Abstract
Purpose.
Connective tissue growth factor (CTGF) is a matricellular protein presumed to be involved in the pathobiology of various fibrotic diseases, including glaucoma. We investigated the effects of Rho GTPase-dependent actin cytoskeletal integrity on CTGF expression and CTGF-induced changes in gene expression profile in human trabecular meshwork (HTM) cells.
Methods.
CTGF levels were quantified by immunoblotting and ELISA. CTGF-induced changes in gene expression, actin cytoskeleton, myosin light chain (MLC) phosphorylation, and extracellular matrix (ECM) proteins were evaluated in trabecular meshwork (TM) cells by cDNA microarray, q-PCR, fluorescence microscopy, and immunoblot analyses. The effects of neuromedin U (NMU) on aqueous humor (AH) outflow were determined in enucleated porcine eyes.
Results.
Expression of a constitutively active form of RhoA (RhoAV14), activation of Rho GTPase by bacterial toxin, or inhibition of Rho kinase by Y-27632 in HTM cells led to significant but contrasting changes in CTGF protein levels that were detectable in cell lysates and cell culture medium. Stimulation of HTM cells with CTGF for 24 hours induced actin stress fiber formation, and increased MLC phosphorylation, fibronectin, and laminin levels, and NMU expression. NMU independently induced actin stress fibers and MLC phosphorylation in TM cells, and decreased AH outflow facility in perfused porcine eyes.
Conclusions.
These data revealed that CTGF influences ECM synthesis, actin cytoskeletal dynamics, and contractile properties in TM cells, and that the expression of CTGF is regulated closely by Rho GTPase. Moreover, NMU, whose expression is induced in response to CTGF, partially mimics the effects of CTGF on actomyosin organization in TM cells, and decreases AH outflow facility, revealing a potentially important role for this neuropeptide in the homeostasis of AH drainage.
Neuromedin-U, a neuropeptide induced by CTGF, mimics partly the effects of CTGF on actin cytoskeleton of TM cells and aqueous humor outflow facility.
This study provides a novel insight into existence of a critical regulatory interaction among Rho/Rho kinase-regulated contractile activity, expression of connective tissue growth factor and Neuromedin-U, and extracellular matrix synthesis in trabecular meshwork cells.
Introduction
Primary open angle glaucoma (POAG) often is described as a chronic and progressive multifactorial optic neuropathy caused by an increased resistance to aqueous humor (AH) drainage through the trabecular meshwork (TM) and Schlemm's canal (SC).1–3 Abnormal resistance to AH drainage leads to an elevated intraocular pressure (IOP), which is a primary risk factor of POAG.3 Overproduction and deposition of extracellular matrix (ECM) in the TM and juxtacanalicular tissue (JCT) is implicated as a causative factor leading to increased resistance to AH drainage through the conventional drainage pathway.4,5 The synthesis and turnover of ECM is regulated by physiologic factors, transforming growth factor (TGF)-beta, cytokines, connective tissue growth factor (CTGF), dexamethasone, mechanical stress, cytoskeletal integrity, and the activity of matrix metalloproteases (MMPs) and tissue inhibitors of metalloproteases (TIMPs).4–7 Further, degradation of ECM by MMPs has been demonstrated to increase AH outflow facility, confirming the direct involvement of ECM in homeostasis of AH drainage.8 Similarly, actin cytoskeletal integrity and myosin II-based contractile tension are thought to influence ECM production and turnover in the TM cells, and AH drainage.9,10 Collectively, these different observations warrant a need for identification of different factors and mechanisms regulating the ECM production, its assembly and turnover in the AH outflow pathway, and etiology of glaucoma.
CTGF (CCN2), a member of the CCN family of proteins, is a cysteine-rich secretory matricellular protein that has a critical role in cell migration, adhesion, proliferation, and matrix production.11–13 Importantly, since CTGF expression is induced potently by TGF-beta, it is presumed that CTGF mediates several of the downstream actions of TGF-beta.13,14 CTGF is characterized as a profibrotic cytokine similar to TGF-beta and both are recognized to have key roles in a variety of fibrotic disorders,11,13 and elevations in aqueous humor CTGF levels have been reported in certain types of glaucoma.15 Other factors, such as Gremlin and BMP7, which influence AH outflow facility and IOP possibly via modulating ECM production, are reported to affect the regulation of CTGF expression in TM cells.7,16,17 Additionally, mechanical stretch, actin cytoskeletal integrity of TM cells, and increased IOP all have been reported to influence the expression of TGF-beta, CTGF, and ECM proteins, suggesting the existence of molecular interaction between mechanical stress, cytoskeletal integrity, CTGF expression, ECM, and AH outflow.6,7,9,18–20
To obtain insight into the cellular mechanisms that link contractile tension and regulation of CTGF expression and outflow facility, we investigated the role of Rho GTPase and Rho kinase activity-mediated effects of actomyosin-based contractile tension on CTGF expression in human trabecular meshwork (HTM) cells. Our study revealed the significance of Rho/Rho kinase-mediated control of actomyosin-based cytoskeletal integrity in the regulation of CTGF expression in TM cells. Additionally, our study identified neuromedin U (NMU) as one of the critical downstream effectors of CTGF in mediating changes in actomyosin-based contractile properties and AH outflow facility.
Materials and Methods
Reagents
Collagenase type IV (Worthington Biochemical Corp., Lakewood, NJ), RNeasy Mini kit (Qiagen, Valencia, CA), Advantage RT-for-PCR kit and Advantage cDNA PCR kit (BD Biosciences Clontech, Palo Alto, CA), iQSYBR Green supermix kit (Bio-Rad Laboratories, Philadelphia, PA), and cell culture media and fetal bovine serum (Gibco-BRL, Gaithersburg, MD) were procured from the respective commercial sources. Tetramethyl rhodamine isothiocyanate (TRITC)–conjugated phalloidin and FITC-conjugated phalloidin were from Sigma-Aldrich (St. Louis, MO). Rabbit anti-fibronectin and laminin antibodies were generous gifts from Harold P. Erickson (Department of Cell Biology, Duke University, Durham, NC). Polyclonal phospho-MLC antibody (Catalog # 3671S) and polyclonal MLC antibody (Catalog # 3672) were from Cell Signaling Technology (Danvers, MA). Enhanced chemiluminescence (ECL) plus detection reagents were from Amersham Pharmacia Biotech (Piscataway, NJ). Protease inhibitor cocktail tablets (complete, Mini, EDTA-free) and phosphatase inhibitor cocktail tablets (Phosphostop) were from Roche (Basel, Switzerland), and Y-27632 was from Calbiochem (Gibbstown, NJ). The Human CTGF ELISA Kit was from Peprotech (Rocky Hill, NJ), porcine NMU was from AnaSpec (Catalog # 23045; Fremont, CA) and human recombinant CTGF was from Cell Sciences (Catalog # CRC022B; Canton, MA). CTGF rabbit polyclonal antibody (Catalog # ab6992) was from Abcam (Cambridge, MA) and cell permeable protein-based Rho GTPase Activator (CN03) was from Cytoskeleton, Inc. (Denver, CO).
Cell Culture
Human primary TM cells were cultured from fresh corneal rings donated by the Duke Ophthalmology Clinical Service. Extracted TM tissue was chopped finely in fetal bovine serum, placed under a glass coverslip in six-well plastic culture plates, and cultured in Dulbecco's modified Eagle medium (DMEM) containing 20% fetal bovine serum (FBS), and penicillin (100 U/500 mL)-streptomycin (100 μg/500 mL)-glutamine (4 mM) at 37°C under 5% CO2. After TM cells had proliferated and reached confluence, they were subcultured and grown in DMEM media containing 10% FBS and penicillin-streptomycin-glutamine. TM cell cultures passaged between 3–6 cycles were used for all experiments. TM cells from freshly enucleated porcine eyes (obtained from the local slaughterhouse) were isolated and cultured as we described previously using collagenase type IV.9
Immunoblotting
Total cellular protein was extracted from treated and control confluent cells with or without respective treatment. Treatments always were performed using serum starved TM cells and the duration of serum starvation for each experiment is detailed in the respective section. Media were collected and centrifuged at 3000 revolutions per minute (rpm) for 15 minutes at 4°C to remove cellular debris. Supernatant was concentrated by centrifugation at 4°C and 2500 rpm using Millipore Centrifugal Filter Units (Millipore, Billerica, MA) with <30 kDa cutoff. Bio-Rad protein assay reagent (Catalog # 500-0006) was used to estimate protein concentration in cell lysate and media supernatant samples. Protein samples were dissolved in Laemmli buffer, boiled for 3 minutes, and separated by SDS-PAGE using 12.5% or 8% gels for detection of CTGF and β-tubulin, and 5.5% gels for fibronectin and laminin. Proteins were transferred to a nitrocellulose membrane, and then were blocked for 1 hour at room temperature in Tris-buffered saline containing 0.1% Tween 20 (TBST) and 5% (wt/vol) nonfat dry milk. Membranes were incubated overnight at 4°C with the respective primary antibodies, and blots were developed as described earlier using horseradish peroxidase-conjugated second antibody and ECL detection.10 Densitometry of scanned films was performed using Adobe Photoshop CS3 Software (Adobe Systems Inc., San Jose, CA), and results expressed in intensity units relative to the loading controls (β-tubulin or myosin light chain).
