Abstract
Purpose.
We analyzed the change in protein expression of tear film proteins in dry eye (DE) and non-DE (NDE) patients using isobaric tag for relative and absolute quantitation (iTRAQ) technology.
Methods.
We categorized 24 participants into NDE, and mild (MDE), moderate-to-severe (MSDE), and mixed (MXDE) DE on the basis of clinical DE tests. Tear samples (n = 6 subjects/group) were collected using Schirmer's strips. Proteins were extracted from strips and were quantified using the Bradford assay. Protein from each sample was pooled as internal standard (IS), and 20 μg protein from each sample and the IS were digested and labeled with different tandem mass tag (TMT) isobaric mass tag labeling reagent. The reaction was quenched and the labeled peptides were mixed. Samples were injected for liquid chromatography-mass spectrometry (LC/MS/MS) analysis on the Orbitrap mass spectrometer. Bioinformatic analyses were performed using protein information resource (PIR).
Results.
Combined results showed a total of 386 proteins in tears as determined by the iTRAQ experiments. An average of 163 proteins was detected in each of 6 biologic replicates. Of those, 55% were detected 6 times and 90% were detected multiple times (>2). In addition to the down-regulation of commonly reported proteins, such as lipocalin-1, lysozyme, and prolactin-inducible protein across all sub groups of DE, a number of proteins were significantly differentially regulated in MSDE and other subgroups of DE. A greater number of proteins were down-regulated in MSDE versus MDE, and the specific functions involved include response to stimulus (8 vs. 6 proteins), immune system process (6 vs. 4), regulation of biologic processes (3 vs. 3), and ion transport (2 vs. 2).
Conclusions.
iTRAQ is one of the newest tools for quantitative mass spectrometry in tear proteome research. Differences in the protein ratios can be detected between normal and DE patients. PIR is a useful resource to interpret pathways and functions of proteins.
This study reports the proteins identified in tears of different sub groups of dry eye patients by using a novel iTRAQ method of quantitative proteomics. This study showed greater number of proteins down-regulated in moderate to severe dry eye group than other sub groups of dry eye. Unique proteins were associated with each sub group of dry eye. iTRAQ technology is a relatively new protein quantification method, especially in tear film which allows the analysis of multiple samples to be obtained simultaneously.
Introduction
The Dry Eye Workshop in 2007 defined dry eye (DE) as a multifactorial ocular surface disease diagnosed by symptoms of discomfort, and signs of visual disturbance, tear film instability, and ocular surface damage, accompanied by increased osmolarity of the tear film and ocular surface inflammation.1 It is evident from the definition that the tear film and ocular surface are altered in DE disease. The tear film serves/performs a variety of functions and is composed of various substances, including proteins, lipids, mucins, salts, and other organic molecules.2 The aqueous component constitutes the majority of the tear layer, and the proteins in the tear film are believed to have a key role in the protection of the external surface from potential pathogens, and also are involved in modulation of wound healing process.3–6
The major source of tear proteins is the secretory acinar cells of the lacrimal gland, including the primary proteins of the tear film: lysozyme, lactoferrin, and lipocalin.7 Numerous proteins have been identified previously in human tears; however, there is an inconsistency in the number of proteins in tears and their specific functions in the existing literature.8 Studies conducted by Gachon et al. a few decades ago identified 60 proteins in the normal tears.9 More recent studies on tear proteins have shown the presence of approximately 500 proteins.8,10–12 To date, limited studies have been performed in which the proteome of abnormal tears, such as in DE disease, have been evaluated and even more so with newer techniques, such as isobaric tag for relative and absolute quantitation (iTRAQ).13
Differential regulation of inflammatory proteins in the tear film has been evaluated using a variety of techniques in several ocular surface conditions, such as meibomian gland dysfunction,14 DE,15 Sjögren's syndrome,13,16–18 and wound healing processes of the ocular surface.6,19 Versura et al., in a recent study, showed that the tear protein changes anticipate the onset of more extensive clinical signs in early stage DE disease.20 Changes of tear protein profile also have been shown to correlate with DE severity,13 and the levels of certain proteins have been correlated with the severity of meibomian gland disease.14
Various factors can influence tear proteomics, including the tear collection methods,7,21–23 storage, and analysis techniques.8 Additional other factors include age of the person,24 severity of dryness in the ocular surface, and contact lens wear.25,26 A recent study conducted in our laboratory demonstrated higher protein concentration with Schirmer's strip collection in comparison with capillary tear collection methods.8 This is important critically in analyses requiring a greater amount of protein, and Schirmer's technique, while providing more protein for analysis, may demonstrate a different protein profile than capillary tears, and should be taken into consideration when proteomic studies are compared.8 Schirmer's tear collection has shown to be a reliable method of tear collection in patients with DE, with the ability to demonstrate the differential protein expression and, hence, in biomarker identification.27
Relative expression quantification of protein is a key aspect of proteomic experiments. Several techniques, such as differential in gel electrophoresis (DIGE),28,29 stable isotope labeling with amino acids in cell culture (SILAC),30,31 isotope-codes affinity tag (iCAT),32,33 iTRAQ,34,35 and absolute protein quantitation (AQUA),36,37 are available for this type of protein identification and quantitation. These techniques offer relative or absolute quantitation by running samples from different sources (control versus tested) in the same gel or mass spectrometric run. Therefore, technical variations caused during sample preparation are minimized and accurate quantitation results are obtained. Among them, iTRAQ uses isotope coded covalent tags that specifically label the N-terminus and side chain amines of peptides from protein digestions. Using iTRAQ, every peptide in the sample is labeled. Therefore, better protein identification and quantitation can be expected. In addition, the iTRAQ method has the ability to compare up to 8 samples in one single experiment. Hence, iTRAQ is useful especially when multiple experimental conditions are studied.
To our knowledge, this is the first study to explore the tear proteomics in different subgroups of DE in human tear film using the iTRAQ quantitative proteomic technique with liquid chromatography-mass spectrometry (LC/MS/MS) analysis on an LTQ Orbitrap mass spectrometer. The aim of our study was to analyze the relative expression of tear film proteins in DE and to interpret the functions of tear proteins in DE patients using protein information resource (PIR) following iTRAQ quantitative proteomics.
Methods
Participants
Approval of this project was granted through the Institutional Review Board at the Ohio State University, and all procedures adhered to the tenets of the Declaration of Helsinki. Participants were recruited and examined at the Ohio State University, College of Optometry. Informed consent was obtained from all participants following explanation of purpose and procedures.
