Abstract
Purpose.
Nonsyndromic high myopia, defined by a refractive error greater than −6 diopters (D), is associated with an increased risk of macular choroidal neovascularization (CNV), a vision-threatening complication. The aim of this study was to investigate whether genetic factors associated with age-related macular degeneration (AMD) are related to myopic CNV.
Methods.
We conducted a case-control study, including 71 cases with myopic CNV and 196 myopic controls without CNV, from Creteil and Toulouse, France, and Boston, MA. Single nucleotide polymorphisms (SNPs) from 15 genes reported to be related to AMD were selected for association testing in this study.
Results.
In univariate analysis, the rs10033900 SNP located in CFI was associated with myopic CNV (P = 0.0011), and a SNP in APOE was also related (P = 0.041). After adjustment for age, sex, and degree of myopia, SNPs in three genes were significantly associated, including CFI (odds ratio [OR] 2.1, 95% confidence interval [CI] 1.3–3.37, P = 0.0023), COL8A1 (OR 1.88, 95% CI 1.18–2.98, P = 0.0076), and CFH (OR 1.65, 95% CI 1.02–2.66, P = 0.04). After correction for multiple testing, only CFI remained significantly related to high myopic CNV (P = 0.045).
Conclusions.
We report the first genetic associations with choroidal neovascularization (CNV) in a high myopic Caucasian population. One SNP (rs10033900) in the CFI gene, which encodes a protein involved in the inflammatory pathway, was significantly associated with myopic CNV in multivariate analysis after correction for multiple testing. This SNP is a plausible biological marker associated with CNV outgrowth among high myopic patients. Results generate hypotheses about potential loci related to CNV in high myopia, and larger studies are needed to expand on these findings.
We report herein the first genetic associations with choroidal neovascularization (CNV) in a high myopic Caucasian population. One SNP in CFI was significantly associated with myopic CNV in multivariate analysis after correction for multiple testing.
Introduction
High myopia or pathologic myopia is defined by an axial length higher than 26 mm or by a refractive error more than −6 diopters (D) with pathological modifications of the posterior pole of the retina, including staphyloma, lacquer cracks, and myopic conus. High myopia is a common vision-threatening disease that affects 0.5% to 5.0% of the worldwide population.1–3 Choroidal neovascularization (CNV) is the most common cause of visual loss related to this disorder, with an estimated prevalence of 4% to 11% among high myopic patients, and there is a 2-fold higher risk among women in some studies.4,5
Genetic factors have been described in nonsyndromic high myopia through linkage analysis, genome-wide association analysis, or candidate gene case-control studies.6–25 However, the genetic factors influencing the risk of CNV in eyes with high myopia have not been extensively investigated.26,27 Several genetic factors have been strongly associated with exudative age-related macular degeneration (AMD), another degenerative retinal disease characterized by a neovascular process developing from the choroid beneath the neurosensory retina located in the macular area of the retina.28–50 Therefore, we hypothesized that genes associated with exudative AMD could be considered as candidate genes for myopic CNV.
Materials and Methods
Participants
High myopic patients with axial myopia more than −6 D and pathologic myopic retinal degeneration were recruited from three different centers (Créteil, Toulouse, and Boston). Cases had high myopia with CNV in one or both eyes. The control group was defined as high myopic patients without CNV with visual acuity of 20/32 or better in both eyes. Only subjects of European/Caucasian ancestry were included. Demographic data and ocular characteristics of cases and controls are shown in Table 1.
Table 1. .
Characteristics of High Myopic Patients with CNV (Cases) and High Myopic Patients without CNV (Controls)
|
Variable |
Cases (n = 71) |
Controls (n = 196) |
P |
| Age at diagnosis, y | 53.9 ± 14.9 (13 to 85) | 40.9 ± 13.9 (19 to 88) | 3.2 × 10−9 |
| Sex, % female (n) | 73.2% (52) | 69.4% (136) | 0.65 |
| Refractive error, diopters | |||
| OD | −12.1 ± 5.3 (−6 to −27) | −9.2 ± 3.0 (−6 to −22) | 1.4 × 10−4 |
| OS | −11.9 ± 5.0 (−6 to −23) | −9.2 ± 3.2 (−6 to −21) | 1.3 × 10−4 |
| Site | |||
| France | 62 (87%) | 182 (91%) | 0.24 |
| United States | 9 (13%) | 14 (7%) |
Values denote means ± SDs and ranges or percentages.
values for age and refractive error are calculated by two-tailed t-test.
values for sex and site are calculated by χ2 test.