ELISA
CTGF levels in the conditioned cell culture media were determined using the Human CTGF ELISA Kit according to the manufacturer's instructions. Equal amounts of total protein from conditioned media were analyzed in triplicate. CTGF protein levels were estimated using a CTGF protein standard-based calibration curve. Results were analyzed for significance using Student's t-test.
Actin Filament Staining
Cells plated on glass coverslips coated with 2% gelatin were cultured to 90% confluence before being exposed to any drug treatment in serum-free media. Cells then were washed twice with 1× PBS, fixed in 4% formaldehyde for 15 minutes at room temperature, permeabilized with 0.5% Triton-X 100 for 15 minutes, and blocked with serum buffer containing 10% FBS. Following this, the cells were incubated with TRITC or FITC-phalloidin for 30 minutes at room temperature as described previously.10 Finally, coverslips were washed and mounted onto glass slides with Aqua Mount (Lerner Laboratories, Pittsburgh, PA), and observed under a Nikon confocal system (C1 Digital Eclipse; Nikon, Melville, NY).
Adenoviral-Mediated Gene Transduction
Replication-defective recombinant adenoviral vectors encoding either GFP alone or constitutively active RhoA (RhoAV14 mutant; provided by Patrick Casey, Department of Pharmacology and Cancer Biology, Duke University School of Medicine) were used in this study.21 The viral vectors were amplified and transfected as we described previously.21 HTM cells derived from passage 3 (P3) of 33-, 45-, and 54-year-old patients, grown either on gelatin-coated glass coverslips or plastic Petri plates, were infected with adenoviral vectors at a multiplicity of infection (MOI) of approximately 50. After confirming an adequate transfection rate (∼80%, as evaluated based on GFP fluorescence during the 24–36-hour period following transfection), cells were serum starved for 24 hours for use in subsequent experiments.
Bacterial Toxin-Stimulated Activation of Rho GTPase
Bacterial cytotoxic necrotizing factor (CNF-1) was used to activate directly Rho GTPase using an independent technique, as opposed to viral vector-based RhoAV14 expression. Serum starved (18 hours) confluent cultures of HTM cells (passage 6) derived from a 57-year-old donor were treated with CN03 (5 μg/mL for 24 hours), which is a cell permeable recombinant CNF that activates Rho GTPase constitutively through deamidation.22 After CN03 treatment, cell lysates and conditioned media from HTM cells were examined for changes in CTGF levels.
Perfusion Studies
Freshly obtained enucleated whole globe porcine eyes were perfused at constant pressure with 10 μM NMU (initially dissolved in dimethyl sulfoxide [DMSO]) in a Dulbecco's phosphate buffered saline (DPBS) perfusion medium containing 5.5 mM dextrose at 37°C as described previously.23 The initial baseline outflow was established at 15 mm Hg for 1 hour. Eyes were paired such that one eye was perfused with 10 μM NMU, while the contralateral eye served as a control and was perfused with perfusion medium containing DMSO (0.1%). The perfusion was carried out for 5 hours and outflow measurements were recorded throughout this time using a PowerLab data acquisition system (ML870/P PowerLab 8/30; AD Instruments, Colorado Springs, CO). Outflow facility was expressed as μL/minutes/mm Hg. The effects of NMU on outflow facility were expressed as percentage change in outflow facility in treated versus untreated paired control eyes from their respective baseline facility values. Values are expressed as mean ± SEM. The paired two-tailed Student's t-test was used to determine significance.
Myosin Light Chain (MLC) Phosphorylation
Changes in MLC phosphorylation were determined by immunoblot analysis using phospho-MLC antibody as we described previously.10 Densitometry of scanned films was performed using Adobe Photoshop CS3 Software, and expressed as relative intensity compared to the loading control (total myosin light chain).
cDNA Microarray Analysis
Human TM cells from 16-year-old donor eyes (passage 4) were cultured to confluence, serum-starved for 48 hours, and treated in duplicate with 50 ng/mL CTGF (prepared in acetate buffer) for 24 hours, with control cells being treated with equal volume of acetate buffer. Total RNA was extracted using the RNeasy Mini kit according to the manufacturer's instructions. Briefly, cells were homogenized using the QiaShredder column and treated with DNAse I to eliminate genomic DNA contamination. Purified RNA was quantitated (A260/280 nm using a Nanodrop 2000 Spectrophotometer; Thermo Scientific, Wilmington, DE) and subjected to cDNA microarray analysis at the core facility of the Duke Institute of Genomic Sciences and Policy (IGSP, Duke University, Durham, NC). Briefly, RNA quality was assessed first using an Agilent 2100 Bioanalyzer G2939A (Agilent Technologies, Santa Clara, CA) and Nanodrop 8000 Spectrophotometer (Thermo Scientific). Hybridization targets were prepared from total RNA using a MessageAmp Premier RNA Amplification Kit (Applied Biosystems/Ambion, Austin, TX), hybridized to GeneChip Human Genome U133 2.0 (Affymetrix, Santa Clara, CA) arrays in Affymetrix GeneChip hybridization Oven 645, washed in Affymetrix GeneChip Fluidics Station 450, and scanned with Affymetrix GeneChip Scanner 7G according to standard Affymetrix GeneChip hybridization, wash, and stain protocols. Raw data were normalized and analyzed using GeneSpring 10 (Silicon Genetics, Wilmington, DE). The genes were filtered by intensity compared to the control channel, and a P value of ≤ 0.05 by the paired Student's t-test was considered significant. A list of genes that were upregulated or down-regulated significantly in both of the CTGF-treated samples analyzed, relative to both of the control samples, was identified based on the magnitude and significance of change.
RT-PCR Analysis
Equal amounts of RNA were reverse transcribed using the Advantage RT-for-PCR kit according to the manufacturer's instructions. A negative control lacking reverse transcriptase (RT) also was run. PCR amplification then was performed on the resultant HTM RT-derived first-strand complementary DNA (cDNA) using the primers listed in Table 1. The amplification was done using GeneAmp Amplification System (Applied Biosciences) with a denaturation step at 94°C for 4 minutes, followed by 94°C for 1 minute, 57°C to 60°C for 60 seconds, and 72°C for 30 seconds. The cycle was repeated 25 to 32 times with a final step at 72°C for 7 minutes. The resulting products were separated by 1% agarose gel electrophoresis and visualized by staining with ethidium bromide using Fotodyne Trans-illuminator (Fotodyne Inc., Hartland, WI). GAPDH (forward: 5′-CCG AGC TGA GCA TAG ACA TT-3′ and reverse: 3′-TCC ACC ACC CTG TTG CTG TA-5′) was run as an internal housekeeping positive control together with a minus RT as negative control for each experiment. GAPDH also was used to normalize the cDNA content of control and CTGF-treated samples in all the PCR reactions. DNA was extracted as per the instructions from the Qiagen Gel Extraction Kit for sequencing.
Table 1. .
Human Oligonucleotide Primers Used in the RT-PCR and Real-Time PCR Amplifications
|
Type |
Sequence |
Size |
| NMU | 5′-GGC ATC CAA CGC ACT GGA GGA GCT T-3′ | 181 bp |
| 5′-GTG AGG AAC GAG CTG CAG CAA CGG A-3′ | ||
| NMU R1 | 5′-GCC ATG CGC ACG CCT ACC AAC TAC T-3′ | 194 bp |
| 5′-GCA GTG ACG TTG AGC ACT GAG GCC A-3′ | ||
| NMU R2 | 5′-ACC TCT TCA GCC TGG CGG TCT CTG A-3′ | 219 bp |
| 5′-GTT TGG CGC GGA ACG GGT GTA GGA T-3′ | ||
| IL6 | 5′-TCC ACA AGC GCC TTC GGT CCA GTT G-3′ | 135 bp |
| 5′-AGA GGT GAG TGG CTG TCT GTG TGG G-3′ | ||
| MMP-1 | 5′-CTC GGC CAT TCT CTT GGA CTC TCC-3′ | 182 bp |
| 5′-GAG CTC AAC TTC CGG GTA GAA GGG-3′ | ||
| STC | 5′-AGT CAG CTC GTG GGT GTG TTT GGG C-3′ | 222 bp |
| 5′-GCC ATG CGC ACG CCT ACC AAC TAC T-3′ |
bp, base pairs.