A total of 24 participants (mean age 42.83 ± 16.92 years, range 23–80 years) was recruited for a single visit study of diagnostic tests in patients suspected to have aqueous deficient DE. Schirmer strips were used to measure aqueous production, and the analysis of the tears from the strips is presented. Of the participants, 58% were females (14 of 24), 92% were Caucasians (22 of 24), and 8% were of Asian origin (2 of 24). Contact lens wearers were excluded from the study. Participants were not diabetic and did not use antidepressants or any antiglaucoma medications. Participants who had undergone any corneal surgery (including LASIK or photorefractive keratectomy [PRK]) and who had diagnosed or suspected Sjögren's syndrome or other autoimmune diseases also were excluded from the study.
Participants completed the validated 12-item Allergan Ocular Surface Disease Index (OSDI) questionnaire, which has been described in detail previously.38 Broadly, the OSDI questionnaire includes 12 questions. The OSDI scoring is based on a 0 to 100 scale, with the highest score representing greater disability. The OSDI questionnaire responses are graded on a scale from 0 to 4, The following formula is used to find the total OSDI score: OSDI = (sum of scores for all questions answered × 100)/(total number of questions answered).38,39 Schirmer's test was performed and scores were recorded. These results also were used, post examination, for participant categorization purposes.
Participant Categorization
Based on symptoms scores and Schirmer's wetting length, participants were categorized into one of the four groups (6 subjects/group), non-DE (NDE), and mild (MDE), moderate–to-severe (MSDE), and mixed (MXDE) DE as described in Table 1. Patient groupings were performed after clinical examinations were complete. While the groupings are not consistent completely with clinical grading schemes and protocols, clustering of these subjects can be described best in four groups. Participants in the NDE group had no symptoms of dryness as recorded in the questionnaire (Table 2) and displayed no significant signs of dryness. MDE participants were mildly symptomatic with aqueous deficiency. MSDE patients were symptomatic of DE and showed aqueous deficiency. MXDE participants presented with variable symptoms and showed a range of Schirmer's scores as described in Table 1. Clinicians often encounter patients who show variable signs in the presence or absence of symptoms and vice versa. For the purposes of research, to have a well-defined group of subjects, this “no correlation” group often is ignored by scientists or is considered as “screen failure.” It is logical to study well-defined groups; however, the MDE group (combination group) exists as they may represent a group faced frequently by clinicians. Also, of significant note, assessment of several clinical components, such as a thorough meibomian gland assessment, was not part of the original study design and could not be incorporated into the groupings. This study was exploratory in nature, and future exploratory and confirmatory studies could use this as well as other grouping schemes.
Table 1. .
Characteristics of Study Patients
|
Groups (n = 6 for Each Group) |
Demographics (Age Expressed as Mean ± SD) |
Schirmer's Scores* |
OSDI Score |
| NDE—No symptoms and sign | Age 29.83 ± 8.10 y (range 24–45) | ≥16 mm | ≤12 |
| M = 2; F = 4 | |||
| MDE—Mildly symptomatic with aqueous deficiency | Age 59.67 ± 15.98 y (range 33–80) | ≤5 mm | ≤23 |
| M = 2; F = 4 | |||
| MSDE—Symptomatic aqueous deficiency | Age 45.17 ± 10.53 y (range 29–58) | ≤5 mm | ≥24 |
| M = 2; F = 4 | |||
| MXDE—Combination group | Age: 36.67 ± 17.02 y (range 23–70) | 6–15 mm | Variable |
| M = 4; F = 2 |
Schirmer's test was the primary criterion for participant categorization and Schirmer values override the OSDI scores in case of conflict in scores.
Table 2. .
Questionnaire and Schirmer's Data of Study Patients
|
|
Schirmer's Test (Wetting Length in mm) Mean ± SD |
OSDI (Scores) Mean ± SD |
| NDE group | 23.16 ± 6.43 | 2.16 ± 4.02 |
| MDE group | 2.833 ± 0.98 | 11.83 ± 8.84 |
| MSDE group | 2.50 ± 1.57 | 36.66 ± 9.79 |
| MXDE group | 11.16 ± 3.18 | 17.83 ± 18.69 |
Tear Sampling
During the study visit, Schirmer's test was performed by placing a strip over the lower lid and the tears collected during this test were used towards tear proteomics. The strip was placed approximately 6 mm from the lateral canthal region of the lower lid. The subject was instructed to close his/her eyes for the 5-minute test duration, and Schirmer's value was recorded by reading the wetting length in millimeters directly from the strip. Schirmer's test was conducted without the administration of anesthetic drops. The strip then was placed in a 1.6 mL Eppendorf tube on ice, and stored at −80°C until further analysis. Gloves were worn by the examiner during the tear collection procedure and by the investigators handling the samples in the laboratory.
Schirmer's Strip Extraction
The Schirmer strips were placed in the spin filter (Part number 5185-5990; Agilent Technologies, Santa Clara, CA) and 200 μL of extraction buffer were added (100 mM triethylammonium bicarbonate [TEAB] with 0.05% ProteaseMAX) to the strip. The strip was pushed down to the bottom of the filter to make sure it was soaked completely in the solution. The filter was put back into a clean tube. The tube was placed on a shaker for 2 hours on ice and then centrifuged at 13,000 revolutions per minute (rpm) for 5 minutes. Another extraction was performed with additional 200 μL of the extraction buffer and the extracted solution was pooled together for quantitation.
Total Protein Quantification
Bradford quantitation was performed using BSA solution as standards. Briefly, Bio-Rad protein assay dye reagent concentrate (catalog number #500-0006; Bio-Rad, Hercules, CA) was diluted 5 times. Each of the 10 μL of the BSA standard solution with concentrations of 0, 50, 100, 150, 200, and 250 μg/mL, and the extracted samples were added to 190 μL of the diluted dye, respectively. Three replicates were used for each standard solution, while 2 replicates were used for the samples. The solution then was mixed and incubated at room temperature (RT) for 20 minutes. The absorbance then was measured at 595 nm. The concentration of each sample was calculated based on the standard curve.
iTRAQ Experiment
For the iTRAQ analysis, 11 μg of protein from each sample was pooled together as an internal standard (IS). Then, 20 μg protein from each sample, including the IS, were reduced, alkylated, and digested with trypsin (enzyme:substrate 1:20) for overnight at 37°C. The peptides then were labeled using TMT isobaric mass tagging labeling reagent (Thermo Scientific, West Palm Beach, FL) for 2 hours at room temperature before quenching with 8% hydroxylamine buffer. The samples were grouped, labeled, and then mixed. The IS group was labeled with labeling reagent 126 to facilitate MASCOT ratio calculation, while the other groups were labeled randomly to eliminate the possible labeling preference.