All patients with myopic CNV underwent complete clinical examination, including visual acuity assessment, dilated fundus examination, and fluorescein angiography. The diagnosis of CNV was based on fundus examination showing a macular scar or a lesion with subretinal hemorrhages in the absence of drusen in either eye, and/or by staining and leakage on early and late phases of fluorescein angiography. An indocyanine green angiography (ICG) scan and an optical coherence tomography (OCT) scan were also performed to confirm the diagnosis of CNV in some cases. On OCT scans, CNV appeared as a hyper-reflective lesion located beneath the neurosensory retina, usually associated with a hyporeflective intraretinal or subretinal accumulation of fluid. On ICG, CNV could be seen as a network in the early phase or as a hyperfluorescent macular lesion in the late phase, sometimes spreading from lacquer cracks or an atrophic patch that appeared hypofluorescent on the late phase. Myopic patients without CNV underwent visual acuity assessment and a dilated fundus examination.
Patients with clinical features of AMD, including drusen or pigmentary changes, or other retinal diseases (i.e., diabetic retinopathy, ocular histoplasmosis syndrome, lacquer cracks due to trauma) related to CNV were excluded. To avoid potential spurious findings due to population admixture, non-Caucasian subjects were also excluded.
Written informed consent was obtained for each individual participating in this study, and all clinical investigations were conducted according to the principles expressed in the Declaration of Helsinki.
SNP Selection
We selected a total of 30 SNPs located in 15 candidate genes or genetic regions related to AMD based on previous reports of whole-genome linkage studies, genome-wide association studies (GWAS), and molecular and functional studies.28–51 We chose SNPs that were reported previously or SNPs tagging each candidate gene/region of transcription.52 Tagging SNPs with a minor allele frequency (MAF) greater than 10% and with a minimum r2 of 0.8 for the tagging region were selected by tagger (http://www.broad.mit.edu/mpg/tagger/) based on the HapMap data from the population of Utah residents with ancestry from northern and western Europe (phase II, http://www.hapmap.org).
Genotyping
Genotyping was performed in the Clinical and Translation Research Center Core Laboratory, Tufts Clinical and Translational Science Institute, Boston, MA, using Applied Biosystem (ABI) BioTrove OpenArray Genotyping Platform and ABI 7900HT Sequence Detection System (Life Technologies, Carlsbad, CA). OpenArray is a new platform designed for medium-throughput genotyping by ABI. SNPs were genotyped on a metal-based array of negatively charged wells in which DNA samples were amplified with TaqMan genotyping chemistry. Per the manufacturer instruction, the samples were loaded onto the assay plate, followed by PCR amplification and imaging on the Open Array NT Imager (Life Technologies). The results were analyzed with TaqMan Genotyper Software (Life Technologies). Three SNPs failed in assay design or genotyping assay on the OpenArray platform and these markers were then genotyped with TaqMan SNP genotyping assay on ABI 7900HT SDS.
Statistical Analyses
Quality control, allele frequency estimation, and tests for association were performed for each SNP using PLINK 1.07.53 SNPs failing the following quality control criteria were excluded from the analyses: missingness rate more than 0.1, minor allele frequency (MAF) less than 0.01, deviant from Hardy-Weinberg equilibrium (HWE) (P < 0.001), or with significantly different missingness in case and control groups (P < 0.001). The resultant SNP set for analysis contained a total of 29 SNPs that complied with the above quality control criteria. Details of these SNPs are shown in Table 2. For each SNP, the genotypes were coded as 0, 1, or 2 by copies of the minor allele based on an additive genetic model. Univariate analysis was performed using logistic regression. Differences in the distribution of each potential confounding factor between cases and controls were tested using a t-test (for continuous age, refractive error) or χ2 test (for sex, site). To adjust for potential confounding factors of age, sex, and degree of myopia, SNPs were further tested using a multivariate logistic regression model. To adjust for multiple testing, corrected P values were calculated by the max (T) procedure (10,000 permutations) in PLINK.53,54 The association between APOE haplotypes and myopic CNV were analyzed by the haplotype-based association tests with logistic models in PLINK.