RT Quantitative PCR (Q-PCR)
Q-PCR was performed as we described previously.21 Briefly, first strand cDNA pools from the treated and control samples first were normalized relative to the housekeeping gene GAPDH. PCR reactions were done in triplicate using the following protocol: 95°C for 2 minutes followed by 50 cycles of 95°C for 15 seconds, 60°C for 15 seconds, and 72°C for 15 seconds, and a PCR Master mix. The PCR master mix (iQ supermix; Bio-Rad) consisted of 1 to 2 μL template cDNA in 20 μL reaction, 2× PCR master mix, 10 nM fluorescein calibration dye (Bio-Rad), 1 μL of a 1:1500 dilution of 10,000× nucleic acid dye, iQSYBR Green, and 500 nM each of a gene-specific oligonucleotide pair. An extension step was used to measure the increase in fluorescence and melting curves obtained immediately after amplification by increasing temperature in 0.4°C increments from 65°C for 85 cycles of 10 seconds each, analyzed (iCycler software; Bio-Rad). The fold difference in NMU, MMP1, and STC-1 expression between control and CTGF-treated samples was normalized to GAPDH and calculated by the comparative threshold (CT) method, as described by the manufacturer (Prism 7700 Sequence Detection System; Applied Biosystem, Inc.).
Results
RhoAV14-Induced Changes in the Expression and Secretion of CTGF in HTM Cells
To explore the cellular mechanisms transducing the effects of actin/myosin-based contractile responses on CTGF expression in TM cells, we evaluated the involvement of Rho GTPase, a crucial regulator of actomyosin assembly, on the expression and secretion of CTGF. For this, we expressed constitutively active RhoA mutant (RhoAV14/GFP) or GFP alone in HTM cells using recombinant adenoviral vectors. Following viral vector infection (typically 24–36 hours following infection, and based on GFP expression), the HTM cells were serum starved for 24 hours, after which the media and cell lysates were immunoblotted using CTGF antibody. RhoAV14-expressing TM cells exhibited an increase in CTGF levels compared to GFP-expressing control cells (Fig. 1A). Densitometric analysis (Fig. 1B) revealed that this increase was significant (n = 4, P < 0.05). Conditioned media derived from the RhoAV14-expressing TM cells also showed increased levels of secreted CTGF based on immunoblot analysis (Fig. 1C). The lower panel of Figure 1C shows a representative Coomassie blue-stained image of equal amounts of protein loads from media samples separated on a SDS-PAGE gel. CTGF protein levels also were noted to be significantly higher (by 3.8-fold) in conditioned media derived from RhoAV14-expressing cells compared to the corresponding GFP controls based on ELISA, as shown in Figure 1D (n = 4, P < 0.05). The diffuse nature of the CTGF bands detected by immunoblotting reflects the nature of this molecule, which is a secreted and glycosylated protein (Figs. 1A, 1C). As we reported previously,21 HTM cells expressing RhoA14 exhibit robust induction of actin stress fibers and a notably stiffened/contractile morphology compared to the GFP expressing cells (data not shown).
Figure 1. .
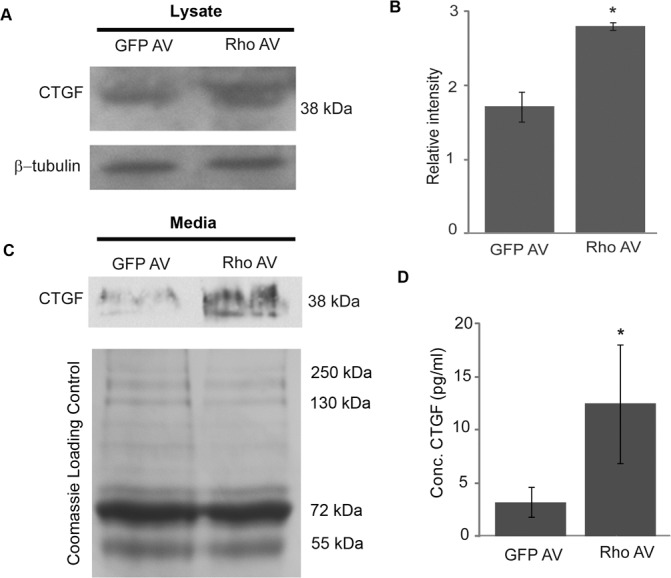
Rho GTPase-induced increase in CTGF protein levels of TM cells. A constitutively active RhoA mutant (RhoAV14) or GFP was overexpressed in HTM cells using recombinant adenovirus. At 24 hours following adenoviral infection, cells were serum starved for another 24 hours. Cell lysates and conditioned media derived from these cells were analyzed by immunoblotting for CTGF (A, C). Compared to the GFP controls, RhoAV14 expressing cells exhibited increased CTGF protein levels in cell lysates and conditioned media. The lower panels in (A) and (C) represent β-tubulin loading controls for the cell lysates, and Coomassie blue stained images of proteins from conditioned media separated by SDS-PAGE to confirm equality of loading, respectively. (B) Densitometric analysis of CTGF protein levels in lysates revealed a significant (P < 0.05, n = 4) increase in TM cells expressing RhoAV14. (D) Significant increases also were noted in the levels of secreted CTGF protein in conditioned media from the RhoAV14/GFP expressing HTM cells, as measured by ELISA, and relative to the levels in conditioned media obtained from GFP-expressing cells (n = 4, *P < 0.05).
Influence of Bacterial Toxin-Induced Rho GTPase Activation on CTGF Expression in HTM Cells
In addition to evaluating the effects of expressing RhoAV14 on CTGF levels in TM cells, we used a cell permeable recombinant bacterial cytotoxin (CN03) to activate directly endogenous Rho GTPase in HTM cells. CN03 contains the catalytic domain of bacterial cytotoxin necrotizing factor-1, which deamidates residues glutamine-63 and constitutively activates Rho GTPase.22 Similar to the effects of RhoAV14, CN03-induced activation of Rho GTPase in HTM cells triggers the appearance of a contractile morphology (not shown) and causes significant (P < 0.05) increase of CTGF levels in the cell lysates (Figs. 2A, 2B; n = 5) and conditioned cell culture media (Figs. 2C, 2D; n = 3), as assessed by immunoblotting. Results for media protein samples were normalized based on the Coomassie blue staining intensities of SDS-PAGE resolved proteins as shown in the bottom panel of Figure 2C.
Figure 2. .
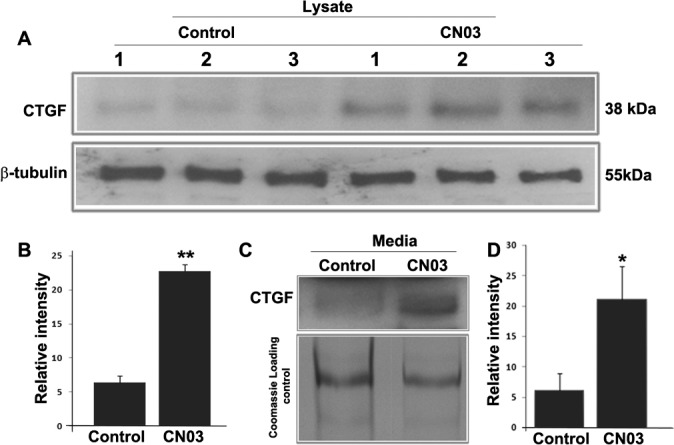
CN03-induces CTGF expression in HTM cells. Treatment of HTM cells treated with the Rho GTPase activator CN03, which is a cell permeable bacterial toxin (5 μg/mL for 24 hours under serum free conditions) leads to significant (*P < 0.05; **P < 0.01) increases in CTGF protein levels in cell lysates (A, B; n = 5) and conditioned medium (C, D; n = 3). Results are based on immunoblotting analysis with subsequent densitometric scanning. The lower panel in (C) indicates equal protein loading of conditioned media samples based on Coomassie blue staining profiles of SDS-PAGE gels.
Effects of Inhibition of Rho Kinase on CTGF Expression in TM Cells
To explore further whether Rho-kinase was engaged as a downstream participant in actomyosin integrity-mediated effects on CTGF expression and secretion, confluent cultures of HTM cells were serum-starved for 24 hours followed by treatment with 5 μM Y-27632 (a selective Rho kinase inhibitor) for 4 hours. After 4 hours, treatment with Y-27632 caused cell shape changes in HTM cells that typically are associated with cellular relaxation (cell-cell separation and rounding) and characterized by decreased actin stress fibers (data not shown).23 CTGF expression was assessed in cell lysates and conditioned media derived from these cells, by immunoblot and ELISA, respectively. Figures 3A and 3B show that there is a decrease in CTGF protein levels (n = 4, P < 0.05) in HTM cell lysates treated with Y-27632, compared to control cells. This effect also was mirrored by CTGF levels based on ELISA in conditioned culture media (Fig. 3C) derived from Y-27632-treated HTM cells (n = 4, P < 0.05).