Each sample was concentrated to a final concentration of approximately 1 μg/μL and 1.5 μL of each sample were injected for LC/MS/MS analysis on the LTQ-Orbitrap-XL mass spectrometer (Thermo Scientific, West Palm Beach, FL) equipped with a microspray source (Michrom Bioresources Inc., Auburn, CA) operated in positive ion mode. Samples were separated on a capillary column (0.2 × 150 mm Magic C18AQ 3 μ 200A; Michrom Bioresources Inc.) using an UltiMate 3000 HPLC system from LC-Packings, A Dionex Co. (Sunnyvale, CA). Each sample was injected into the μ-Precolumn Cartridge (Dionex) and desalted with 50 mM acetic acid for 10 minutes. The injector port then was switched to inject and the peptides were eluted off of the trap onto the column. Mobile phase A was 0.1% formic acid in water and 0.1% formic acid in acetonitrile was used as mobile phase B. Flow rate was set at 2 μL/min. Typically, mobile phase B was increased from 2% to 50% in 140 minutes. Mobile B then was increased from 50% to 90% in 5 minutes and then kept at 90% for another 5 minutes before being brought back quickly to 2% in 1 minute. The column was equilibrated at 2% of mobile phase B (or 98% A) for 30 minutes before the next sample injection. MS/MS data were acquired with a spray voltage of 2 KV and a capillary temperature of 175°C was used. The scan sequence of the mass spectrometer was based on the data dependent TopTen method: the analysis was programmed for a full scan recorded at 300 to 2000 Da and a MS/MS scan to generate product ion spectra to determine amino acid sequence in consecutive scans of the 10 most abundant peaks in the spectrum. Cytokines are low abundance proteins occurring at pg/mL levels, and low molecular weight proteins with high turnover that are directed better by antibody arrays. Labeling methods, such as iTRAQ, require nanogram levels of protein for successful tagging and quantification of relative abundance. The resolution of full scan was set at 30,000 to achieve high mass accuracy MS determination. The collision-induced dissociation (CID) fragmentation energy was set to 35%. Dynamic exclusion is enabled with a repeat count of 30 seconds, exclusion duration of 350 seconds, and a low mass width of 0.50 and high mass width of 1.50 Da. Multiple MS/MS detection of the same peptide was excluded after detecting it three times.
Sequence information from the MS/MS data was processed by converting the raw files (.raw) into a merged file (.mgf) using an in-house program, RAW2MZXML_n_MGF_batch (merge.pl, a Perl script). The resulting .mgf files were searched using Mascot Daemon by Matrix Science version 2.2.2 (Boston, MA) and the database searched against the full SwissProt human database. The mass tolerance of the precursor ions was set to 1.2 Da to include the accidental pick of 13C peaks and the fragment mass tolerance was set to 0.8 Da. Considered modifications (variable) were methionine oxidation and carbamidomethyl cysteine. Three missed cleavages for the enzyme were permitted. The significance identity threshold was set at P < 0.05 for valid protein identification. A decoy database was searched to determine the false discovery rate (FDR). Peptides were filtered according to the FDR, which is less than 0.3% for peptide that matches above identity threshold for all the groups. Proteins with a MASCOT Mowse score of 40 and higher were accepted. The valid identification also requires the presence of only bold red peptides. (A bold red match is the highest scoring match to a particular query listed under the highest scoring protein containing that match. This means that protein hits with many peptide matches that are bold and red are the most likely assignments.) Protein exchange ratios between clinical groups and internal standard were obtained from MASCOT search. Briefly, the ratio of a protein is calculated based on the ratios obtained from the peptides generated from this protein. Only unique peptides identified for the protein are used for the calculation and a minimal of 2 peptides with detectable ratios is required for protein ratio calculation. Outliers were filtered out automatically. The ratio between DE groups and the normal group was calculated as the ratio between DE/IS and normal/IS. Average ratios were used for calculation and statistics to reflect the true ratio between DE and normal groups.
The bioinformatics software was used specifically to analyze iTRAQ data. As such, patient-specific data (i.e., age and sex) were not included in the bioinformatics analysis in this small sample pilot study. Likewise, the P values have not been adjusted for multiple comparisons, as would be necessary in future studies.
Bioinformatics
The functions of the proteins identified by the iTRAQ technique were interpreted using the PIR (information available online at http://pir.georgetown.edu/pirwww/search/). PIR was used on differentially expressed protein within each DE category. Protein IDs were mapped to the iProClass database, and sorted by their biologic processes and molecular functions using gene ontology (GO) annotation.40 iProClass provides reports for all UniProt sequences linked to over 90 biologic databases.
Statistics
Only ratios detected in all six biologic replicates were used as “definite values” for statistics calculation. First, the average, SD, and interquartile range for each group were calculated. For each protein with a detectable ratio, the P value was calculated by comparing the actual ratio (DE/control) of that protein with 1. Ratios with a P < 0.05 were considered as significant. In this experiment, proteins with ratios (DE/control) less than 0.6 were considered underexpressed and proteins with ratios (DE/control) larger than 1.6 were considered overexpressed.
Results
Combined results showed a total of 386 proteins in tears as determined by the iTRAQ experiments. An average of 163 proteins was detected in each of the six biologic replicates. Of those, 55% were detected six times and 90% were detected multiple times (>2). The MSDE group showed greater number of down-regulated proteins (18 proteins) than the MDE (15 proteins) and MXDE (10 proteins) groups. Down-regulation of proteins across all subgroups of DE is shown in Tables 3–5. Ratios ranging from 0.6 to 0.7 and 1.5 to 1.6 in the down-regulated and upregulated groups, respectively, also are indicated in Tables 3–5 if the P values are significant.
Table 3. .