Table 2. .
Candidate SNPs Tested for Association with Myopic CNV
|
SNP |
Chr |
Genomic Location (hg19) |
Gene |
Minor Allele Frequencies |
OR* (95% CI) |
P Value* |
Corrected P Value† |
OR (95% CI) Adjusted for Covariates‡ |
P Value Adjusted for Covariates‡ |
Corrected P Value Adjusted for Covariates§ |
||
|
Cases |
Controls |
Allele |
||||||||||
| A. SNPs significantly associated with myopic CNV in 1 or more analyses | ||||||||||||
| rs10033900 | 4 | 110,659,067 | CFI | 0.51 | 0.35 | T | 1.95 (1.31–2.92) | 0.0011 | 0.021 | 2.1 (1.3–3.37) | 0.0023 | 0.045 |
| rs669676 | 3 | 99,448,852 | COL8A1 | 0.56 | 0.47 | A | 1.42 (0.96–2.08) | 0.08 | 0.86 | 1.88 (1.18–2.98) | 0.0076 | 0.15 |
| rs1061170 | 1 | 196,659,237 | CFH | 0.42 | 0.34 | C | 1.47 (0.98–2.21) | 0.064 | 0.79 | 1.65 (1.02–2.66) | 0.04 | 0.63 |
| rs769455 | 19 | 45,412,040 | APOE | 0.039 | 0.005 | T | 5.44 (1.07–27.56) | 0.041 | 0.61 | 4.54 (0.91–22.75) | 0.066 | 0.82 |
| B. Other SNPs tested for association with myopic CNV | ||||||||||||
| rs10737680 | 1 | 196,679,455 | CFH | 0.37 | 0.42 | C | 0.78 (0.51–1.18) | 0.24 | 1 | 0.77 (0.48–1.26) | 0.3 | 1 |
| rs1410996 | 1 | 196,696,933 | CFH | 0.38 | 0.43 | A | 0.76 (0.5–1.17) | 0.21 | 1 | 0.77 (0.48–1.25) | 0.29 | 1 |
| rs7645305 | 3 | 99,363,985 | COL8A1 | 0.28 | 0.22 | G | 1.39 (0.89–2.16) | 0.15 | 0.98 | 1.26 (0.76–2.09) | 0.37 | 1 |
| rs793494 | 3 | 99,508,768 | COL8A1 | 0.29 | 0.31 | A | 0.91 (0.6–1.38) | 0.64 | 1 | 1.14 (0.71–1.82) | 0.6 | 1 |
| rs9332739 | 6 | 31,903,804 | C2 | 0.028 | 0.043 | C | 0.63 (0.2–1.93) | 0.41 | 1 | 0.46 (0.14–1.53) | 0.2 | 1 |
| rs641153 | 6 | 31,914,180 | CFB | 0.103 | 0.14 | A | 0.72 (0.4–1.3) | 0.27 | 1 | 0.59 (0.3–1.16) | 0.12 | 0.97 |
| rs699947 | 6 | 43,736,389 | VEGFA | 0.5 | 0.49 | A | 1.06 (0.71–1.56) | 0.79 | 1 | 1.11 (0.72–1.72) | 0.63 | 1 |
| rs13207351 | 6 | 43,737,794 | VEGFA | 0.51 | 0.49 | A | 1.1 (0.75–1.63) | 0.62 | 1 | 1.18 (0.77–1.81) | 0.44 | 1 |
| rs735286 | 6 | 43,744,621 | VEGFA | 0.27 | 0.29 | T | 0.89 (0.57–1.4) | 0.61 | 1 | 0.82 (0.49–1.36) | 0.44 | 1 |
| rs2146323 | 6 | 43,745,095 | VEGFA | 0.35 | 0.35 | A | 0.98 (0.65–1.48) | 0.92 | 1 | 1.03 (0.64–1.64) | 0.92 | 1 |
| rs3025021 | 6 | 43,749,163 | VEGFA | 0.32 | 0.34 | T | 0.88 (0.58–1.34) | 0.54 | 1 | 0.91 (0.56–1.46) | 0.69 | 1 |
| rs3025039 | 6 | 43,752,536 | VEGFA | 0.13 | 0.11 | T | 1.22 (0.67–2.2) | 0.52 | 1 | 1.22 (0.61–2.44) | 0.57 | 1 |
| rs4711751 | 6 | 43,828,582 | VEGFA | 0.48 | 0.46 | C | 1.07 (0.72–1.59) | 0.75 | 1 | 1.17 (0.73–1.85) | 0.52 | 1 |
