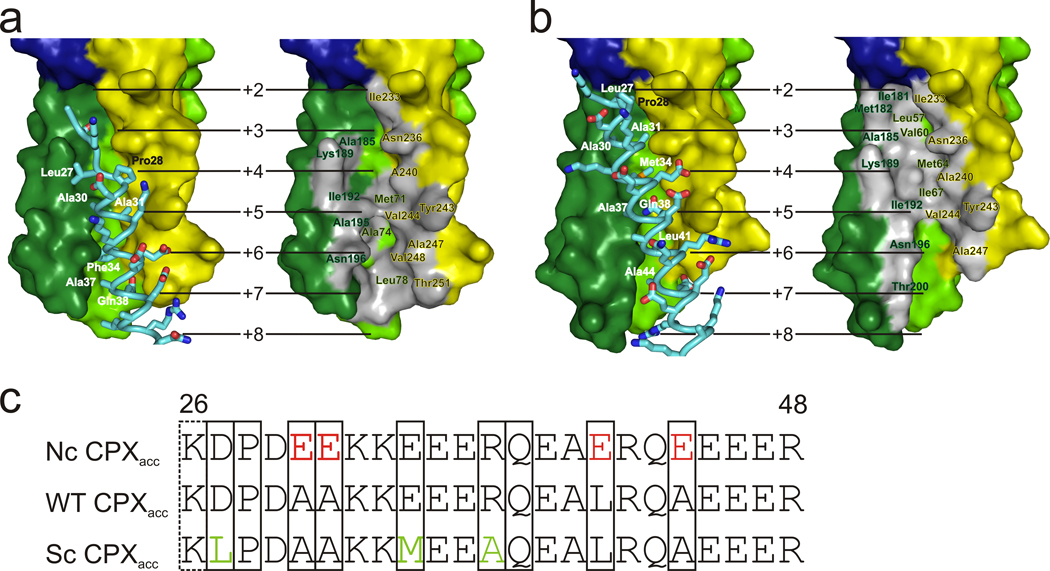
Residues at the N-terminal end of CPXacc (Leu27, Ala30, Ala31, Phe34, and Ala37) form a hydrophobic surface which binds to the t-SNARE in a second SNARE complex in a site normally occupied by the C-terminus of the VAMP2 helix in post-fusion state, which was deleted from the mimetic used here (Figure 2a–c, Supplementary Figure 3). In crystals of scCPX-F34M–SNARE, the interactions between CPXacc and the t-SNARE groove are as just described for the scCPX–SNARE crystals in four of eight crystallographically distinct complexes. In the remaining four complexes, the binding site on the t-SNARE is shifted by approximately two helical turns, so that the interface between CPXacc and the t-SNARE is larger (~1000 Å2 versus ~715 Å2, Figure 2b, Supplementary Figure 3), additionally involving CPX residues Leu41, Ala44 and Arg48. Because a single mutation in the CPXacc sequence allows for two different binding modes, we expect that the high sequence variability in CPXacc of different complexins (isoforms 1–4 and in different organisms) results in slight variations of SNARE-bridging interactions and strength. The recurrence of the zig-zag arrangement of CPX–SNARE complexes in two different crystal forms, however, supports the notion that this arrangement may be physiologically relevant.
Figure 2.
Interacting surfaces of CPXacc and the t-SNAREs. (a) Interacting residues of scCPX are labeled in left panels; the binding site on the t-SNARE is outlined as grey patch and labeled on right panels. (b) For scCPX-F34M, CPXacc can bind to the t-SNARE groove as in (a) or as shown here. (c) Sequence of the accessory helix of wild-type (WT) CPX and the non-clamping (nc) and superclamp (sc) mutants. Residues of CPX interacting with the t-SNARE in the crystal structures are boxed. The side chain of Lys26 is disordered in our structure, but functional data19 suggest that it has a role in clamping. It may interact with the VAMP2 C-terminus absent in our structure.
Notably, for both scCPX and scCPX-F34M, residues that were mutated to make the superclamp CPX (D27L, E34F or E34M, R37A) are an integral part of the hydrophobic interface with the SNARE complex. The ability to bind the t-SNARE surface via a more extended hydrophobic interface may explain why the superclamp sequences have a higher affinity for pre-fusion SNARE complexes than wild-type CPX (shown below) and why superclamp CPXacc clamps more effectively in vitro and vivo19,38.
Solution studies confirm the CPXacc–SNAREΔ60 interaction
Thus, in both the CPX–SNARE structures, we find CPXacc interacting with the t-SNAREs in such a way that CPX, linked by its central helix to one SNARE complex, blocks binding of the VAMP2 C-terminus to another complex, cross-linking the SNARE complexes into an array in the process.