Upregulated and Down-Regulated Proteins in MSDE Patients versus NDE Patients
|
Upregulated (n of Detections = 6) |
Average Ratio |
SD |
P Value |
Down-Regulated (n of Detections = 6) |
Average Ratio |
SD |
P Value |
| Apolipoprotein A* | 1.747 | 0.923 | 0.0020 | Cystatin-S | 0.442 | 0.118 | 0.0001 |
| Ezrin* | 1.880 | 0.645 | 0.0206 | Ig alpha-1 chain C* | 0.614 | 0.207 | 0.0060 |
| Ig gamma-3 chain C region* | 2.467 | 1.510 | 0.0028 | Ig alpha-2 chain C* | 0.612 | 0.215 | 0.0069 |
| Vitamin D–binding protein* | 2.045 | 0.954 | 0.0043 | Immunoglobulin J chain | 0.551 | 0.171 | 0.0013 |
| Peroxiredoxin | 1.535 | 0.442 | 0.0314 | Ig lambda chain C regions | 0.613 | 0.154 | 0.0017 |
| Extracellular glycoprotein lacritin | 0.242 | 0.090 | <0.0001 | ||||
| Putative lipocalin 1-like protein | 0.395 | 0.092 | <0.0001 | ||||
| Lipocalin-1 | 0.337 | 0.103 | <0.0001 | ||||
| Lysozyme | 0.324 | 0.175 | 0.0002 | ||||
| Polymeric immunoglobulin receptor | 0.455 | 0.148 | 0.0003 | ||||
| Prolactin-inducible protein-1 | 0.396 | 0.117 | 0.0001 | ||||
| Proline-rich protein-1* | 0.431 | 0.171 | 0.0005 | ||||
| Proline-rich protein-4 | 0.393 | 0.205 | 0.008 | ||||
| Secretoglobin family | 0.229 | 0.121 | <0.0001 | ||||
| Mammaglobin-B | 0.280 | 0.116 | <0.0001 | ||||
| Lactotransferrin | 0.394 | 0.234 | 0.0014 | ||||
| Zinc-alpha-2-glycoprotein | 0.390 | 0.163 | 0.0003 | ||||
| Zymogen granule protein-16* | 0.537 | 0.354 | 0.0239 |
Represents unique IDs.
Table 5. .
Upregulated and Down-Regulated Proteins in MXDE versus NDE Patients
|
Upregulated (n of Detections = 6) |
Average Ratio |
SD |
P Value |
Down-Regulated (n of Detections = 6) |
Average Ratio |
SD |
P Value |
| Ezrin | 1.551 | 0.508 | 0.0450 | Cystatin-S | 0.640 | 0.237 | 0.0137 |
| Ig lambda chain C regions | 0.687 | 0.169 | 0.0062 | ||||
| Extracellular glycoprotein lacritin | 0.556 | 0.263 | 0.0091 | ||||
| Putative lipocalin 1-like protein | 0.538 | 0.220 | 0.0036 | ||||
| Lipocalin-1 | 0.498 | 0.284 | 0.0075 | ||||
| Prolactin-inducible protein-1 | 0.639 | 0.276 | 0.0248 | ||||
| Polymeric immunoglobulin receptor | 0.648 | 0.335 | 0.0495 | ||||
| Secretoglobin family | 0.466 | 0.230 | 0.0023 | ||||
| Mammaglobin-B | 0.502 | 0.299 | 0.0095 | ||||
| Zinc-alpha-2-glycoprotein | 0.648 | 0.288 | 0.0303 |
Table 4. .
Upregulated and Down-Regulated Proteins in MDE versus NDE Patients
|
Upregulated (n of Detections = 6) |
Average Ratio |
SD |
P Value |
Down-Regulated (n of Detections = 6) |
Average Ratio |
SD |
P Value |
| Aldehyde dehydrogenase* | 2.585 | 1.121 | 0.0146 | Cystatin-S | 0.528 | 0.160 | 0.0008 |
| Haptoglobin* | 2.264 | 1.117 | 0.0393 | Immunoglobulin J chain | 0.661 | 0.261 | 0.0245 |
| Complement C3 | 1.355 | 0.2882 | 0.0296 | Ig lambda chain C regions | 0.665 | 0.120 | 0.0010 |
| Extracellular glycoprotein lacritin | 0.321 | 0.153 | 0.0001 | ||||
| Putative lipocalin 1-like protein | 0.459 | 0.185 | 0.0008 | ||||
| Lipocalin-1 | 0.364 | 0.195 | 0.0005 | ||||
| Lysozyme | 0.357 | 0.164 | 0.0002 | ||||
| Polymeric immunoglobulin receptor | 0.584 | 0.371 | 0.0404 | ||||
| Prolactin-inducible protein-1 | 0.371 | 0.109 | <0.0001 | ||||
| Proline-rich protein-4 | 0.467 | 0.179 | 0.0008 | ||||
| Secretoglobin family | 0.446 | 0.172 | 0.0005 | ||||
| Mammaglobin-B | 0.324 | 0.170 | 0.0002 | ||||
| Transcobalamin-1* | 0.389 | 0.196 | 0.0006 | ||||
| Lactotransferrin | 0.364 | 0.163 | 0.0002 | ||||
| Zinc-alpha-2-glycoprotein | 0.430 | 0.162 | 0.0004 |
Represents unique IDs.