| rs25648 | 6 | 43,846,955 | VEGFA | 0.2 | 0.17 | T | 1.21 (0.74–2) | 0.45 | 1 | 1.35 (0.75–2.42) | 0.32 | 1 |
| rs1999930 | 6 | 116,387,134 | FRK/COL10A1 | 0.36 | 0.33 | G | 1.14 (0.77–1.68) | 0.53 | 1 | 1.07 (0.67–1.68) | 0.79 | 1 |
| rs12196141 | 6 | 116,489,550 | COL10A1 | 0.35 | 0.29 | G | 1.27 (0.85–1.91) | 0.25 | 1 | 1.17 (0.73–1.86) | 0.52 | 1 |
| rs1883025 | 9 | 107,664,301 | ABCA1 | 0.33 | 0.30 | T | 1.16 (0.77–1.74) | 0.48 | 1 | 1.28 (0.8–2.05) | 0.3 | 1 |
| rs10490924 | 10 | 124,214,448 | ARMS2 | 0.23 | 0.19 | T | 1.3 (0.81–2.08) | 0.28 | 1 | 1.42 (0.83–2.42) | 0.2 | 1 |
| rs10468017 | 15 | 58,678,512 | LIPC | 0.24 | 0.3 | T | 0.71 (0.45–1.13) | 0.15 | 0.98 | 0.81 (0.48–1.36) | 0.42 | 1 |
| rs493258 | 15 | 58,687,880 | LIPC | 0.43 | 0.49 | C | 0.79 (0.54–1.17) | 0.24 | 1 | 0.76 (0.48–1.19) | 0.22 | 1 |
| rs3764261 | 16 | 56,993,324 | CETP | 0.25 | 0.31 | A | 0.76 (0.49–1.17) | 0.21 | 1 | 0.68 (0.41–1.12) | 0.13 | 0.97 |
| rs2230199 | 19 | 6,718,387 | C3 | 0.21 | 0.17 | C | 1.28 (0.78–2.08) | 0.33 | 1 | 1.32 (0.76–2.28) | 0.33 | 1 |
| rs429358 | 19 | 50,103,781 | APOE | 0.11 | 0.13 | C | 0.86 (0.46–1.61) | 0.64 | 1 | 1.02 (0.5–2.06) | 0.96 | 1 |
| rs7412 | 19 | 50,103,919 | APOE | 0.09 | 0.06 | T | 1.53 (0.76–3.06) | 0.23 | 1 | 1.41 (0.62–3.18) | 0.41 | 1 |
| rs9621532 | 22 | 33,084,511 | TIMP3 | 0.06 | 0.049 | C | 1.17 (0.48–2.83) | 0.73 | 1 | 1.02 (0.38–2.75) | 0.97 | 1 |
values in bold are significant (P < 0.05).
Chr, chromosome; OR, odds ratios per allele comparing allele frequencies between myopic patients with CNV with myopic patients without CNV.
ORs and P values for univariate logistic regression model.
P values for univariate analysis corrected for multiple testing by 10,000 permutations.
ORs and P values for multivariate logistic regression model adjusting for age, sex, and spherical equivalent of worse eye.
P values for multivariate analysis corrected for multiple testing by 10,000 permutations.
Results
As shown in Table 1, cases included 71 patients with high myopia in both eyes with a refractive error greater than −6 D in both eyes and CNV in one or both eyes. Controls consisted of 196 individuals with high myopia not complicated by CNV. Cases tended to be older than controls: mean ± SD age at diagnosis for cases was 53.9 ± 14.9 years for cases, and 40.9 ± 13.9 years for controls (P = 3.2 × 10−9). Sex distributions were balanced between cases and controls (P = 0.65). Cases had a higher degree of myopia than controls (P = 1.4 × 10−4 for OD and P = 1.3 × 10−4 for OS). On average, there was a difference of 3D between cases and controls.