Figure 3. .
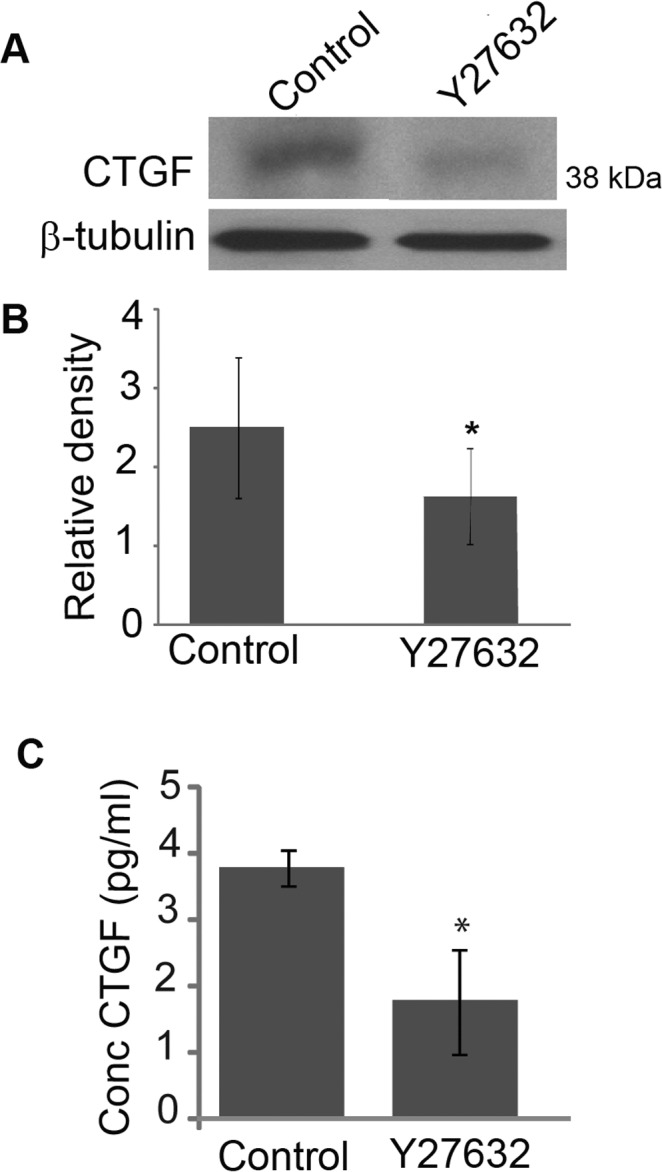
Effects of Rho Kinase inhibition on CTGF protein levels in HTM cells. Serum starved HTM cells treated for 4 hours with Rho kinase inhibitor Y-27632 (5 μM) revealed decreased levels of CTGF in cell lysates (A) and conditioned media (C), a response that was confirmed to be significant based on densitometric analysis (B) and ELISA quantification (C) as compared to controls (*P < 0.05, n = 4).
Effect of CTGF on Actin Cytoskeleton and ECM Proteins in HTM Cells
To explore the influence of actin cytoskeletal dynamics in CTGF-regulated ECM synthesis, serum-starved (for 48 hours) HTM cells grown on 2% gelatin-coated glass coverslips and plastic Petri dishes, and derived from 18- (passage 5) and 74 (passage 3)-year-old donor eyes, were treated with 50 ng/mL CTGF. After 24 hours of treatment, TM cells treated with CTGF exhibited a stiffened and contractile morphology, with notable changes in cell shape compared to the sham (sodium acetate)-treated control cells (Fig. 4A). This change in cell morphology was associated with an increase in actin stress fibers based on FITC-phalloidin staining (Fig. 4B). Additionally, quantitative immunoblot analysis of cells collected by scraping (which results in acquiring ECM components associated with the cell monolayer) confirmed an increase in the levels of fibronectin and laminin in CTGF-treated HTM cells compared to control cells (Figs. 4C–4F). A significant increase in the levels of phosphorylated MLC (P < 0.05, n = 4), a regulatory subunit of myosin II, also was noted in CTGF-treated cells, based on immunoblot analysis (Figs. 4G, 4H).
Figure 4. .
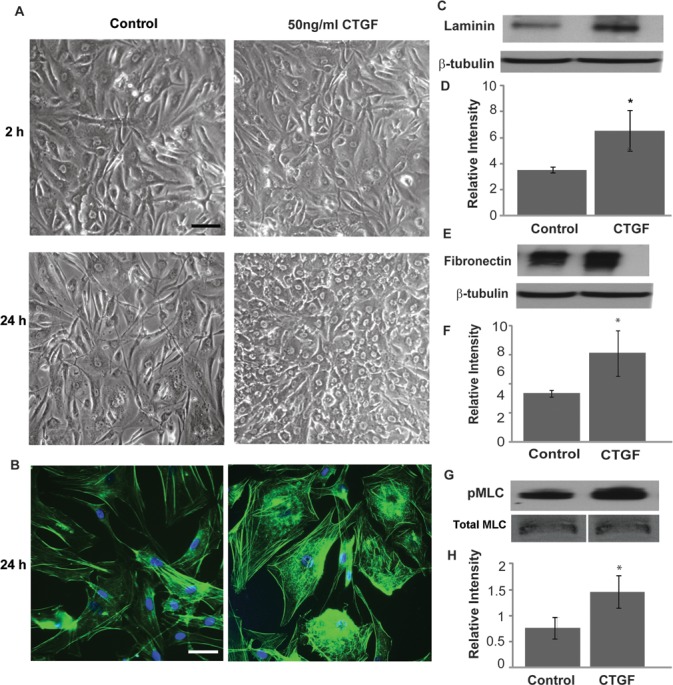
CTGF-induced changes in actin cytoskeletal organization, MLC phosphorylation, and ECM synthesis in HTM cells. Serum-starved HTM cells were treated with CTGF (50 ng/mL) or acetate buffer (control) for 24 hours and examined for changes in cell morphology, actin stress fibers, pMLC, and ECM proteins (fibronectin and laminin). (A) Phase contrast images revealed notable changes in cell morphology characterized by stiffened and contractile nature following treatment with CTGF after 24 hours compared to 2 hours of treatment and controls. This change in cell morphology was associated with increased actin stress fibers (phalloidin staining) in CTGF-treated TM cells (B). CTGF treatment for 24 hours also led to an increase in the levels of laminin (C, D) and fibronectin (E, F), and in the levels of phosphorylated MLC in HTM cells (G, H). Bars: 10 μm. β-tubulin and total MLC were probed as loading controls for ECM and phospho-MLC, respectively. *Denotes significant changes (P < 0.05, based on n = 4).
CTGF-Induced Changes in Gene Expression Profile in HTM Cells
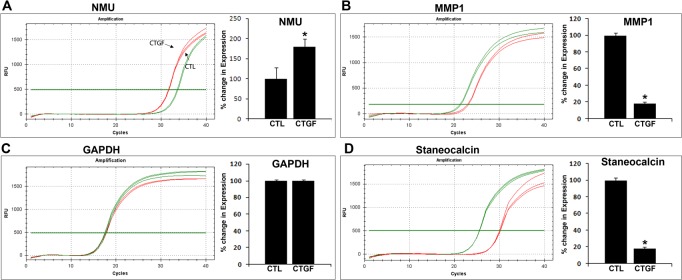
To obtain broader insight regarding how CTGF affects TM cell physiology, we evaluated changes in global gene expression profile in HTM cells treated with CTGF by cDNA microarray analysis using an Affymetrix human gene chip containing over 47,000 gene transcripts. We used duplicate sets of control and CTGF-stimulated HTM cells (derived from passage 4 of cells from a 16-year-old donor). Only genes that showed a consistent and significant trend in both sets are discussed in this report. Microarray data revealed a number of genes that either were upregulated or down-regulated significantly in CTGF treated cells (summarized in Tables 2 and 3, respectively). To our surprise, only a limited number of genes showed consistent and significant differences between control and CTGF treated cells. Some of the upregulated genes include tumor-associated calcium signal transducer 2 (TACSTD2), several of melanoma antigen family genes, NMU, interleukin-6, interleukin-32, C/EBP (delta) transcription factor, tripartite motif-containing 58 and Ras-related GTP binding D. There was a slightly higher number of down-regulated genes in CTGF-treated HTM cells, including stanniocalcin-1, endothelial cell specific molecule-1 (ESM1/Endocan), matrix metallopeptidase-1 (MMP1), interleukin-8, pentraxin-related gene (PTX3), BMP-2, interleukin-13 receptor, angiopoietin-like-4, coronin, endothelin receptor type A, phorbol-12-myristate-13-acetate-induced protein 1 (PMAIP1), tumor necrosis factor (TNF)-alpha-induced protein (TSG-6), dual specific phosphatase-6 (DUSP6) and hyaluronan synthase-2. Of the differentially expressed genes, a few representative upregulated and down-regulated genes were evaluated further independently by Q-PCR and semi-quantitative PCR. For this, we used the same source of RNA used to conduct the cDNA microarray analysis described above. The results for NMU, MMP1, and stanniocalcin-1 were consistent with the results of cDNA microarray analysis (Fig. 5). GAPDH expression was quantified in treated and untreated specimens along with the above described genes to confirm normalization of the test and control single strand cDNA specimens (Fig. 5).