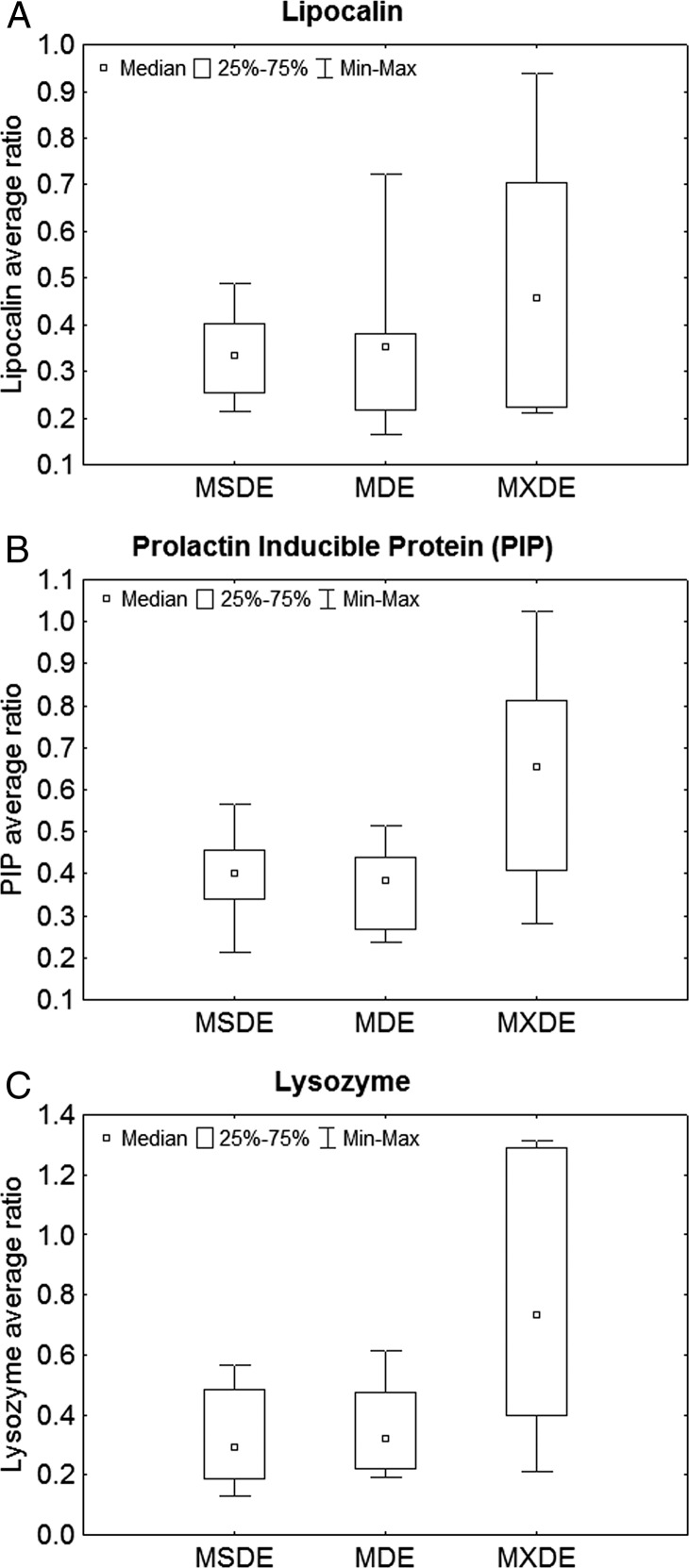
The average, SD, and interquartile range for commonly down-regulated proteins, such as lipocalin, prolactin inducible protein (PIP), and lysozyme, for each of the DE subgroups are shown in Figures 1A–1C. These Figures show trends of decrease in average protein ratios in MSDE in comparison with the other subgroups. Few proteins were found to be upregulated in MDE and MSDE (Tables 3–5). Although 90% of the proteins were detected multiple times, these Tables report the proteins detected all six times during the experiment. The unique proteins IDs that are upregulated and down-regulated in MSDE and MDE are highlighted by an asterisk in Tables 3–5. An example showing the spectrum for peptides labeled with TMT isobaric mass tag labeling reagent is represented in Figure 2.
Figure 1. .
(A–C) Box plot of the median ratios with the interquartile ranges of commonly down-regulated proteins, such as (A) lipocalin, (B) PIP, and (C) lysozyme in MSDE, MDE, and MXDE groups. Inner boxes represent median. Outer box represents the 25% to 75% ranges. Whiskers represent minimum–maximum values.
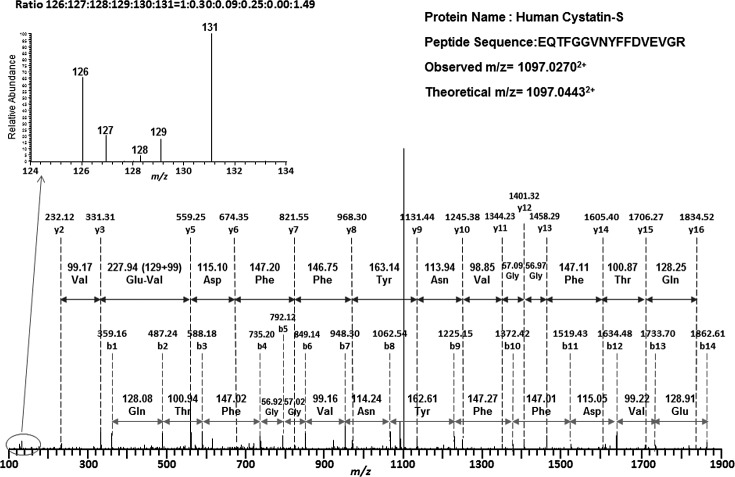
Figure 2. .
An example spectrum for peptides labeled with TMT isobaric mass tag labeling reagent. The MSMS fragmentations were used to sequence the peptide. Based on the amino acid ladder, the peptide was identified as EQTFGGVNYFFDVEVGR with the N-terminus modified by TMT isobaric mass tag labeling reagent. This peptide belongs to human cystatin-S (CTYS). Mass tags (126–131) observed in the lower m/z region (inserted figure) indicate the relative abundance of this peptide in each group. The relative intensity (ratios) was normalized considering peak 126 as the base peak. The samples were labeled in the following order: IS (126), NDE (131), MDE (128), MSDE (127), and Mix (129). Therefore, the ratios suggested that CTYS was down-regulated in the diseased groups.
Ezrin, apolipoprotein, Ig gamma-3 chain C region, vitamin D-binding protein, and peroxiridoxin were detected to be upregulated in MSDE. The unique protein identifications of upregulated proteins in MDE were aldehyde dehydrogenase and haptoglobin. Ezrin was found to be upregulated in the MXDE group.