Table 2 shows the allele frequencies, odds ratios (OR) and P values for the candidate SNPs and associations with high myopic CNV. In univariate logistic regression analyses, two of the SNPs were significantly associated with high myopic CNV: rs10033900 in the CFI gene (P = 0.0011, OR = 1.95 [95% confidence interval (CI) 1.31–2.92]) and rs769455 in the APOE gene (P = 0.041, OR = 5.44 [95% CI 1.07–27.56]). Only CFI rs10033900 remained significantly associated with myopic CNV after correction for multiple testing (P = 0.021) in the univariate model.
In the multivariate model, with adjustment for age, sex, and degree of myopia, the T allele of rs10033900 in the CFI gene was significant (P = 0.0023, OR = 2.1 [95% CI 1.3–3.37]) and remained significant after correction for multiple testing (P = 0.045). Two other SNPs, rs669676 in COL8A1 (P = 0.0076, OR = 1.88 [95% CI 1.18–2.98]) and rs1061170 in CFH (P = 0.04, OR = 1.65 [95% CI 1.02–2.66]) were associated with myopic CNV in the multivariate analysis but were not significant after correction for multiple testing. Rs769455 in APOE was significant in the uncorrected univariate analysis, but was no longer significant (P = 0.066) after adjustment for the covariates. The APOE haplotypes (E2, E3, E4) were not significantly associated with myopic CNV in either univariate or multivariate analysis (see Supplementary Material and Supplementary Table S1, http://www.iovs.org/content/53/8/5004/suppl/DC1).
Discussion
Despite some similarities between AMD and myopic CNV, including macular CNV, subretinal location of the CNV outgrowth, and some degree of atrophy, no genetic variants have been previously reported to be associated with myopic CNV among Caucasians. In this study, we evaluated several AMD genetic variants that could also be involved in myopic CNV development. Following a candidate gene approach, this case-control study identified one SNP in the CFI gene significantly associated with myopic CNV even after adjusting for confounding factors and multiple testing. Other SNPs that showed suggestive evidence for association are worthy of further exploration as well.
The T allele of rs10033900, which is located 2781 bp upstream of the 3′ untranslated region of the complement factor I gene (CFI), has been associated with increased risk of exudative AMD in Caucasians.44,47 Both complement factors I and H are complement regulatory proteins. CFH encodes factor H, the most important alternative pathway discriminator that binds C3b and prevents the formation of C3 convertase and acts as a cofactor of factor I to cleave C3b in iC3b.49 CFI is expressed by hepatocytes, macrophages, lymphocytes, endothelial cells, and fibroblasts and encodes factor I, a regulator protein of the three complement pathways.50 By cleaving of C3b and C4b, factor I reduces the formation of the C3 and C5 convertase enzymes.50 Other genes in the complement cascade pathway are associated with exudative AMD, including CFH, C2, CFB, and C3.34–36,39–43 Interestingly, CFH (rs1061170, P = 0.04) and CFI (rs10033900, P = 0.0023) appeared to be related to myopic CNV in this study after adjustment for age, sex, and degree of myopia, and CFI remained significant after adjustment for multiple testing. The possible difference in effect of CFI compared with CFH on myopic CNV risk may be related to the fact that factor I is involved in the classic, lectin and alternative pathways, while factor H is only involved in the regulation of the alternative pathway.55 It is interesting to note that the gene on chromosome 10, ARMS2/HTRA1, was not related to myopic CNV in this study even though this gene is more strongly associated with CNV compared with geographic atrophy in AMD47,56 and also more strongly associated with all AMD subtypes when compared with the CFH at-risk common variant.57 It is also noteworthy that the SNPs in the VEGF gene were not related to myopic CNV, given that VEGF rs4711751 is related to advanced dry and exudative AMD.47
The intronic SNP rs669676 in the COL8A1 gene was associated with myopic CNV after adjustment for age, sex, and degree of myopia (P = 0.0076). This gene encodes one of the two alpha chains of type VIII collagen, a major component of basement membranes of Bruch's membrane and choroidal stroma.58 The intronic SNP rs13095226 in this gene is associated with advanced AMD in our previous studies.45,47 The SNP rs669676 of COL8A1 might lead to direct or indirect structural alterations of the Bruch's membrane as frequently observed during high myopia (Fig.), which is a risk factor for myopic CNV.59
Figure. .