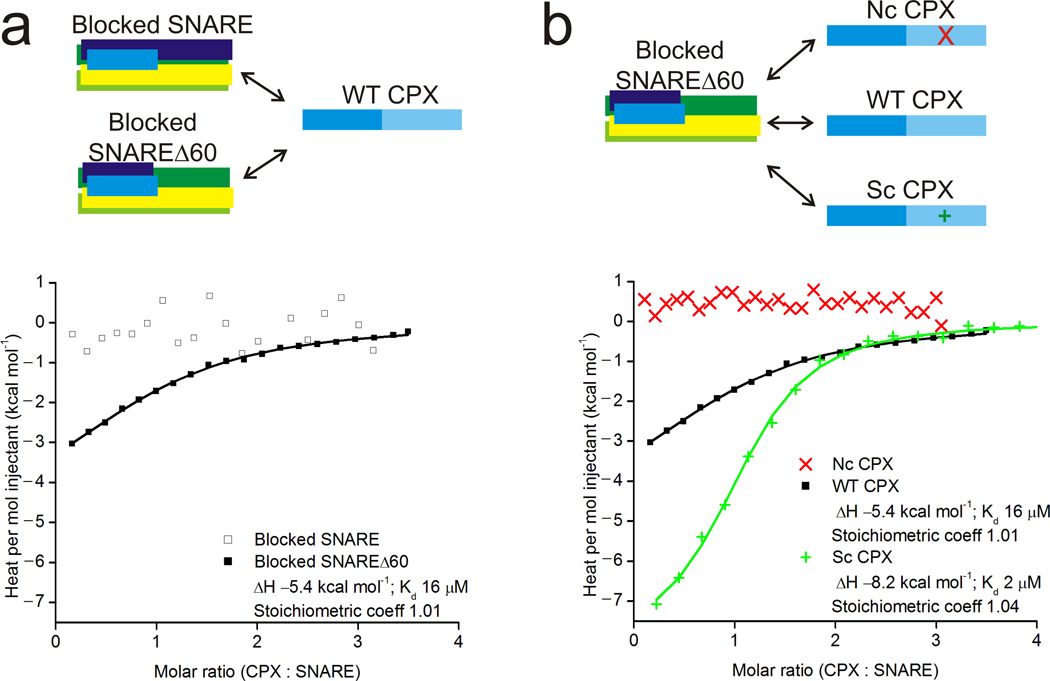
We used isothermal titration calorimetry (ITC) experiments to confirm that CPXacc interacts with the t-SNARE in pre-fusion SNARE complexes. In these experiments, we used a complexin construct comprising both the central and accessory helices (residues 26–83) rather than a peptide corresponding to the accessory helix alone. Our rationale was that the accessory peptide does not fold into an alpha helix, as monitored by circular dichroism (CD), and thus does not fold as in the full length protein, where it has high helical propensity (CD and 39). The longer complexin construct was chosen to avoid complications in binding measurements resulting from folding energetics. To observe the interaction between only CPXacc and the SNARE complex, we blocked the CPXcen binding site on either a fully-assembled post-fusion SNARE or SNAREΔ60 by pre-binding CPXcen (residues 48–134). Various CPX constructs were then titrated in to derive interaction affinities. As predicted from the post-fusion SNARE–CPX crystal structure, we find no additional interaction between wild-type complexin (wtCPX, residues 26–83) and the blocked post-fusion SNAREpin (Figure 3a). In contrast, wt-CPX interacts with blocked SNAREΔ60 with Kd ~10 µM affinity, consistent with an additional binding site present only pre-fusion and the finding that CPX competes with the VAMP2 C-terminus for binding19. Further, the interaction affinity can be modulated by mutating residues in CPXacc, as expected if CPXacc participates in the interaction (Figure 3b). We used CPX mutants where residues at the CPXacc–SNARE interface in the crystal structure were altered. In addition to scCPX (D27L, E34F, R37A) we designed a non-clamping CPX mutant (ncCPX: A30E, A31E, L41E, A44E), where hydrophobic residues at the CPXacc–t-SNARE interface in the crystal structure were replaced by charged residues (Figure 2c). As expected, the binding affinity for scCPX is ~8-fold stronger than wild-type, consistent with the difference in activity observed in both in vitro and in vivo assays19,38, whereas ncCPX no longer interacts with blocked SNAREΔ60 (Figure 3b). Thus, binding studies corroborate an interaction between CPXacc – both wild-type and superclamp – and the pre-fusion SNARE complex as observed in the crystal structure.
Figure 3.
Characterization of the interaction of CPXacc with SNARE complexes by isothermal titration calorimetry. (a) A groove in the t-SNARE is a second binding site for CPX distinct from the central helix binding site. When the central helix binding site on the SNARE complex is blocked, CPX still binds to the SNARE complex once the C-terminal half of VAMP2 is removed in the pre-fusion SNARE mimetic. (b) Binding to the t-SNARE groove is mediated by CPXacc. Mutations in the accessory helix of CPX modulate the binding affinity to the t-SNARE positively (scCPX) or negatively (ncCPX) as expected from the crystal structure.
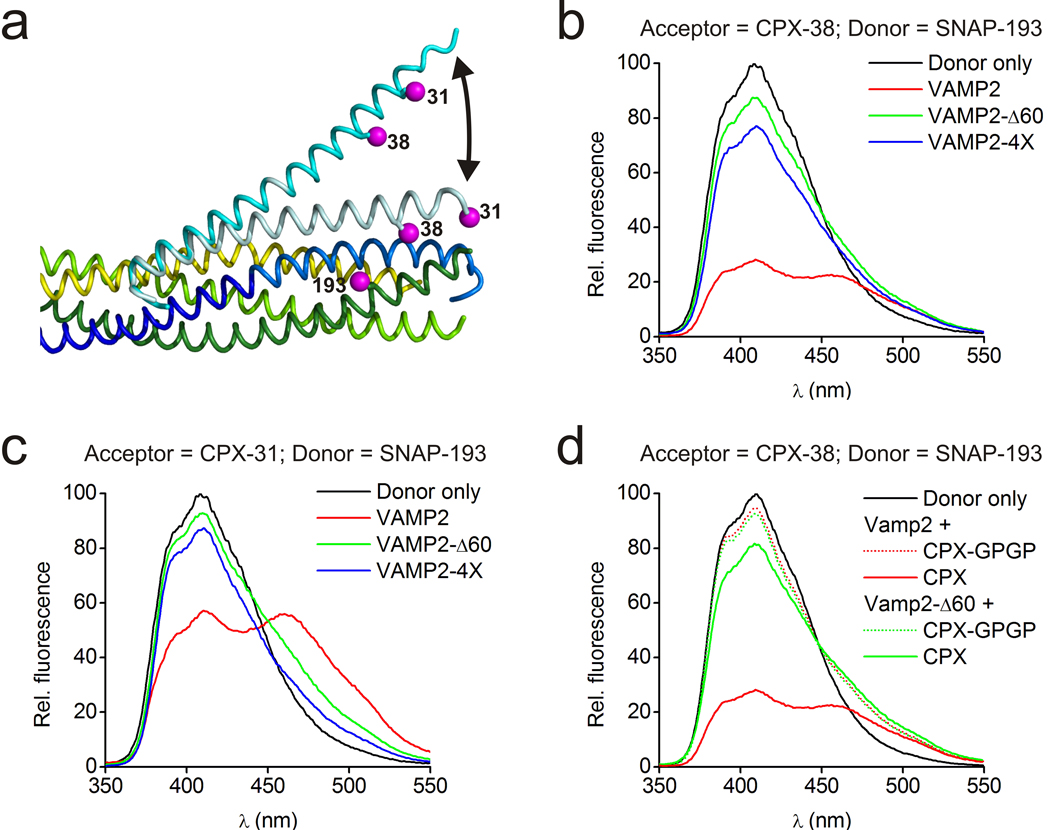
We used Förster resonance energy transfer (FRET) experiments to establish that the angled conformation of CPX also occurs in solution and therefore is not dictated by crystal packing. The donor dye (stilbene) was attached to SNAP-25 residue 193, with the acceptor dye (bimane) positioned either at residue 31 or 38 of superclamp CPX (Figure 4a). (Note that acceptor positions are placed so that they would interfere with CPXacc–t-SNARE cross-linking interactions, enabling monodisperse CPX–SNARE complexes to be studied.) Distances estimated via quenching of donor fluorescence for CPX bound to the fully zippered SNARE complex correspond closely to distances observed in the crystal structure of the post-fusion CPX–SNARE complex (PDB 1KIL), where CPXacc runs parallel to the SNARE complex (Figure 4b,c, Supplementary Table 3).