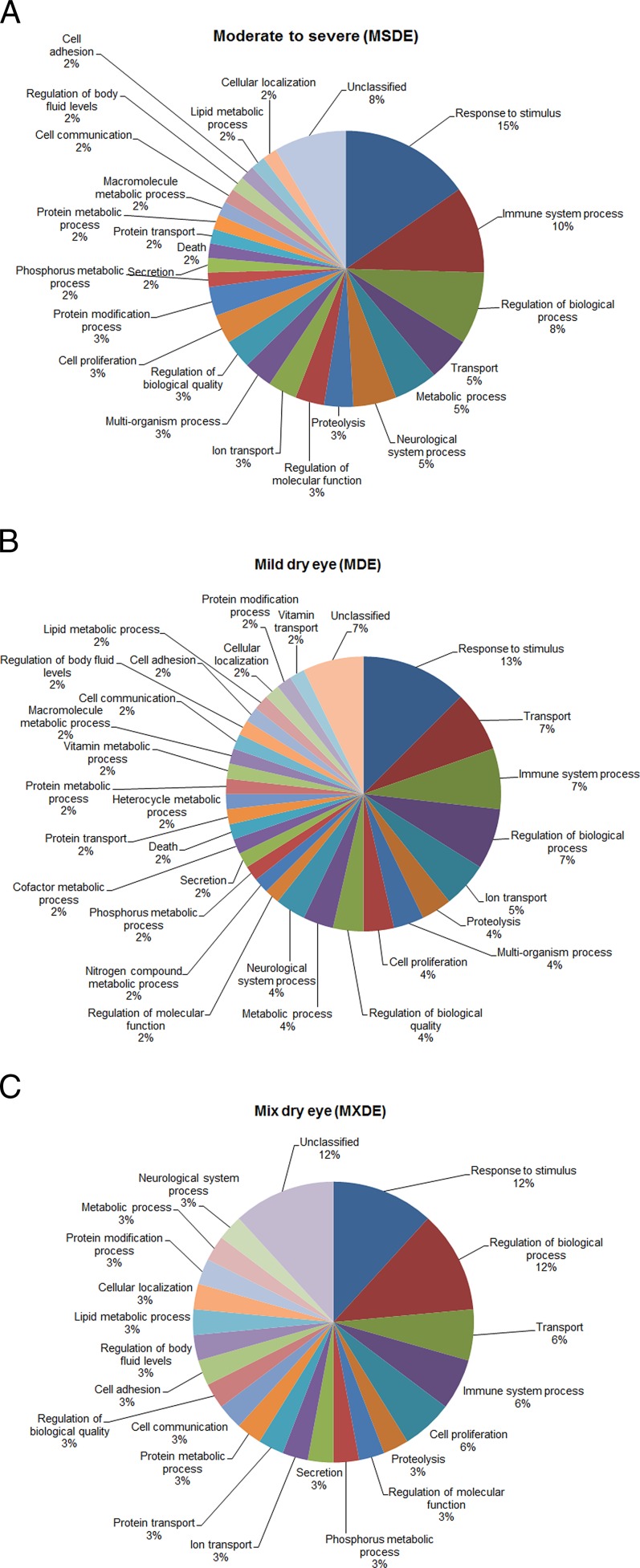
By GO analysis, all five upregulated proteins in the MSDE category were represented in the “response to stimulus process” with apolipoprotein involved in “cytokine production,” and peroxiredoxin associated with “lipid metabolic process” and “oxidation-reduction process.” Similarly in the MDE category, all upregulated proteins were represented in the “response to stimulus” with complement C3 involved in “cytokine production.” The distribution of down-regulated proteins based on their cellular processes is presented in Figures 3A–3C. The most represented biologic process terms were related to response to stimulus (15% in MSDE, 13% in MDE, 12% in MXDE), followed by immune system process, regulation of biologic process, and transport. Down-regulation of commonly reported tear proteins (lysozyme, lipocalin-1, lactotransferrin, IgA-alpha) in all subgroups of DE indicates there is an impairment of regulation, transportation, and immune system response in DE conditions.
Figure 3. .
GO analysis representing the distribution (percentage of occurrences) of the biologic processes of down-regulated proteins. (A) The MSDE category. (B) The MDE category. (C) The MXDE category.
Discussion
Our study reports the proteins identified in tears of different subgroups of DE patients by using a novel method of quantitative proteomics. Statistically significant differences in the protein ratios were detected between normal and DE groups. Our study showed a greater number of proteins down-regulated in MSDE than other DE groups (Table 3). Unique proteins were associated with each subgroup of DE. iTRAQ technology is a relatively new protein quantification method, especially in tear film, that allows the analysis of multiple samples to be obtained simultaneously.
To date, there are very few studies in the literature on tears proteomics in humans using this novel quantitative proteomic method. Zhou et al.13 and Tong et al.14 have employed iTRAQ technique to study proteins in human tears. Zhou et al. identified tear biomarkers in DE patients in a recent study that demonstrated 4 down-regulated proteins (PIP, lipocalin-1, lactoferrin, and lysozyme) in DE patients.13 These results are comparable to our study in which three (PIP, lipocalin-1, and lysozyme) of the four proteins were detected. However, the number of upregulated proteins is different in both studies.
A comparison of defensin (NP-1 and NP-2), levels in tear film in rabbits before and after corneal wounding (days 1–3) was conducted by Zhou et al. with iTRAQ experiments.19 Similarly, iTRAQ also has been used to quantify the relative difference in tear protein N-glycosylation levels between controls and patients with climatic droplet keratopathy.41 Tong et al. concluded that unique tear proteins (S100A8 and S100A9) are associated with meibomian gland dysfunction (MGD) in DE patients, and the level of certain proteins, such as S100A8, S100A9, and lipocalin-1, correlated significantly with grittiness, and symptoms of redness, transient blurring, heaviness of the eyelids, and tearing, respectively.14 Our results need exploration to correlate individual protein levels with specific symptomatology of DE.
Mass spectrometry analysis by Versura P et al., conducted in tear samples of 60 evaporative DE patients, detected the following proteins in tears, including lactotransferrin, serum albumin, lipocalin-1, lipophilin-c, extracellular glycoprotein lacritin, proline-rich protein-4, lysozyme C, lipophilin-a, prolactin-inducible protein, proline-rich protein-1, Ig gamma-1 chain C region, serotransferrin, and zinc-α-2-glycoprotein.20 A statistically significant decrease in lactoferrin, lipophilin, and lipocalin amount was found in patients versus controls; however, our study did not show a change in lysozyme and zinc-alpha-2-glycoprotein amounts. Tear proteomics on contact lens wearers,42 Sjögren's syndrome patients,13,16–18 and keratoconus patients43,44 have shown upregulation and down-regulation of unique proteins.
Our study has shown two upregulated proteins, namely aldehyde dehydrogenase and haptoglobin, in the MDE group. These two upregulated proteins did not appear in the MSDE group. At this stage, it is unclear as to why there was an upregulation noted only in the MDE group. Interestingly, a recent report by Joseph et al. examined epithelial and stromal proteins from keratoconus and normal cornea using label-free Nano-ESI-LC MS (MS)2, and showed that there was an upregulation of aldehyde dehydrogenase and haptoglobin.45
There are mixed views in the literature on the levels of certain proteins in different age groups. For upregulated tear proteins, Zhou et al. reported that higher levels of proteins, such as S100A8 and S100A9, were associated significantly with increased signs of dryness,13 and this association still remained in all age groups. No significant correlation was found for the rest of the tear biomarker candidates that were assessed. A previous study reported that, due to the reduction in secretion function of the lacrimal glands in the elderly, there was a negative correlation between age and some tear protein levels, such as lysozyme and lactoferrin.24 However, another recent study conducted by Zhou et al. showed that age correlation assessments did not demonstrate significant association between age, and the levels of lysozyme and lactoferrin.13
The biologic functions of commonly reported down-regulated tear proteins in DE, such as lysozyme, lactotransferrin, and lipocalin, include antibacterial protection of the ocular surface2 and general protection factor of epithelial cell surfaces, respectively.46 PIP was found to be decreased in patients with MGD, which is involved in water transport function.14 A recently conducted study in Sjögren's syndrome tears also showed a few similar proteins, which were detected in our current study, including Ig alpha-2 chain C, Ig alpha-1 chain C, zinc-alpha-2 glycoprotein, and polymeric-immunoglobulin.47 The determination of biologic functions of other proteins highlighted in results in relation to DE needs further research.