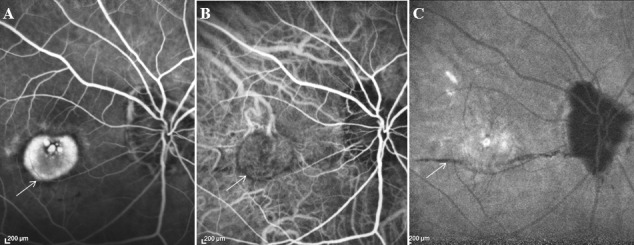
High myopic patient (−12 D) with CNV in the right eye. (A) Fluorescein angiography showing CNV (white arrow). (B) Indocyanine green angiography showing the CNV in the early phase (white arrow). (C) Indocyanine green angiography showing a lacquer crack in the late phase (white arrow).
The association between the E4 APOE haplotype and a reduced risk of AMD has been described in two independent case-control studies,32,33 and supported by other studies or in meta-analyses.60–62 The lipid component of soft drusen observed in AMD and the genotypic correlations between APOE and macular pigment63 could possibly support the hypothesis of a genetic association between this gene and CNV due to high myopia or AMD. However, associations between AMD and APOE are not consistent48 and the APOE gene is known to be linked to human longevity.64 In a murine model, apoE4 mice showed a more severe AMD-like pathological phenotype and also developed marked CNV, a hallmark of exudative AMD.65 In this study, although we did not find significant evidence supporting the association between E2, E3, and E4 APOE haplotypes and myopic CNV (see Supplementary Material and Supplementary Table S1, http://www.iovs.org/content/53/8/5004/suppl/DC1), we found suggestive evidence of association between myopic CNV and another SNP (rs769455), which is a rare variant of the APOE gene previously implicated with type III hyperlipoproteinemia.51 This was not significant after correction for multiple testing, however. Additional studies are required to replicate this result and elucidate the roles of this gene in myopic CNV etiology.
A limitation of this study is the relatively small sample size, especially for cases with myopic CNV; however, the study is strengthened by at least two factors. First, all participants came from a similar ethnic background, which reduced the chances of heterogeneity in different populations. Second, both the cases and controls were high myopic patients with high myopic genetic profile, which enhanced the ability to detect susceptible loci for myopic CNV. In contrast, comparing myopic CNV patients with nonmyopic controls may be confounded by the genetic and environmental factors influencing the risk of myopia.
To our knowledge, this study is the first to explore specific genetic effects influencing risk of CNV in high myopic patients compared with controls who are also highly myopic. This study suggests that the inflammatory pathway may be associated with myopic CNV, a vision-threatening complication of high myopia, through CFI. Larger studies are needed to analyze this gene and other candidate loci for this important vision-threatening complication of high myopia.
Supplementary Material
Acknowledgments
We thank Joelle Dumas, Jérôme Barré, and Patrick Ledudal for providing technical assistance and Mark Daly for helpful suggestions.
Footnotes
Supported in part by Grant RO1-EY11309 from the National Institutes of Health, Bethesda, Maryland; Massachusetts Lions Eye Research Fund, Inc., New Bedford, Massachusetts; an unrestricted grant from Research to Prevent Blindness, Inc., New York, New York to Tufts University School of Medicine; and the Macular Degeneration Research Fund of the Ophthalmic Epidemiology and Genetics Service, New England Eye Center, Tufts Medical Center, Tufts University School of Medicine, Boston, Massachusetts.
These authors contributed equally to the work presented here and should therefore be regarded as equivalent authors.
Presented at the annual meeting of the Association for Research in Vision and Ophthalmology in Fort Lauderdale, Florida, May 9, 2012.