Table 2. .
CTGF-Induced Upregulation of Gene Expression in Human TM Cells
|
Symbol |
Gene Title |
Transcript ID |
Ratio |
| PLAC8 | Placenta-specific-8 | NM_001,130,715 | 3.943 |
| TACSTD2 | Tumor-associated calcium signal transducer-2 | NM_002,353 | 3.899 |
| MAGEA12 | Melanoma antigen family A, 12 | NM_005,367 | 2.366 |
| MAGEB2 | Melanoma antigen family B, 2 | NM_002,364 | 2.217 |
| NMU | Neuromedin U | NM_006,681 | 1.799 |
| XAGE1A | X antigen family, member 1A | NM_001,097,591 | 1.768 |
| RRAGD | Ras-related GTP binding D | NM_021,244 | 1.636 |
| MEIS2 | Meis homeobox-2 | NM_002,399 | 1.610 |
| CEBPD | CCAAT/enhancer binding protein (C/EBP), delta | NM_005,195 | 1.600 |
| SLC7A1 | Solute carrier family 7, member 1 | NM_003,045 | 1.569 |
| IL6 | Interleukin-6 (interferon, beta-2) | NM_000,600 | 1.562 |
| TRIM58 | Tripartite motif-containing-58 | NM_015,431 | 1.557 |
| IL32 | Interleukin-32 | NM_001,012,631 | 1.540 |
| ID1 | Inhibitor of DNA binding-1 | NM_002,165 | 1.533 |
Table 3. .
CTGF-Induced Down-Regulation of Gene Expression in Human TM Cells
|
Symbol |
Gene Title |
Transcript ID |
Ratio |
| HAS2 | Hyaluronan synthase-2 | NM_005,328 | −1.503 |
| TNFAIP6 | TNF alpha-induced protein-6 | NM_007,115 | −1.511 |
| PMAIP1 | Phorbol-12-myristate-13-acetate-induced protein-1 | NM_021,127 | −1.513 |
| DUSP6 | Dual specificity phosphatase-6 | NM_001,946 | −1.550 |
| EDNRA | Endothelin receptor type A | NM_001,957 | −1.557 |
| ANPEP | Alanyl (membrane) aminopeptidase | NM_001,150 | −1.608 |
| CORO2B | Coronin, actin binding protein-2B | NM_006,091 | −1.624 |
| ANGPTL4 | Angiopoietin-like-4 | NM_001,039,667 | −1.650 |
| TMEM158 | Transmembrane protein-158 | NM_015,444 | −1.770 |
| IL13RA2 | Interleukin-13 receptor, alpha-2 | NM_000,640 | −1.772 |
| BMP2 | Bone morphogenetic protein-2 | NM_001,200 | −1.854 |
| PTX3 | Pentraxin-related gene | NM_002,852 | −1.857 |
| HMGA2 | High mobility group AT-hook-2 | NM_003,483 | −1.898 |
| IL8 | Interleukin-8 | NM_000,584 | −1.907 |
| MMP1 | Matrix metallopeptidase-1 | NM_001,145,938 | −1.969 |
| ESM1 | Endothelial cell-specific molecule-1 | NM_001,135,604 | −2.060 |
| STC1 | Stanniocalcin-1 | NM_003,155 | −2.253 |
Figure 5. .
Q-PCR-based confirmation of CTGF-induced changes in expression of selected genes in HTM cells. Q-PCR was used to confirm independently the results of cDNA microarray analyses, which had been performed to detect relative changes in gene expression induced by CTGF stimulation of HTM cells. To this end, a portion of the same RNA used to conduct the cDNA microarray experiments (and derived from CTGF-stimulated HTM cells) was reverse-transcribed and used for Q-PCR analyses. Real-time quantification of expression of selected genes was normalized to the cycle value (Cycles) of GAPDH. Left panels: plots of log fluorescence units versus cycle number. Right panels: relative fold change in gene expression upon CTGF treatment of HTM cells. Fold changes were calculated based on the mean values from triplicate analyses of individual samples. Consistent with cDNA microarray-based results, NMU expression was significantly upregulated (A), while MMP1 (B) and STC (D) exhibited decreased expression in CTGF treated TM cells. (C) GAPDH was used to normalize the cDNA content of control and CTGF treated samples.
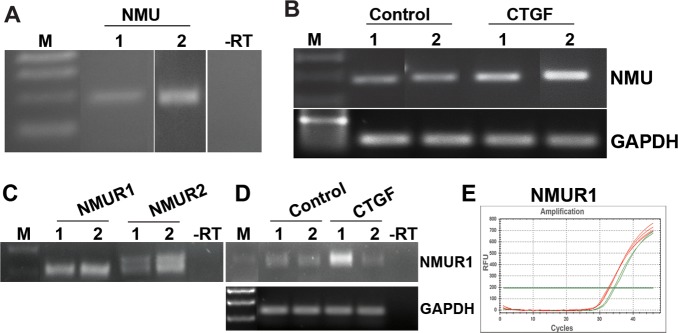
Since NMU was one of the few genes upregulated in response to CTGF treatment of HTM cells (Table 2), and there was no known information regarding NMU or its receptors (NMU-R1 and -R2) in TM cells, we evaluated the expression of NMU, NMU-R1, and NMU-R2 in two additional HTM cell strains derived from 18-year-old and 74-year-old donors, using RT-PCR analysis and confirmation of sequence identity of the RT-PCR products (Figs. 6A, 6C). NMU expression was confirmed in more than three independent HTM cell strains (data shown for two independent samples), and also confirmed to be upregulated in both HTM cell libraries derived from CTGF-treated cells, based on semi-quantitative RT-PCR (Fig. 6B). Expression of NMU-R1 also was upregulated in one of the CTGF-treated samples based on a semi-quantitative PCR analysis (Fig. 6D) and Q-PCR analysis (Fig. 6E).
Figure 6. .
Expression profiles of NMU and its receptors (NMU-R1 and NMU-R2) in HTM cells. In addition to cDNA microarray data, expression of NMU (A) and its receptors (C) was confirmed by RT-PCR analysis of 2 independent HTM cell lines (lanes 1 and 2). (B) Semi-quantitative RT-PCR analysis (in duplicate) revealed increased NMU expression in CTGF-treated TM cells compared to acetate buffer treated controls. (D) Expression of NMU-R1 also was upregulated in one of the cDNA libraries generated from the CTGF-treated TM cells. (E) The increase in NMU-R1 expression was confirmed further by Q-PCR analysis.
Effect of Neuromedin U on Actin Cytoskeleton and Phosphorylated MLC in HTM Cells
NMU is a well-characterized structurally conserved neuropeptide involved in smooth muscle contraction.24–26 Since the contractile activity of TM is known to influence aqueous humor outflow,27,28 we investigated the effects of NMU on TM cell actin cytoskeletal organization. Serum starved (for 18 hours) porcine TM cells treated with 1 μM porcine NMU-8 peptide for 1 hour exhibited stiffened and contractile morphology with notable change in cell shape compared to control cells (Fig. 7A, top panel). This change in cell morphology was associated with a robust increase in actin stress fibers based on phalloidin staining (Fig. 7B). Further, NMU treatment led to a significant increase in the levels of phosphorylated MLC in porcine TM cells as determined by immunoblot analysis (Figs. 7C, 7D). Interestingly, these effects were very similar to those induced by CTGF in TM cells (Fig. 4). These experiments were performed using PTM cells to obtain a relevant set of data for subsequent work using enucleated porcine eyes for perfusion studies on the effects of porcine NMU-8 on AH outflow facility.
Figure 7. .