Figure 4.
FRET experiments probing CPX orientation in pre- and post-fusion CPX–SNARE complexes. (a) Superposition of pre- and post-fusion CPX–SNARE complexes, where pre-fusion CPX is cyan and post-fusion CPX is pale cyan. As indicated (magenta), SNAP25 was labeled with stilbene at position 193, and CPX was labeled with bimane at positions 31 or 38. (b) Fluorescence emission spectra of stilbene only (black) and stilbene or bimane labeled CPX–SNARE complexes containing VAMP2 (residues 25–96, red), VAMP2-Δ60 (residues 25–60, green), or VAMP2–4X (residues 25–96 with mutations L70D, A74R, A81D, L84D to preclude zippering of the VAMP2 C-terminus, blue). CPX is labeled with bimane at residue 38. (c) As in (b), but CPX is labeled with bimane at residue 31. These data were used to calculate distances shown in Supplementary Table 3. (d) FRET of a “flexible” CPX mutant (CPX-GPGP) in comparison to wild-type (WT) CPX when bound to pre-fusion (VAMP2-Δ60) or post-fusion (VAMP2) SNARE complexes. When the accessory helix is uncoupled from the central helix by a helix-breaking GPGP insertion, there is a complete loss of FRET signal with both SNARE complexes, different from the partial change in FRET observed with intact CPX. Thus, it is unlikely that differences between the FRET signals observed with intact CPX are due to random motion in CPXacc.
In contrast, residues nearer the CPXacc N-terminus appear to move increasingly away from the SNAREΔ60 complex used for crystallization (so that the dye at CPX residue 31 is farther from the donor than the dye at CPX residue 38) and the distances estimated via quenching of donor fluorescence (see Supplementary Figure 4 for comparable data for acceptor fluorescence increase and with a second FRET pair) agree with the angled conformation in the crystal structure (Supplementary Table 3, Figure 4b,c). Use of a “flexible” CPX construct (CPX-GPGP), where a helix-breaking GPGP linker was inserted between the central and accessory helices of CPX, discounts the possibility that the change in the FRET signal reflects random motion in CPXacc due to increased CPX flexibility rather than a discrete change in CPX conformation (Figure 4d). In contrast to the experiments with the undisrupted CPX constructs, there was no detectable FRET signal for CPX-GPGP bound to either SNAREΔ60 or to the post-fusion SNARE, consistent with random motion in CPX-GPGP but not for the intact CPX. To rule out that the angled conformation in solution results from VAMP2 truncation, we also studied and obtained similar results (Supplementary Table 3, Figure 4b,c) for a complex containing the entire VAMP2 SNARE motif, but harboring mutations in its C-terminal hydrophobic layers (L70D, A74R, A81D, L84D) that prevent assembly of this region with syntaxin1 and SNAP25 and eliminate fusion activity40. These experiments indicate that when bound to a half-zippered form of the SNARE complex, it is the intrinsic property of CPXacc to extend away from the complex. Because this conformation is maintained in solution, it determines how the complex crystallizes, and not vice versa.
Thus, as CPX rigidly extends away from the half-zippered SNARE complex, the only plausible way for both its central and accessory helices to interact with the SNAREpin is if CPX can interact with two different pre-fusion SNAREs, cross-linking SNAREs into an array like the zig-zag observed in the crystals.
Mutations in the CPXacc binding surface affect clamping
Further support that the CPXacc–t-SNARE binding interface observed in the crystal structure represents biologically relevant interactions comes from in vitro clamping assays. In these experiments, “flipped” SNARE proteins are expressed on the cell surface, and the effects of CPX and synaptotagmin constructs on cell-cell fusion are monitored. These flipped-SNARE cell-cell fusion assays were initially developed to demonstrate clamping by CPX and clamp release by synaptotagmin4,18 and the effects of CPX mutations (including the superclamp mutations) in these assays are consistent with their effects in vivo38.