Disclosure: N. Leveziel, None; Y. Yu, None; R. Reynolds, None; A. Tai, None; W. Meng, None; V. Caillaux, None; P. Calvas, None; B. Rosner, None; F. Malecaze, None; E.H. Souied, None; J.M. Seddon, None
References
- 1.Sawada A, Tomidokoro A, Araie M, Iwase A, Yamamoto T. Refractive errors in an elderly Japanese population: the Tajimi study. Ophthalmology. 2008;115:363–370.e3 [DOI] [PubMed] [Google Scholar]
- 2.Vitale S, Ellwein L, Cotch MF, Ferris FL III, Sperduto R. Prevalence of refractive error in the United States, 1999–2004. Arch Ophthalmol. 2008;126:1111–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noble KG, Carr RE. Pathologic myopia. Ophthalmology. 1982;89:1099–1100 [DOI] [PubMed] [Google Scholar]
- 4.Avila MP, Weiter JJ, Jalkh AE, Trempe CL, Pruett RC, Schepens CL. Natural history of choroidal neovascularization in degenerative myopia. Ophthalmology. 1984;91:1573–1581 [DOI] [PubMed] [Google Scholar]
- 5.Vongphanit J, Mitchell P, Wang JJ. Prevalence and progression of myopic retinopathy in an older population. Ophthalmology. 2002;109:704–711 [DOI] [PubMed] [Google Scholar]
- 6.Li YJ, Goh L, Khor CC, et al. Genome-wide association studies reveal genetic variants in CTNND2 for high myopia in Singapore Chinese. Ophthalmology. 2011;118:368–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo X, Xiao X, Li S, Wang P, Jia X, Zhang Q. Nonsyndromic high myopia in a Chinese family mapped to MYP1: linkage confirmation and phenotypic characterization. Arch Ophthalmol. 2010;128:1473–1479 [DOI] [PubMed] [Google Scholar]
- 8.Ma JH, Shen SH, Zhang GW, et al. Identification of a locus for autosomal dominant high myopia on chromosome 5p13.3-p15.1 in a Chinese family. Mol Vis. 2010;16:2043–2054 [PMC free article] [PubMed] [Google Scholar]
- 9.Khor CC, Fan Q, Goh L, et al. Support for TGFB1 as a susceptibility gene for high myopia in individuals of Chinese descent. Arch Ophthalmol. 2010;128:1081–1084 [DOI] [PubMed] [Google Scholar]
- 10.Metlapally R, Ki CS, Li YJ, et al. Genetic association of insulin-like growth factor-1 polymorphisms with high-grade myopia in an international family cohort. Invest Ophthalmol Vis Sci. 2010;51:4476–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin HJ, Wan L, Tsai Y, Chen WC, Tsai SW, Tsai FJ. The association between lumican gene polymorphisms and high myopia. Eye (Lond). 2010;24:1093–1101 [DOI] [PubMed] [Google Scholar]
- 12.Nakanishi H, Yamada R, Gotoh N, et al. A genome-wide association analysis identified a novel susceptible locus for pathological myopia at 11q24.1. PLoS Genet. 2009;5:e1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanovitch T, Li YJ, Metlapally R, Abbott D, Viet KN, Young TL. Hepatocyte growth factor and myopia: genetic association analyses in a Caucasian population. Mol Vis. 2009;15:1028–1035 [PMC free article] [PubMed] [Google Scholar]
- 14.Metlapally R, Li YJ, Tran-Viet KN, et al. COL1A1 and COL2A1 genes and myopia susceptibility: evidence of association and suggestive linkage to the COL2A1 locus. Invest Ophthalmol Vis Sci. 2009;50:4080–4086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li YJ, Guggenheim JA, Bulusu A, et al. An international collaborative family-based whole-genome linkage scan for high-grade myopia. Invest Ophthalmol Vis Sci. 2009;50:3116–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paget S, Julia S, Vitezica ZG, Soler V, Malecaze F, Calvas P. Linkage analysis of high myopia susceptibility locus in 26 families. Mol Vis. 2008;14:2566–2574 [PMC free article] [PubMed] [Google Scholar]