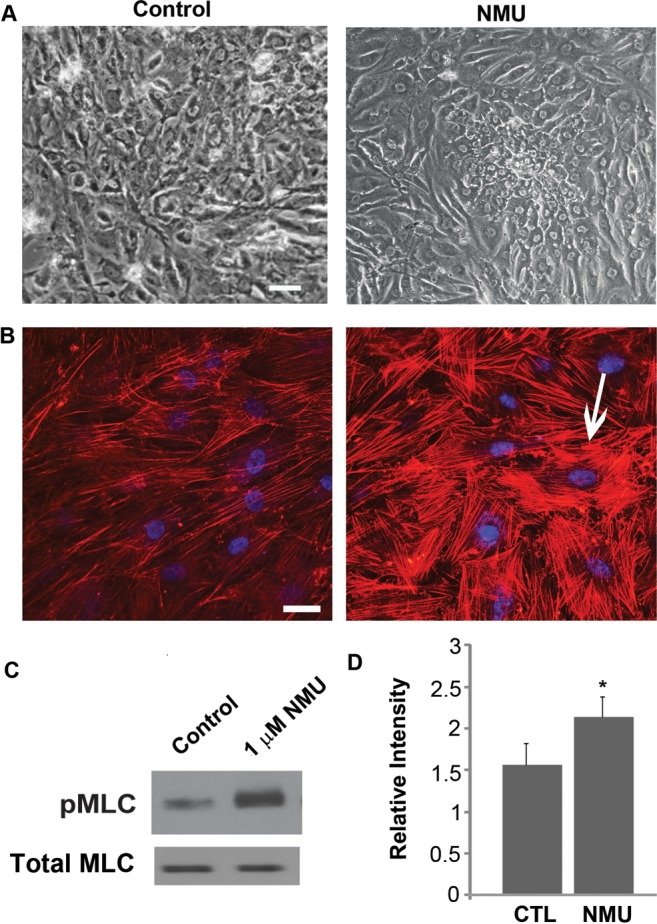
NMU-induced changes in actin cytoskeletal organization and phospho-MLC in porcine TM cells. Treatment of serum-starved porcine TM cells with 1 μM NMU for 1 hour (A) induced a contractile morphology (recorded using a phase contrast microscope) and (B) led to formation of increased actin stress fibers, as shown by the phalloidin staining (arrow) compared to serum-starved control cells (left panels). (C) NMU-treated porcine TM cells also exhibited increased MLC phosphorylation compared to control cells, as determined by immunoblot analysis. Total MLC was immunoblotted as a loading control. (D) The increase in pMLC levels was significant based on densitometric analysis. Bar: 10 μm. n = 4, *P < 0.05.
Effects of Perfusion of NMU on Aqueous Outflow Facility in the Enucleated Porcine Eye Perfusion Model
Based on the ability of NMU to induce changes in actin cytoskeletal organization and MLC phosphorylation, we hypothesized that NMU also might influence AH outflow facility. To explore this possibility, we tested the effects of NMU on AH outflow facility in enucleated porcine eyes (Fig. 8). Freshly obtained contralateral porcine eyes perfused under constant pressure (15 mm Hg) with either 10 μM porcine NMU-8 peptide or media alone at 37°C showed a significant decrease in outflow facility with NMU-8 relative to baseline facility after 1 hour (48%, P < 0.006, n = 6). However, this change in outflow facility returned back to baseline facility by the second hour of perfusion (Fig. 8). In contrast, the sham-treated eyes showed no significant difference in outflow facility during a 5-hour perfusion period.
Figure 8. .
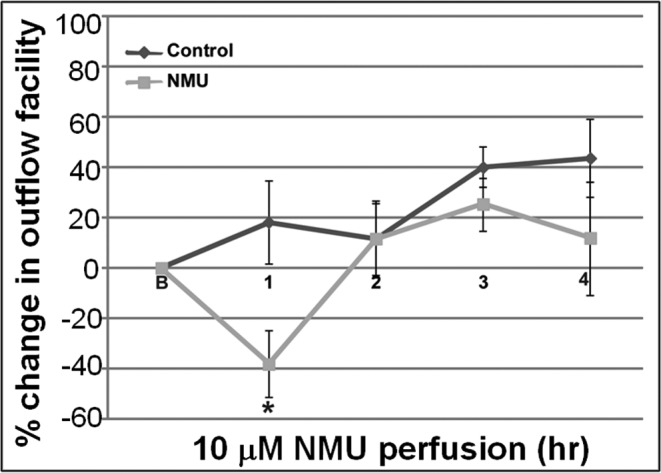
Effects of NMU perfusion on aqueous outflow facility in enucleated porcine eyes. Freshly enucleated porcine eyes were perfused for 5 hours with either medium containing 10 μM NMU or with medium alone, at a constant pressure of 15 mm Hg. Baseline facility was obtained with perfusion media at 37°C. There was a significant decrease (48%) in outflow facility relative to baseline facility after 1 hour of perfusion (*P < 0.006, n = 6) in eyes perfused with NMU, compared to the sham-perfused eyes. However, the outflow facility in the NMU-perfused eyes reverted back to baseline facility after this point, and was comparable to control baseline facility thereafter for the entire course of the experiment.
Discussion
Homeostasis of aqueous humor drainage is presumed to be influenced partly by mechanobiology of the cells of the conventional outflow pathway.29 Actomyosin-based contractile force, cell-ECM interactions via focal adhesions and ECM homeostasis are some of the key components of mechanotransduction and regulate various cellular processes.30–33 Since CTGF has an important role in ECM synthesis, in our study we explored whether the expression of CTGF is regulated by Rho GTPase-controlled actomyosin organization in TM cells. Our study demonstrated that the activities of Rho GTPase and Rho kinase regulate CTGF expression in TM cells, possibly via controlling actomyosin–based contraction and the levels of free G-actin. Additionally, the CTGF-induced changes in actomyosin organization and myosin II activity were associated with increased levels of fibronectin and laminin in TM cells. Interestingly, NMU, a neuropeptide whose expression is induced by CTGF in TM cells, showed a transient impact on AH outflow facility plausibly by mimicking the CTGF-induced actomyosin-based contractile properties in TM cells. Collectively, these observations revealed the existence of a close regulatory and functional interaction between Rho/Rho kinase pathway activity, CTGF expression, and cell contractile properties in TM cells, and supported the involvement of such network interactions in the homeostasis of ECM synthesis and AH drainage through the trabecular pathway (Fig. 9).
Figure 9. .
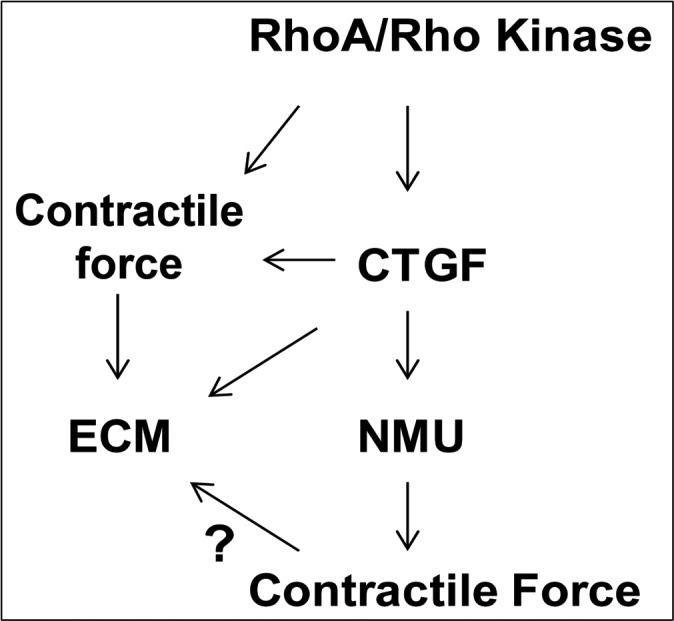
Schematic representation of potential interaction between the Rho GTPase signaling pathway, CTGF, and NMU in regulation of actomyosin-based contractile force and ECM protein profiles in TM cells.
In previous perfusion studies, several physiologic compounds, including TGF-beta, LPA, S1P, thrombin, and endothelin-1, have been reported to decrease AH outflow facility in different species,10,34–40 with this response being associated with increased Rho GTPase activation, actomyosin-based contraction, focal adhesions, and increased synthesis of ECM proteins in TM cells. Increased cyclic stretch, elevated intraocular pressure, and some of the physiologic compounds mentioned above also have been shown to induce the expression of CTGF,18,19 a regulator of ECM production in TM cells and tissue, suggesting a possible regulatory interaction among CTGF, ECM, Rho GTPase signaling, and actomyosin-based contractile force.10,18,19 To seek the mechanistic basis for such a possibility, we studied the role of Rho GTPase and Rho kinase activities on CTGF expression in TM cells in the context of actomyosin-based contractile activity.