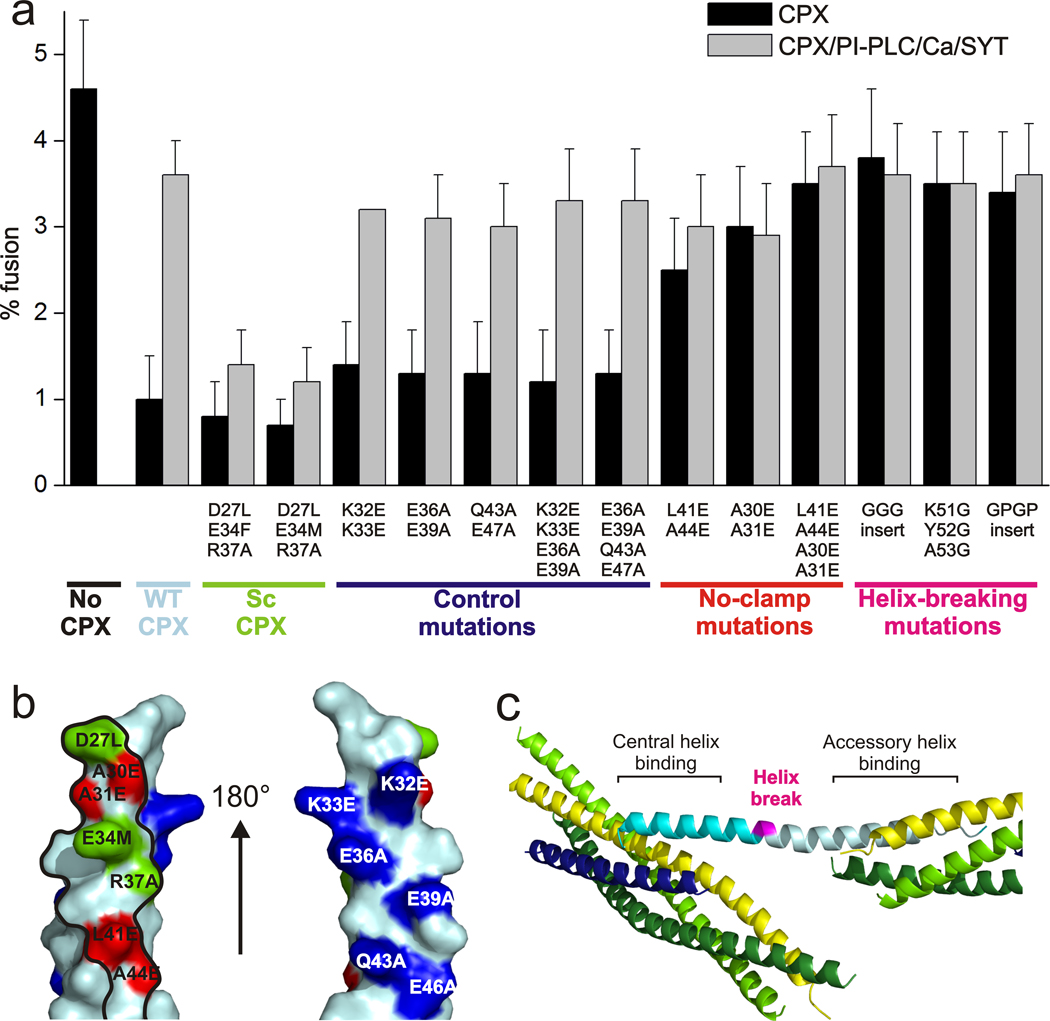
We systematically tested the effect of the mutations introduced in CPXacc and mapped residues that do or do not affect clamping onto the surface of CPX (Figure 5a,b). Mutations which are located at the CPXacc–SNARE interface observed in the structure all alter clamping efficiency. As expected, clamping is affected positively by scCPX mutations and negatively by ncCPX mutations (Figure 5). As a control, mutations that are oriented away from the interface on the opposite side of CPX have no effect on clamping (Figure 5). These findings strongly support that the interface observed in the crystal contact is relevant for the physiological function of complexin and verify the rational of our mutant design for ITC.
Figure 5.
Effects of CPX and VAMP2 mutations on clamping in cell-cell fusion assays. (a) Mutational analysis of CPX accessory helix mutations in the cell-cell fusion assay. (b) Mapping of the mutational analysis of the CPXacc–t-SNARE interface. CPXacc is shown with the surface that interacts with the t-SNARE in the crystal structures outlined in black. Mutations in CPX that affect clamping positively (green) or negatively (red) are at the interface. Mutations that do not affect clamping (blue) are on the opposite side of CPX. (c) Location of the helix breaking mutations (magenta) between central and accessory helix in the CPX–SNARE pre-fusion crystal structure.
We note, however, that although ITC and the in vitro clamping assays validate that CPXacc interacts with pre-fusion t-SNAREs using a surface similar to that identified from the crystal structure, it is likely—given the differences between the superclamp and wild-type sequences—that the details of the interaction differ for wild-type CPX. But as discussed earlier, due to low sequence conservation in the accessory helix, there may be also variability in the interactions of CPXacc from different organisms.
The observations from the crystal structure and their agreement with FRET from mono-disperse solutions suggest that the CPX accessory helix rigidly bends away from the SNARE complex. To test whether the rigidity of CPX is important for clamping, we again used the flipped SNARE cell-cell fusion assays. We used CPX mutants (CPX-GPGP, CPX-GGG) which had a helix-breaking linkers (GPGP and GGG, respectively) inserted after residue 50, between CPXcen and CPXacc, as well as a construct where residues 51–53 at the central-accessory helix junction were replaced by glycines to disrupt the long CPX helix (see Figure 5c). Clamping should be affected if the continuity and hence rigidity of the CPX helix is mechanistically important. We found that clamping indeed was reduced in all three cases, consistent with the requirement for a continuous helix (Figure 5a).
Discussion
Model for clamping
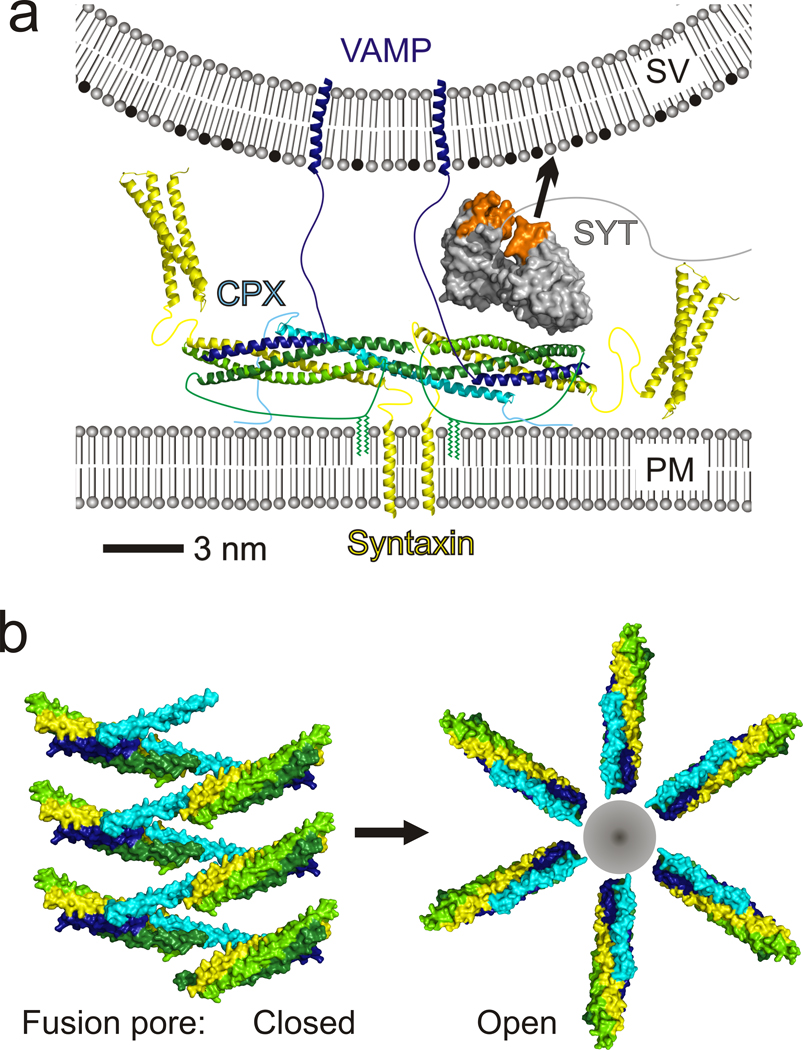
Binding, fluorescence and functional studies all corroborate the conformation of CPX as observed in the crystal structure as well the novel interaction identified between CPXacc and the t-SNARE. Based on the crystal structure, we therefore propose that CPX directly cross-links pre-synaptic, pre-fusion SNARE complexes and further that the arrangement of CPX–SNARE complexes in the clamped state is similar to the zig-zag observed in the crystal lattice. Such an arrangement is plausible given the length of linkers that anchor the t- and v-SNAREs to the membranes, as the linkers for syntaxin1 and the half-zippered VAMP2 are longer than 10 (~ 37 Å) and 30 residues (>100 Å), respectively (Figure 6a). The number of CPX–SNARE complexes in the zig-zag would be limited due to curvature in the vesicle, which increases the distance between the vesicle and plasma membranes with increasing distance from the fusion site (close to the zig-zag center), so that polymer extension beyond a certain distance is untenable. Experiments suggest that for optimal fusion rates, there are 5–10 SNARE complexes in a fusion pore14, allowed by our model.