In our study, several proteins were identified multiple times (greater than 2 times, but less than 6 times); however, these proteins were not reported here, which may be of relevance. Differential protein expression can be detected between normal and DE patients using iTRAQ.
The sample size of our study was small and warrants further work to show consistency in the number of proteins expressed, and the ratio of upregulated and down-regulated proteins in larger samples. Tear collection methods, patient grouping techniques, and tear processing techniques have a major role in tear proteomics. In our study, a full battery of DE diagnostic tests was not performed, thus common DE diagnostic elements, aqueous production, and symptoms, were used to classify patients. Our study was exploratory in nature, and future exploratory and confirmatory studies could use this as well as other grouping schemes, including additional tests, such as the evaluation of the status of the meibomian glands. Schirmer's test is particularly problematic and variable, and may be influenced by reflex tearing to some extent, so its value does not reflect merely basal aqueous production. In this classification, this fact is particularly relevant as some of the mixed category may have the measured Schirmer's partly as a result of reflex tearing.
Consistent approaches to tear collection and analysis additionally are required. There also is a need for the standardization of the number of replicates while performing tear proteomics. Research data obtained using new proteomics tools, such as iTRAQ, must be validated with comprehensive bioinformatic tools, such as PIR. The use of protein ID mapping supports the functional inference of down-regulated or upregulated proteins in relation to the severity of DE. Continued work to characterize involved pathways is needed. Future work to validate findings using Western blot and ELISA assays are planned.
Acknowledgments
This work was conducted at The Ohio State University, College of Optometry, Columbus, Ohio. The authors have no proprietary interest in any of the products mentioned in this manuscript.
Footnotes
Presented at the annual meetings of the Association for Research in Vision and Ophthalmology, May 2–6, 2010, Fort Lauderdale, Florida, and the American Academy of Optometry, November 17–20, 2010, San Francisco, California.
Supported by National Institutes of Health Grant NIH R01 EY015519 (KN, KGB).
Disclosure: S. Srinivasan, None; M. Thangavelu, None; L. Zhang, None; K.B. Green, None; K.K. Nichols, None
References
- 1.Report of the 2007 International Dry Eye WorkShop (DEWS) Glossary. Ocul Surf. 2007;5:73–74 [Google Scholar]
- 2.Donald RK, Jennifer C, Michael D, Jean-Pierre G, George S, Alan T. The Tear Film. Oxford: Butterworth-Heinemann; 2002:51–81 [Google Scholar]
- 3.Vesaluoma M, Teppo AM, Gronhagen-Riska C, Tervo T. Platelet-derived growth factor-BB (PDGF-BB) in tear fluid: a potential modulator of corneal wound healing following photorefractive keratectomy. Curr Eye Res. 1997;16:825–831 [DOI] [PubMed] [Google Scholar]
- 4.Tervo T, Vesaluoma M, Bennett GL, et al. Tear hepatocyte growth factor (HGF) availability increases markedly after excimer laser surface ablation. Exp Eye Res. 1997;64:501–504 [DOI] [PubMed] [Google Scholar]
- 5.Lembach M, Linenberg C, Sathe S, et al. Effect of external ocular surgery and mode of post-operative care on plasminogen, plasmin, angiostatins and alpha(2)-macroglobulin in tears. Curr Eye Res. 2001;22:286–294 [DOI] [PubMed] [Google Scholar]
- 6.Zhou L, Beuerman RW, Barathi A, Tan D. Analysis of rabbit tear proteins by high-pressure liquid chromatography/electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom. 2003;17:401–412 [DOI] [PubMed] [Google Scholar]
- 7.Fullard RJ, Tucker D. Tear protein composition and the effects of stimulus. Adv Exp Med Biol. 1994;350:309–314 [DOI] [PubMed] [Google Scholar]
- 8.Green-Church KB, Nichols KK, Kleinholz NM, Zhang L, Nichols JJ. Investigation of the human tear film proteome using multiple proteomic approaches. Mol Vis. 2008;14:456–470 [PMC free article] [PubMed] [Google Scholar]
- 9.Gachon AM, Verrelle P, Betail G, Dastugue B. Immunological and electrophoretic studies of human tear proteins. Exp Eye Res. 1979;29:539–553 [DOI] [PubMed] [Google Scholar]
- 10.Jacob JT, Ham B. Compositional profiling and biomarker identification of the tear film. Ocul Surf. 2008;6:175–185 [DOI] [PubMed] [Google Scholar]
- 11.Li N, Wang N, Zheng J, et al. Characterization of human tear proteome using multiple proteomic analysis techniques. J Proteome Res. 2005;4:2052–2061 [DOI] [PubMed] [Google Scholar]
- 12.de Souza GA, Godoy LM, Mann M. Identification of 491 proteins in the tear fluid proteome reveals a large number of proteases and protease inhibitors. Genome Biol. 2006;7:R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou L, Beuerman RW, Chan CM, et al. Identification of tear fluid biomarkers in dry eye syndrome using iTRAQ quantitative proteomics. J Proteome Res. 2009;8:4889–4905 [DOI] [PubMed] [Google Scholar]
- 14.Tong L, Zhou L, Beuerman RW, Zhao SZ, Li XR. Association of tear proteins with Meibomian gland disease and dry eye symptoms. Br J Ophthalmol. 2011;95:848–852 [DOI] [PubMed] [Google Scholar]
- 15.Mackie IA, Seal DV. Diagnostic implications of tear protein profiles. Br J Ophthalmol. 1984;68:321–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koo BS, Lee DY, Ha HS, Kim JC, Kim CW. Comparative analysis of the tear protein expression in blepharitis patients using two-dimensional electrophoresis. J Proteome Res. 2005;4:719–724 [DOI] [PubMed] [Google Scholar]