Human TM cells expressing a constitutively active form of RhoA (RhoaV14 mutant) exhibited induction of actin stress fiber formation, focal adhesions, and MLC phosphorylation, together with an increase in CTGF levels in TM cell lysates and cell-conditioned culture media, revealing a direct influence of Rho GTPase activity on the regulation of CTGF expression. This observation also was consistent with our earlier report on RhoAV14-induced CTGF expression in TM cells based on cDNA microarray analysis,21 along with Rho activator CN03-induced CTGF expression demonstrated in this study (Fig. 2). Consistent with the effects of RhoAV14 and CN03 on CTGF expression, inhibition of Rho kinase, which is a critical downstream kinase of RhoA and regulates actomyosin assembly and myosin II-based contractile force in TM cells,41 led to a decrease in CTGF protein levels in cell lysates and cell-conditioned culture media. Inhibition of Rho kinase was associated with a dramatic decrease in actin stress fibers, focal adhesions and MLC phosphorylation, as we demonstrated previously.23 Rho GTPase-induced focal adhesions are critical focal points where actin stress fibers interact with integrins, which in turn tether to ECM molecules.42,43 These adhesion sites have a crucial role in mechanotransduction by transducing mechanical signals derived from ECM stiffness/rigidity (outside to inside) and inside-out from actomyosin-induced contractile force.30,33,44 Moreover, mechanotransduction regulated by Rho GTPase has been thought to be crucial for CTGF expression in various other cell types.45
Having found evidence for the direct role of Rho GTPase and Rho kinase activity in regulating CTGF expression, we also examined whether CTGF induces any effects on actin cytoskeletal organization and contractile activity in TM cells. Recombinant CTGF-stimulated actin stress fiber formation and increased MLC phosphorylation in association with induction of a contractile cell morphology in TM cells, revealing a direct influence on actomyosin organization and contractile activity. TGF-beta, which also induces CTGF and ECM expression, has been reported to activate Rho GTPase activity and actomyosin-based contractile activity in different cell types, including TM cells.10,46–48 These observations suggest that similar to TGF-beta, CTGF, which regulates ECM synthesis, also controls actomyosin-based contractile activity. Importantly, as has been demonstrated in different cell types, CTGF (Fig. 4) and Rho GTPase induce ECM synthesis in TM cells.10,19
Additionally, to understand the broader impact of CTGF in TM cell biology, the cDNA microarray analysis of HTM cells treated with recombinant CTGF (for 24 hours) revealed increased expression of certain selected genes. Among the upregulated genes, IL-6, IL-32, and transcriptional regulator-C/EBP (delta) were the only ones related directly to the expression of ECM proteins. Intriguingly, many of the genes involved either in ECM synthesis or inflammation, including hyaluronan synthase-2, TSG-6 (TNF-alpha–induced protein-6), BMP2, IL-13, PTX3, MMP-1, IL-8, endocan, HMGA2, and angiopoietin-like-4 genes, were down-regulated by CTGF in TM cells. However, we did not determine the immediate effects of CTGF treatment on the expression profile of these genes. It appears that chronic exposure (24 hours) to CTGF triggers some negative feedback response on the expression of certain ECM regulatory genes, indicating the significance of CTGF in homeostasis of ECM synthesis. Consistent to our observation on the increased levels of fibronectin and laminin by CTGF, Junglas et al. also reported increased levels of various ECM proteins in TM cells with different doses of recombinant CTGF.19 Significantly, recent publication has demonstrated that Rho kinase inhibitors reduce CTGF-induced intraocular pressure and ECM protein expression in TM cells, further supporting the functional interaction between the Rho/Rho kinase pathway and CTGF.49
Interestingly, NMU, a structurally conserved neuropeptide involved in smooth muscle contraction, blood pressure, and blood flow was upregulated by CTGF stimulation of TM cells. This neuropeptide exists in one of 2 forms: a 23 to 25 amino acid peptide or an alternate form that is shorter by 15/17 amino acid residues. NMU mediates its effects through two G-protein coupled receptors (NMU-R1 and NMU-R2).26,50 NMU-R1 is known to be distributed in different tissues, whereas NMU-R2 was thought to be expressed predominantly in brain tissue.26 HTM cells derived from several donor eyes revealed expression of NMU and its receptors. CTGF appears to induce expression of NMU-R1 in TM cells in addition to NMU. NMU has been shown to regulate calcium and phosphoinositide signaling, and generate arachidonic acid by coupling to Gαq/11 and Gαi G-proteins.26,50,51 Interestingly, porcine NMU-8 peptide was found to induce actin stress fibers and MLC phosphorylation in TM cells, with the effects being more robust than those of CTGF. Further, consistent with earlier observations documented for agents that activate TM contraction (LPA, SIP, TGF-beta, and endothelin-1),10,27,34–36,40 perfusion of porcine eyes with NMU decreased AH outflow facility. However, unlike LPA, SIP, and TGF-beta, NMU induced transient changes in aqueous humor outflow facility that reverted back to normal within a short time, indicating its rapid influence on the homeostatic regulatory mechanisms to control the changes in AH outflow facility. Similar to NMU, CTGF also induces actin stress fibers and MLC phosphorylation in TM cells (Fig. 4), and has been reported to increase IOP.49 These similarities suggested that NMU partly mimics the CTGF response on AH outflow by regulating the actomyosin organization and contractile properties of TM cells. The possible influence of NMU on ECM proteins and MMPs is not known and yet to be determined in future studies. NMU-R1 and R2 also are targets for neuromedin-S (NMS), which exhibits some structural and functional similarity to NMU.52
In conclusion, our study provided a novel insight into existence of a critical regulatory interaction among Rho/Rho kinase-regulated contractile activity, expression of CTGF and NMU, and ECM synthesis in TM cells, and the possible significance of such interaction in homeostasis of AH humor outflow and IOP.
Footnotes
Supported by R01 Grant (EY018590) from the National Institutes of Health (NIH).
Disclosure: P. Iyer, None; R. Maddala, None; P.P. Pattabiraman, None; P.V. Rao, None
References
- 1.Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90:262–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kwon YH, Fingert JH, Kuehn MH, Alward WL. Primary open-angle glaucoma. N Engl J Med. 2009;360:1113–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gabelt BT, Kaufman PL. Changes in aqueous humor dynamics with age and glaucoma. Prog Retin Eye Res. 2005;24:612–637 [DOI] [PubMed] [Google Scholar]
- 4.Keller KE, Aga M, Bradley JM, Kelley MJ, Acott TS. Extracellular matrix turnover and outflow resistance. Exp Eye Res. 2009;88:676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuchshofer R, Tamm ER. Modulation of extracellular matrix turnover in the trabecular meshwork. Exp Eye Res. 2009;88:683–688 [DOI] [PubMed] [Google Scholar]
- 6.Acott TS, Kelley MJ. Extracellular matrix in the trabecular meshwork. Exp Eye Res. 2008;86:543–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sethi A, Jain A, Zode GS, Wordinger RJ, Clark AF. Role of TGFbeta/Smad signaling in gremlin induction of human trabecular meshwork extracellular matrix proteins. Invest Ophthalmol Vis Sci. 2011;52:5251–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradley JM, Vranka J, Colvis CM, et al. Effect of matrix metalloproteinases activity on outflow in perfused human organ culture. Invest Ophthalmol Vis Sci. 1998;39:2649–2658 [PubMed] [Google Scholar]
- 9.Sanka K, Maddala R, Epstein DL, Rao PV. Influence of actin cytoskeletal integrity on matrix metalloproteinase-2 activation in cultured human trabecular meshwork cells. Invest Ophthalmol Vis Sci. 2007;48:2105–2114 [DOI] [PubMed] [Google Scholar]