Figure 6.
Molecular models for CPX clamping. (a) Model for the clamp at the synapse. CPX–SNARE complexes with half-zippered VAMP2 are cross-linked by CPX into a zig-zag topology incompatible with fusion (see text). The plane of the zig-zag is normal to the vertical direction. For clarity, only two of the CPX–SNARE complexes in the zig-zag are shown. Palmitoylation on SNAP25 is indicated and restrains the distance between the CPX–SNARE zig-zag and the plasma membrane (PM). The distance between the zig-zag plane and the vesicle (SV) must be less than ~110 Å, the maximum distance spanned by the v-SNARE linker. The calcium sensor synaptotagmin (grey with Ca2+-binding loops orange), which relieves CPX clamping, is accommodated by this model and is positioned according to FRET analysis45. Its Ca2+-binding loops are juxtaposed to the vesicle membrane, which is rich in anionic lipids like phosphatidyl-serine (black), well positioned for interactions with this membrane in response to Ca2+ stimulus. (b) Model of the CPX–SNARE assembly in the clamped state when the fusion pore is “closed” (left). The fusion pore can open only once the zig-zag clamped array has disassembled (right). Complexes in “open” state are modeled on PDBID 1KIL.
The crystal structure naturally suggests several synergistic mechanisms by which CPX might stabilize the pre-fusion state and inhibit fusion (Figure 6):
First, CPXacc binds the t-SNAREs in a site occupied by C-terminal portions of the VAMP2 SNARE motif in post-fusion SNARE complexes, competitively blocking the completion of zippering by VAMP2 as proposed previously from biochemical studies19,34, but with the critical modification that this interaction occurs inter-molecularly.
Second, close apposition of SNAREpins by cross-linking at their zippering ends should prevent further zippering which, if it occurred would cause them to clash sterically (~2 turns of the N-terminal SNAP25 SNARE motifs are not folded in our structure).
Third, the linker regions of Syntaxin1 and VAMP2 on different sides of the zig-zag mid-line emerge on opposite sides of the zig-zag plane, again sterically interfering with complete zippering.
Fourth, the fusion pore cannot form as it is blocked by the CPX–SNARE zig-zag array, which is interposed between the vesicle and plasma membranes. Our finding from functional assays that flexibility in CPX interferes with clamping suggests that the CPX–SNARE zig-zag must be rigid at least to some extent. The requirement for rigidity is consistent with a role as a barrier between membranes that are poised for fusion.
And fifth, cross-linking the SNARE complexes into a zig-zag prevents them from forming the circular arrangement needed to accommodate either a hemi-fusion stem41 or a fusion pore42, precluding their formation. Notably, though, even in the zig-zag, the orientation of the SNAREs is very similar to that in a fusion-competent arrangement (compare panels in Figure 6b), except that the cross-links must dissolve in order for fusion to take place. As the zig-zag clamp disassembles and SNAREs zipper, steric repulsion would push the SNAREs radially away from the zig-zag mid-line to form a circular arrangement (Figure 6b), now enclosing a nascent fusion pore which opens progressively as zippering completes43.
Though clearly vital for clamping19, each pairwise interaction between CPXacc and t-SNARE complexes seems of relatively low affinity (Kd ~ 10 µM, corresponding to ~6.8 kcal mol−1). Nonetheless, binding with this affinity is likely to occur physiologically because the concentrations of CPX and SNARE proteins in the region local to fusion between apposed bilayers (~20 nm × ~20 nm × ~20 nm (Figure 6a) containing 5–10 CPX–SNARE complexes14) are likely to be in the 1–2 mM range (incidentally, an order of magnitude higher than their concentration in the crystallization mixture). And due to entropy considerations, polymerization would be more favored for proteins constrained to two dimensions, as at the synapse, than in solution.
The clamp may be further stabilized by the CPX N-terminus, which is absent in our structure, and which can interact with membrane proximal portions of SNAREs20,44. And functional assays show that synaptotagmin (included in Figure 6a according to45), in its calcium-free conformation, stabilizes the clamped state produced by CPX18,46, although how this occurs is currently unclear.
SNARE activation
As noted previously, in addition to its inhibitory role, CPX also has an important positive role in promoting fusion21. Some of the domains shown to be required for this mode of action21,24 are not present in our structure. It has been speculated, however, that one positive contribution may result from the binding of CPXcen to the VAMP2–syntaxin1 interface, which would stabilize initial SNARE assembly and zippering27,29. Our studies now suggest a similar role for the accessory helix: its interaction with the t-SNARE groove newly identified by us indicates that CPX might facilitate t-SNARE folding by binding to the C-terminal part of their SNARE motifs prior to VAMP2 binding.