- 17.Grus FH, Podust VN, Bruns K, et al. SELDI-TOF-MS ProteinChip array profiling of tears from patients with dry eye. Invest Ophthalmol Vis Sci. 2005;46:863–876 [DOI] [PubMed] [Google Scholar]
- 18.Tomosugi N, Kitagawa K, Takahashi N, Sugai S, Ishikawa I. Diagnostic potential of tear proteomic patterns in Sjögren's syndrome. J Proteome Res. 2005;4:820–825 [DOI] [PubMed] [Google Scholar]
- 19.Zhou L, Beuerman RW, Huang L, et al. Proteomic analysis of rabbit tear fluid: defensin levels after an experimental corneal wound are correlated to wound closure. Proteomics. 2007;7:3194–3206 [DOI] [PubMed] [Google Scholar]
- 20.Versura P, Nanni P, Bavelloni A, et al. Tear proteomics in evaporative dry eye disease. Eye (Lond). 2010;24:1396–1402 [DOI] [PubMed] [Google Scholar]
- 21.Fullard RJ, Snyder C. Protein levels in nonstimulated and stimulated tears of normal human subjects. Invest Ophthalmol Vis Sci. 1990;31:1119–1126 [PubMed] [Google Scholar]
- 22.Fullard RJ, Tucker DL. Changes in human tear protein levels with progressively increasing stimulus. Invest Ophthalmol Vis Sci. 1991;32:2290–2301 [PubMed] [Google Scholar]
- 23.Baguet J, Claudon-Eyl V, Sommer F, Chevallier P. Normal protein and glycoprotein profiles of reflex tears and trace element composition of basal tears from heavy and slight deposits on soft contact lenses. Clao J. 1995;21:114–121 [PubMed] [Google Scholar]
- 24.McGill JI, Liakos GM, Goulding N, Seal DV. Normal tear protein profiles and age-related changes. Br J Ophthalmol. 1984;68:316–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sack RA, Sathe S, Hackworth LA, Willcox MD, Holden BA, Morris CA. The effect of eye closure on protein and complement deposition on Group IV hydrogel contact lenses: relationship to tear flow dynamics. Curr Eye Res. 1996;15:1092–1100 [DOI] [PubMed] [Google Scholar]
- 26.Kijlstra A, Polak BC, Luyendijk L. Transient decrease of secretory IgA in tears during rigid gas permeable contact lens wear. Curr Eye Res. 1992;11:123–126 [DOI] [PubMed] [Google Scholar]
- 27.Saijyothi AV, Angayarkanni N, Syama C, et al. Two dimensional electrophoretic analysis of human tears: collection method in dry eye syndrome. Electrophoresis. 2010;31:3420–3427 [DOI] [PubMed] [Google Scholar]
- 28.Marouga R, David S, Hawkins E. The development of the DIGE system: 2D fluorescence difference gel analysis technology. Anal Bioanal Chem. 2005;382:669–678 [DOI] [PubMed] [Google Scholar]
- 29.Minden J. Comparative proteomics and difference gel electrophoresis. Biotechniques. 2007;43:739, 741, 743 passim [DOI] [PubMed] [Google Scholar]
- 30.Ong SE, Foster LJ, Mann M. Mass spectrometric-based approaches in quantitative proteomics. Methods. 2003;29:124–130 [DOI] [PubMed] [Google Scholar]
- 31.Ong SE, Blagoev B, Kratchmarova I, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386 [DOI] [PubMed] [Google Scholar]
- 32.Gygi SP, Rist B, Griffin TJ, Eng J, Aebersold R. Proteome analysis of low-abundance proteins using multidimensional chromatography and isotope-coded affinity tags. J Proteome Res. 2002;1:47–54 [DOI] [PubMed] [Google Scholar]
- 33.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999 [DOI] [PubMed] [Google Scholar]
- 34.Chen X, Sun L, Yu Y, Xue Y, Yang P. Amino acid-coded tagging approaches in quantitative proteomics. Expert Rev Proteomics. 2007;4:25–37 [DOI] [PubMed] [Google Scholar]
- 35.Ross PL, Huang YN, Marchese JN, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169 [DOI] [PubMed] [Google Scholar]
- 36.Kirkpatrick DS, Gerber SA, Gygi SP. The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications. Methods. 2005;35:265–273 [DOI] [PubMed] [Google Scholar]
- 37.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci U S A. 2003;100:6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schiffman RM, Christianson MD, Jacobsen G, Hirsch JD, Reis BL. Reliability and validity of the Ocular Surface Disease Index. Arch Ophthalmol. 2000;118:615–621 [DOI] [PubMed] [Google Scholar]
- 39.Walt JG, Rowe MM, Stern KL. Evaluating the functional impact of dry eye: the Ocular Surface Disease Index. Drug Inf J. 1997;31:1436 [Google Scholar]
- 40.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lei Z, Beuerman RW, Chew AP, et al. Quantitative analysis of N-linked glycoproteins in tear fluid of climatic droplet keratopathy by glycopeptide capture and iTRAQ. J Proteome Res. 2009;8:1992–2003 [DOI] [PubMed] [Google Scholar]
- 42.Nichols JJ, Green-Church KB. Mass spectrometry-based proteomic analyses in contact lens-related dry eye. Cornea. 2009;28:1109–1117 [DOI] [PubMed] [Google Scholar]
- 43.Lema I, Brea D, Rodriguez-González R, Diez-Feijoo E, Sobrino T. Proteomic analysis of the tear film in patients with keratoconus. Mol Vis. 2010;16:2055–2061 [PMC free article] [PubMed] [Google Scholar]
- 44.Pannebaker C, Chandler HL, Nichols JJ. Tear proteomics in keratoconus. Mol Vis. 2010;16:1949–1957 [PMC free article] [PubMed] [Google Scholar]
- 45.Joseph R, Srivastava OP, Pfister RR. Differential epithelial and stromal protein profiles in keratoconus and normal human corneas. Exp Eye Res. 2011;92:282–298 [DOI] [PubMed] [Google Scholar]
- 46.Flanagan JL, Willcox MD. Role of lactoferrin in the tear film. Biochimie. 2009;91:35–43 [DOI] [PubMed] [Google Scholar]
- 47.Cojocaru VM, Ciurtin C, Uyy E, Antohe F. Nano-LC mass spectrometry proteomic tear secretion analysis in patients with secondary Sjögren's syndrome. Digest J Nanomaterials Biostructures. 2011;6:491–498 [Google Scholar]