- 10.Pattabiraman PP, Rao PV. Mechanistic basis of Rho GTPase-induced extracellular matrix synthesis in trabecular meshwork cells. Am J Physiol Cell Physiol. 2010;298:C749–C763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cicha I, Goppelt-Struebe M. Connective tissue growth factor: context-dependent functions and mechanisms of regulation. Biofactors. 2009;35:200–208 [DOI] [PubMed] [Google Scholar]
- 12.Perbal B. CCN proteins: multifunctional signalling regulators. Lancet. 2004;363:62–64 [DOI] [PubMed] [Google Scholar]
- 13.Ihn H. Pathogenesis of fibrosis: role of TGF-beta and CTGF. Curr Opin Rheumatol. 2002;14:681–685 [DOI] [PubMed] [Google Scholar]
- 14.Leask A, Abraham DJ. TGF-beta signaling and the fibrotic response. FASEB J. 2004;18:816–827 [DOI] [PubMed] [Google Scholar]
- 15.Browne JG, Ho SL, Kane R, et al. Connective tissue growth factor is increased in pseudoexfoliation glaucoma. Invest Ophthalmol Vis Sci. 2011;52:3660–3666 [DOI] [PubMed] [Google Scholar]
- 16.Fuchshofer R, Yu AH, Welge-Lüssen U, Tamm ER. Bone morphogenetic protein-7 is an antagonist of transforming growth factor-beta2 in human trabecular meshwork cells. Invest Ophthalmol Vis Sci. 2007;48:715–726 [DOI] [PubMed] [Google Scholar]
- 17.Wordinger RJ, Fleenor DL, Hellberg PE, et al. Effects of TGF-beta2, BMP-4, and gremlin in the trabecular meshwork: implications for glaucoma. Invest Ophthalmol Vis Sci. 2007;48:1191–1200 [DOI] [PubMed] [Google Scholar]
- 18.Chudgar SM, Deng P, Maddala R, Epstein DL, Rao PV. Regulation of connective tissue growth factor expression in the aqueous humor outflow pathway. Mol Vis. 2006;12:1117–1126 [PubMed] [Google Scholar]
- 19.Junglas B, Yu AH, Welge-Lüssen U, Tamm ER, Fuchshofer R. Connective tissue growth factor induces extracellular matrix deposition in human trabecular meshwork cells. Exp Eye Res. 2009;88:1065–1075 [DOI] [PubMed] [Google Scholar]
- 20.Liton PB, Liu X, Challa P, Epstein DL, Gonzalez P. Induction of TGF-beta1 in the trabecular meshwork under cyclic mechanical stress. J Cell Physiol. 2005;205:364–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M, Maddala R, Rao PV. Novel molecular insights into RhoA GTPase-induced resistance to aqueous humor outflow through the trabecular meshwork. Am J Physiol Cell Physiol. 2008;295:C1057–C1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt G, Sehr P, Wilm M, Selzer J, Mann M, Aktories K. Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature. 1997;387:725–729 [DOI] [PubMed] [Google Scholar]
- 23.Rao PV, Deng PF, Kumar J, Epstein DL. Modulation of aqueous humor outflow facility by the Rho kinase-specific inhibitor Y-27632. Invest Ophthalmol Vis Sci. 2001;42:1029–1037 [PubMed] [Google Scholar]
- 24.Minamino N, Kangawa K, Matsuo H. Neuromedin U-8 and U-25: novel uterus stimulating and hypertensive peptides identified in porcine spinal cord. Biochem Biophys Res Commun. 1985;130:1078–1085 [DOI] [PubMed] [Google Scholar]
- 25.Westfall TD, McCafferty GP, Pullen M, et al. Characterization of neuromedin U effects in canine smooth muscle. J Pharmacol Exp Ther. 2002;301:987–992 [DOI] [PubMed] [Google Scholar]
- 26.Brighton PJ, Szekeres PG, Willars GB., Neuromedin U. and its receptors: structure, function, and physiological roles. Pharmacol Rev. 2004;56:231–248 [DOI] [PubMed] [Google Scholar]
- 27.Wiederholt M, Thieme H, Stumpff F. The regulation of trabecular meshwork and ciliary muscle contractility. Prog Retin Eye Res. 2000;19:271–295 [DOI] [PubMed] [Google Scholar]
- 28.Rao VP, Epstein DL. Rho GTPase/Rho kinase inhibition as a novel target for the treatment of glaucoma. BioDrugs. 2007;21:167–177 [DOI] [PubMed] [Google Scholar]
- 29.WuDunn D. Mechanobiology of trabecular meshwork cells. Exp Eye Res. 2009;88:718–723 [DOI] [PubMed] [Google Scholar]
- 30.Bershadsky AD, Balaban NQ, Geiger B. Adhesion-dependent cell mechanosensitivity. Annu Rev Cell Dev Biol. 2003;19:677–695 [DOI] [PubMed] [Google Scholar]
- 31.Ingber DE. Cellular mechanotransduction: putting all the pieces together again. FASEB J. 2006;20:811–827 [DOI] [PubMed] [Google Scholar]
- 32.Clark K, Langeslag M, Figdor CG, van Leeuwen FN. Myosin II and mechanotransduction: a balancing act. Trends Cell Biol. 2007;17:178–186 [DOI] [PubMed] [Google Scholar]
- 33.Chiquet M, Gelman L, Lutz R, Maier S. From mechanotransduction to extracellular matrix gene expression in fibroblasts. Biochim Biophys Acta. 2009;1793:911–920 [DOI] [PubMed] [Google Scholar]
- 34.Mettu PS, Deng PF, Misra UK, Gawdi G, Epstein DL, Rao PV. Role of lysophospholipid growth factors in the modulation of aqueous humor outflow facility. Invest Ophthalmol Vis Sci. 2004;45:2263–2271 [DOI] [PubMed] [Google Scholar]
- 35.Gottanka J, Chan D, Eichhorn M, Lutjen-Drecoll E, Ethier CR. Effects of TGF-beta2 in perfused human eyes. Invest Ophthalmol Vis Sci. 2004;45:153–158 [DOI] [PubMed] [Google Scholar]
- 36.Fleenor DL, Shepard AR, Hellberg PE, Jacobson N, Pang IH, Clark AF. TGFbeta2-induced changes in human trabecular meshwork: implications for intraocular pressure. Invest Ophthalmol Vis Sci. 2006;47:226–234 [DOI] [PubMed] [Google Scholar]
- 37.Kumar J, Epstein DL. Rho GTPase-mediated cytoskeletal organization in Schlemm's canal cells play a critical role in the regulation of aqueous humor outflow facility. J Cell Biochem. 2011;112:600–606 [DOI] [PubMed] [Google Scholar]
- 38.Wiederholt M, Bielka S, Schweig F, Lütjen-Drecoll E, Lepple-Wienhues A. Regulation of outflow rate and resistance in the perfused anterior segment of the bovine eye. Exp Eye Res. 1995;61:223–234 [DOI] [PubMed] [Google Scholar]
- 39.Stamer WD, Read AT, Sumida GM, Ethier CR. Sphingosine-1-phosphate effects on the inner wall of Schlemm's canal and outflow facility in perfused human eyes. Exp Eye Res. 2009;89:980–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rao PV, Deng P, Sasaki Y, Epstein DL. Regulation of myosin light chain phosphorylation in the trabecular meshwork: role in aqueous humour outflow facility. Exp Eye Res. 2005;80:197–206 [DOI] [PubMed] [Google Scholar]
- 41.Rao PV, Deng P, Maddala R, Epstein DL, Li CY, Shimokawa H. Expression of dominant negative Rho-binding domain of Rho-kinase in organ cultured human eye anterior segments increases aqueous humor outflow. Mol Vis. 2005;11:288–297 [PubMed] [Google Scholar]
- 42.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635 [DOI] [PubMed] [Google Scholar]
- 43.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701 [DOI] [PubMed] [Google Scholar]
- 44.Schoenwaelder SM, Burridge K. Bidirectional signaling between the cytoskeleton and integrins. Curr Opin Cell Biol. 1999;11:274–286 [DOI] [PubMed] [Google Scholar]
- 45.Chaqour B, Goppelt-Struebe M. Mechanical regulation of the Cyr61/CCN1 and CTGF/CCN2 proteins. FEBS J. 2006;273:3639–3649 [DOI] [PubMed] [Google Scholar]
- 46.Nakamura Y, Hirano S, Suzuki K, Seki K, Sagara T, Nishida T. Signaling mechanism of TGF-beta1-induced collagen contraction mediated by bovine trabecular meshwork cells. Invest Ophthalmol Vis Sci. 2002;43:3465–3472 [PubMed] [Google Scholar]
- 47.Bhowmick NA, Ghiassi M, Bakin A, et al. Transforming growth factor-beta1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol Biol Cell. 2001;12:27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maddala R, Reddy VN, Epstein DL, Rao V. Growth factor induced activation of Rho and Rac GTPases and actin cytoskeletal reorganization in human lens epithelial cells. Mol Vis. 2003;9:329–336 [PubMed] [Google Scholar]
- 49.Junglas B, Kuespert S, Seleem AA, et al. Connective tissue growth factor causes glaucoma by modifying the actin cytoskeleton of the trabecular meshwork. Am J Pathol. 2012;180:2386–2403 [DOI] [PubMed] [Google Scholar]
- 50.Mitchell JD, Maguire JJ, Davenport AP. Emerging pharmacology and physiology of neuromedin U and the structurally related peptide neuromedin S. Br J Pharmacol. 2009;158:87–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brighton PJ, Szekeres PG, Wise A, Willars GB. Signaling and ligand binding by recombinant neuromedin U receptors: evidence for dual coupling to Galphaq/11 and Galphai and an irreversible ligand-receptor interaction. Mol Pharmacol. 2004;66:1544–1556 [DOI] [PubMed] [Google Scholar]
- 52.Mori K, Miyazato M, Ida T, et al. Identification of neuromedin S and its possible role in the mammalian circadian oscillator system. EMBO J. 2005;24:325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]