Most importantly, we note that the assembled clamp itself might promote fusion by simply setting the stage: multiple SNARE complexes are gathered in orientations close to that required for fusion pore formation (compare panels in Figure 6b) even as their cross-linking impedes it, and they are already half-zippered. This alone will allow for fast, efficient fusion as soon as the clamp is released upon stimulus.
The mechanism of clamp disassembly is further explored in an accompanying manuscript40, as is the finding that clamp release is intrinsically coupled to a conformational change in CPX, where CPX switches from the angled conformation observed in the CPX–SNAREΔ60 structure to that in the post-fusion CPX–SNARE complex28,29.
Methods
Protein Expression, Purification and Complex Assembly
Recombinant fusion proteins were expressed in E.coli BL21 (DE3) cells by induction with 0.5 mM IPTG for 4 h at 37 °C. Selenomethionine substituted CPX-L27M and CPX-F34M were expressed according to Doublie47. Proteins were purified with either glutathione-Sepharose (GE) or Ni-NTA-agarose (Qiagen) resin, and tags were cleaved according to manufacturer instructions. Complexes were reconstituted by mixing proteins, followed by gel filtration on a HiLoad Superdex 75 (16/60, GE Healthcare). See Supplementary Methods for detailed protocols.
Crystallization and Data Collection
Purified complex was concentrated to ~10 mg ml−1, and crystallized at 20 °C using the hanging drop vapor diffusion method. The best crystals were obtained when the SNARE complex was mixed with 6× molar excess of CPXacc peptide (residues 26–35, purchased from Biosynthesis) and equilibrated against 0.05 M calcium acetate, 27% (v/v) 2-methyl-2,4-pentanediol, 0.1 M sodium cacodylate pH 6.5–7.0. The crystals were loop-mounted from mother liquor and plunged into liquid nitrogen for cryopreservation.
Crystals of the CPX–SNARE complex were obtained by equilibration against a solution containing 13–15% (w/v) polyethyleneglycol (PEG) 5000MME, 0.2 M ammonium sulfate, 0.01 M EDTA, and 0.1 M Tris pH 7.5. Crystallization conditions for the CPX-L27M–SNARE and CPX-F34M–SNARE complexes were similar. Crystals were transferred into buffer supplemented with 15% (w/v) PEG 400 prior to flash-freezing in liquid nitrogen.
Data were collected at NSLS (National Synchrotron Light Source, Brookhaven) beamline X29 or APS (Advanced Photon Source, Argonne) beamline ID-24C and processed with HKL200048.
Structure determination
For all crystals, phases were obtained by the molecular replacement method as implemented in Phaser49. Models were build in Coot50 and refined with Refmac51. For the truncated SNARE complex we used a search model based on PDB 1KIL. TLS groups and non-crystallographic symmetry (NCS) restraints were used in refinement to 2.1 Å resolution. The CPXacc peptide (26–35) in the crystallization solution is not bound to the SNARE complex.
Crystals of the CPX–SNARE complex belong to space group C2 and diffract to 3.5 Å resolution. The truncated SNARE complex served as the search model. Complexin was manually built into difference density as a continuous α-helix. For refinement, TLS groups and H-bond restraints for α-helical secondary structure derived from the high resolution SNARE complex structure were used.
The selenomethionine substituted CPX-F34M–SNARE crystals diffract to 3.8 Å resolution. The crystals have P21 pseudosymmetry but we were able to refine to reasonable R values only in P1. As there are eight CPX-F34M–SNARE complexes in the P1 asymmetric unit, we used the thin shell method in choosing the Rfree set in order to avoid bias from non-crystallographic symmetry (NCS)52. The truncated SNARE complex structure was used as a search model in molecular replacement. A Fourier anomalous difference map was calculated using CNS53, allowing us to unambiguously locate the positions of the Se atoms of residues 34 as well as 55, a methionine in the wild-type sequence in CPX-F34M. NCS restraints were used in refinement.
Composite simulated-annealed omit maps calculated in CNS53 confirm the CPX–SNARE models (Figure S1).
Crystals of the selenomethionine substituted CPX-L27M–SNARE complex belong to space group P1 and diffract to 4.5 Å. A molecular replacement solution using the CPX–SNARE complex as search model was found, identifying 4 complexes in the asymmetric unit. Although data resolution did not allow for refinement, we could determine the position of the Se atoms in residues Met27 and Met55 in an anomalous difference map calculated using phases from the molecular replacement solution53. The positions are consistent with register in the CPX–SNARE complex.
All figures were prepared with Pymol (DeLano Scientific LLC). Data collection and refinement statistics are summarized in Table 1.
Isothermal titration calorimetry (ITC) analysis
ITC experiments are described in detail in the Supplementary Methods. Briefly, measurements were carried out with a ITC200 instrument (Microcal). CPX constructs (200–600 µM) were titrated into a solution of SNARE complexes in the sample cell (10–30 µM), and thermodynamic parameters were calculated using the Microcal Origin ITC200 software package assuming a “one-set-of-sites” binding model.
FRET analysis
Positions Asp193 on SNAP25 and Gln38 or Ala31 on scCPX (hCpx1 residues1–134 carrying superclamp mutations D27L, E34F and R37A) were mutated into cysteines using the Stratagene QuikChange Kit. SNAP25 D193C was labeled with the donor probe, Stilbene (4-acetamido-4’-((iodoacetyl)amino)-stilbene-2,2’-disulfonic acid, disodium salt, Invitrogen) and either CPX Q38C or A31C was labeled with the acceptor Bimane (Monochlorobimane, Invitrogen), using 10× molar excess of dye overnight at 4°C in 50 mM Tris Buffer, pH 7.4, containing 150 mM NaCl, 10 % (w/v) glycerol and 1 mM tris(2-carboxyethyl)phosphine (TCEP). Excess dye was separated from the labeled proteins using a NAP desalting column (GE Healthcare). Double-labeled CPX–SNARE complexes were assembled overnight at 4°C and purified by gel-filtration on a Superdex 75 (10/30, GE Healthcare) gel filtration column. Fluorescence data were obtained on a Perkin-Elmer LS55 luminescence spectrometer at 25°C. Excitation and emission slits of 5 nm were used in all measurements. Fluorescence emission spectra were measured over the range of 350–550 nm with the excitation wavelength set at 335 nm. The donor probe concentration was adjusted to 2 µM in all samples. We used fluorescence resonance energy transfer to calculate the distance between the two fluorophores with a R0 of 27.5 Å for the Stilbene-Bimane FRET pair54. See Supplementary Methods for detailed experimental procedures.
Cell-Cell fusion assay
The flipped SNARE cell-cell fusion assay was performed as described before4,18,19,30. In brief, HeLa cell lines were transiently transfected with flipped VAMP2 (wt or 3xDA), DsRed2-NES and either with or without CPX mutants and synaptotagmin as indicated (v-cells). After one day, transfected v-cells were seeded onto glass coverslips containing cells stably co-expressing flipped syntaxin1, flipped SNAP-25 and CFP-NLS (t-cells). The following day, cells were fixed with 4% (v/v) paraformaldehyde directly or after treatment with recovery solution (1 U ml−1 Phosphatidylinositol Specific Phospholipase-C, 20 µg ml−1 laminin, with or without 1.8 mM EGTA), washed and mounted with Prolong Antifade Gold mounting medium (Molecular Probes). Confocal images were acquired on a Zeiss 510-Meta confocal microscope and processed using Adobe Photoshop software.
Supplementary Material
Acknowledgements
We wish to thank the staffs of X29 at the NSLS and of NE-CAT at APS for their help in data collection, and Lavan Khandan (Yale) and Stephanie Baguley (Yale) for technical assistance, and Dr. Jeff Coleman (Yale) for advice. We are grateful to Dr. Erdem Karatekin (Yale) and Professor David W. Rodgers (University of Kentucky) for discussions regarding this manuscript. This work was supported by grants from the NIH to KMR (R01GM080616) and to JER, an ANR PCV grant to FP, and a grant from the DFG to DK.
Footnotes
Accession Codes. Coordinates and structure factors for the structures described in this manuscript have been deposited in the PDB (accession codes 3RK2, 3RK3, 3RLO).
Author Contributions. DK coordinated the experiments in this paper. He is responsible for structure analysis and designed constructs for the functional analyses. SSK and DTR performed the FRET experiments; FL performed the ITC analysis, and CCG carried out the cell-cell fusion experiments. FP contributed to the analysis of the FRET and ITC data. DK, JER, and KMR analyzed data and wrote this manuscript.
Competing Interests Statement
The authors declare no competing financial interests.
References
- 1.Fatt P, Katz B. Spontaneous subthreshold activity at motor nerve endings. J Physiol. 1952;117:109–128. [PMC free article] [PubMed] [Google Scholar]
- 2.Palade GE, Palay SL. Electron microscope observations of interneuronal and neuromuscular synapses. Anat. Record. 1954;118:335–336. [Google Scholar]
- 3.Sollner T, et al. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 4.Hu C, et al. Fusion of cells by flipped SNAREs. Science. 2003;300:1745–1749. doi: 10.1126/science.1084909. [DOI] [PubMed] [Google Scholar]
- 5.Weber T, et al. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 6.McNew JA, et al. Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature. 2000;407:153–159. doi: 10.1038/35025000. [DOI] [PubMed] [Google Scholar]
- 7.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 8.Perin MS, Fried VA, Mignery GA, Jahn R, Sudhof TC. Phospholipid binding by a synaptic vesicle protein homologous to the regulatory region of protein kinase C. Nature. 1990;345:260–263. doi: 10.1038/345260a0. [DOI] [PubMed] [Google Scholar]
- 9.Brose N, Petrenko AG, Sudhof TC, Jahn R. Synaptotagmin: a calcium sensor on the synaptic vesicle surface. Science. 1992;256:1021–1025. doi: 10.1126/science.1589771. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez-Chacon R, et al. Synaptotagmin I functions as a calcium regulator of release probability. Nature. 2001;410:41–49. doi: 10.1038/35065004. [DOI] [PubMed] [Google Scholar]
- 11.Geppert M, et al. Synaptotagmin I: a major Ca2+ sensor for transmitter release at a central synapse. Cell. 1994;79:717–727. doi: 10.1016/0092-8674(94)90556-8. [DOI] [PubMed] [Google Scholar]
- 12.Pang ZP, Shin OH, Meyer AC, Rosenmund C, Sudhof TC. A gain-of-function mutation in synaptotagmin-1 reveals a critical role of Ca2+-dependent soluble N-ethylmaleimide-sensitive factor attachment protein receptor complex binding in synaptic exocytosis. J Neurosci. 2006;26:12556–12565. doi: 10.1523/JNEUROSCI.3804-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domanska MK, Kiessling V, Stein A, Fasshauer D, Tamm LK. Single vesicle millisecond fusion kinetics reveals number of SNARE complexes optimal for fast SNARE-mediated membrane fusion. J Biol Chem. 2009;284:32158–32166. doi: 10.1074/jbc.M109.047381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karatekin E, et al. A fast, single-vesicle fusion assay mimics physiological SNARE requirements. Proc Natl Acad Sci U S A. 2010;107:3517–3521. doi: 10.1073/pnas.0914723107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu T, Tucker WC, Bhalla A, Chapman ER, Weisshaar JC. SNARE-driven, 25-millisecond vesicle fusion in vitro. Biophys J. 2005;89:2458–2472. doi: 10.1529/biophysj.105.062539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishizuka T, Saisu H, Odani S, Abe T. Synaphin: a protein associated with the docking/fusion complex in presynaptic terminals. Biochem Biophys Res Commun. 1995;213:1107–1114. doi: 10.1006/bbrc.1995.2241. [DOI] [PubMed] [Google Scholar]
- 17.McMahon HT, Missler M, Li C, Sudhof TC. Complexins: cytosolic proteins that regulate SNAP receptor function. Cell. 1995;83:111–119. doi: 10.1016/0092-8674(95)90239-2. [DOI] [PubMed] [Google Scholar]
- 18.Giraudo CG, Eng WS, Melia TJ, Rothman JE. A clamping mechanism involved in SNARE-dependent exocytosis. Science. 2006;313:676–680. doi: 10.1126/science.1129450. [DOI] [PubMed] [Google Scholar]
- 19.Giraudo CG, et al. Alternative zippering as an on-off switch for SNARE-mediated fusion. Science. 2009;323:512–516. doi: 10.1126/science.1166500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maximov A, Tang J, Yang X, Pang ZP, Sudhof TC. Complexin controls the force transfer from SNARE complexes to membranes in fusion. Science. 2009;323:516–521. doi: 10.1126/science.1166505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xue M, et al. Tilting the balance between facilitatory and inhibitory functions of mammalian and Drosophila Complexins orchestrates synaptic vesicle exocytosis. Neuron. 2009;64:367–380. doi: 10.1016/j.neuron.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho RW, Song Y, Littleton JT. Comparative analysis of Drosophila and mammalian complexins as fusion clamps and facilitators of neurotransmitter release. Mol Cell Neurosci. 2010 doi: 10.1016/j.mcn.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huntwork S, Littleton JT. A complexin fusion clamp regulates spontaneous neurotransmitter release and synaptic growth. Nat Neurosci. 2007;10:1235–1237. doi: 10.1038/nn1980. [DOI] [PubMed] [Google Scholar]
- 24.Xue M, et al. Distinct domains of complexin I differentially regulate neurotransmitter release. Nat Struct Mol Biol. 2007;14:949–958. doi: 10.1038/nsmb1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hobson RJ, Liu Q, Watanabe S, Jorgensen EM. Complexin Maintains Vesicles in the Primed State in C. elegans. Curr Biol. 2011;21:106–113. doi: 10.1016/j.cub.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin JA, Hu Z, Fenz KM, Fernandez J, Dittman JS. Complexin has opposite effects on two modes of synaptic vesicle fusion. Curr Biol. 2011;21:97–105. doi: 10.1016/j.cub.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sudhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bracher A, Kadlec J, Betz H, Weissenhorn W. X-ray structure of a neuronal complexin-SNARE complex from squid. J Biol Chem. 2002;277:26517–26523. doi: 10.1074/jbc.M203460200. [DOI] [PubMed] [Google Scholar]
- 29.Chen X, et al. Three-dimensional structure of the complexin/SNARE complex. Neuron. 2002;33:397–409. doi: 10.1016/s0896-6273(02)00583-4. [DOI] [PubMed] [Google Scholar]
- 30.Giraudo CG, et al. Distinct domains of complexins bind SNARE complexes and clamp fusion in vitro. J Biol Chem. 2008;283:21211–21219. doi: 10.1074/jbc.M803478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hua SY, Charlton MP. Activity-dependent changes in partial VAMP complexes during neurotransmitter release. Nat Neurosci. 1999;2:1078–1083. doi: 10.1038/16005. [DOI] [PubMed] [Google Scholar]
- 32.Reim K, et al. Complexins regulate a late step in Ca2+-dependent neurotransmitter release. Cell. 2001;104:71–81. doi: 10.1016/s0092-8674(01)00192-1. [DOI] [PubMed] [Google Scholar]
- 33.Tang J, et al. A complexin/synaptotagmin 1 switch controls fast synaptic vesicle exocytosis. Cell. 2006;126:1175–1187. doi: 10.1016/j.cell.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 34.Lu B, Song S, Shin YK. Accessory alpha-helix of complexin I can displace VAMP2 locally in the complexin-SNARE quaternary complex. J Mol Biol. 2010;396:602–609. doi: 10.1016/j.jmb.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melia TJ, et al. Regulation of membrane fusion by the membrane-proximal coil of the t-SNARE during zippering of SNAREpins. J Cell Biol. 2002;158:929–940. doi: 10.1083/jcb.200112081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walter AM, Wiederhold K, Bruns D, Fasshauer D, Sorensen JB. Synaptobrevin N-terminally bound to syntaxin-SNAP-25 defines the primed vesicle state in regulated exocytosis. J Cell Biol. 2010;188:401–413. doi: 10.1083/jcb.200907018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellena JF, et al. Dynamic structure of lipid-bound synaptobrevin suggests a nucleation-propagation mechanism for trans-SNARE complex formation. Proc Natl Acad Sci U S A. 2009;106:20306–20311. doi: 10.1073/pnas.0908317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang X, Kaeser-Woo YJ, Pang ZP, Xu W, Sudhof TC. Complexin clamps asynchronous release by blocking a secondary Ca(2+) sensor via its accessory alpha helix. Neuron. 2010;68:907–920. doi: 10.1016/j.neuron.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pabst S, et al. Selective interaction of complexin with the neuronal SNARE complex. Determination of the binding regions. J Biol Chem. 2000;275:19808–19818. doi: 10.1074/jbc.M002571200. [DOI] [PubMed] [Google Scholar]
- 40.Krishnakumar SS, et al. A Conformational Switch in Complexin is Required for Synaptotagmin to Trigger Synaptic Fusion. Nat Struct Mol Biol. 2011 doi: 10.1038/nsmb.2103. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuzmin PI, Zimmerberg J, Chizmadzhev YA, Cohen FS. A quantitative model for membrane fusion based on low-energy intermediates. Proc Natl Acad Sci U S A. 2001;98:7235–7240. doi: 10.1073/pnas.121191898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chernomordik LV, Zimmerberg J, Kozlov MM. Membranes of the world unite! J Cell Biol. 2006;175:201–207. doi: 10.1083/jcb.200607083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xue M, et al. Binding of the complexin N terminus to the SNARE complex potentiates synaptic-vesicle fusogenicity. Nat Struct Mol Biol. 2010;17:568–575. doi: 10.1038/nsmb.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi UB, et al. Single-molecule FRET-derived model of the synaptotagmin 1-SNARE fusion complex. Nat Struct Mol Biol. 2010;17:318–324. doi: 10.1038/nsmb.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chicka MC, Hui E, Liu H, Chapman ER. Synaptotagmin arrests the SNARE complex before triggering fast, efficient membrane fusion in response to Ca2+ Nat Struct Mol Biol. 2008;15:827–835. doi: 10.1038/nsmb.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doublie S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 48.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CW Jr, Sweet RM, editors. Methods in Enzymology. Vol. 276. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 49.McCoy AJ, et al. Phaser Crystallography Software. J. Appl. Chrystallog. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 52.Kleywegt GJ, Jones TA. Where freedom is given, liberties are taken. Structure. 1995;3:535–540. doi: 10.1016/s0969-2126(01)00187-3. [DOI] [PubMed] [Google Scholar]
- 53.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 54.Lakowicz JR. Principles of fluorescence spectroscopy. xxvi. New York ; Berlin: Springer; 2006. p. 954. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.