- 17.Han W, Leung KH, Fung WY, et al. Association of PAX6 polymorphisms with high myopia in Han Chinese nuclear families. Invest Ophthalmol Vis Sci. 2009;50:47–56 [DOI] [PubMed] [Google Scholar]
- 18.Nishizaki R, Ota M, Inoko H, et al. New susceptibility locus for high myopia is linked to the uromodulin-like 1 (UMODL1) gene region on chromosome 21q22.3. Eye (Lond). 2009;23:222–229 [DOI] [PubMed] [Google Scholar]
- 19.Lam CY, Tam PO, Fan DS, et al. A genome-wide scan maps a novel high myopia locus to 5p15. Invest Ophthalmol Vis Sci. 2008;49:3768–3778 [DOI] [PubMed] [Google Scholar]
- 20.Tsai YY, Chiang CC, Lin HJ, Lin JM, Wan L, Tsai FJA. PAX6 gene polymorphism is associated with genetic predisposition to extreme myopia. Eye (Lond). 2008;22:576–581 [DOI] [PubMed] [Google Scholar]
- 21.Chen CY, Stankovich J, Scurrah KJ, et al. Linkage replication of the MYP12 locus in common myopia. Invest Ophthalmol Vis Sci. 2007;48:4433–4439 [DOI] [PubMed] [Google Scholar]
- 22.Zhang Q, Guo X, Xiao X, Jia X, Li S, Hejtmancik JF. A new locus for autosomal dominant high myopia maps to 4q22-q27 between D4S1578 and D4S1612. Mol Vis. 2005;11:554–560 [PubMed] [Google Scholar]
- 23.Paluru PC, Nallasamy S, Devoto M, Rappaport EF, Young TL. Identification of a novel locus on 2q for autosomal dominant high-grade myopia. Invest Ophthalmol Vis Sci. 2005;46:2300–2307 [DOI] [PubMed] [Google Scholar]
- 24.Farbrother JE, Kirov G, Owen MJ, Pong-Wong R, Haley CS, Guggenheim JA. Linkage analysis of the genetic loci for high myopia on 18p, 12q, and 17q in 51 UK families. Invest Ophthalmol Vis Sci. 2004;45:2879–2885 [DOI] [PubMed] [Google Scholar]
- 25.Paluru P, Ronan SM, Heon E, et al. New locus for autosomal dominant high myopia maps to the long arm of chromosome 17. Invest Ophthalmol Vis Sci. 2003;44:1830–1836 [DOI] [PubMed] [Google Scholar]
- 26.Nakanishi H, Gotoh N, Yamada R, et al. ARMS2/HTRA1 and CFH polymorphisms are not associated with choroidal neovascularization in highly myopic eyes of the elderly Japanese population. Eye (Lond). 2010;24:1078–1084 [DOI] [PubMed] [Google Scholar]
- 27.Fernandez-Robredo P, Maestre SR, Zarranz-Ventura J, Mulero HH, Salinas-Alaman A, Garcia-Layana A. Myopic choroidal neovascularization genetics. Ophthalmology. 2008;115:1632, 1632.e1 [DOI] [PubMed] [Google Scholar]
- 28.Haddad S, Chen CA, Santangelo SL, Seddon JM. The genetics of age-related macular degeneration: a review of progress to date. Surv Ophthalmol. 2006;51:316–363 [DOI] [PubMed] [Google Scholar]
- 29.Seddon JM, Sobrin L. Epidemiology of age-related macular degeneration. In: Ryan SJ.ed Retina. St. Louis, MO: C.V. Mosby; 2012. In press [Google Scholar]
- 30.Katta S, Kaur I, Chakrabarti S. The molecular genetic basis of age-related macular degeneration: an overview. J Genet. 2009;88:425–449 [DOI] [PubMed] [Google Scholar]
- 31.Edwards AO. Genetics of age-related macular degeneration. Adv Exp Med Biol. 2008;613:211–219 [DOI] [PubMed] [Google Scholar]
- 32.Klaver CC, Kliffen M, van Duijn CM, et al. Genetic association of apolipoprotein E with age-related macular degeneration. Am J Hum Genet. 1998;63:200–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Souied EH, Benlian P, Amouyel P, et al. The epsilon4 allele of the apolipoprotein E gene as a potential protective factor for exudative age-related macular degeneration. Am J Ophthalmol. 1998;125:353–359 [DOI] [PubMed] [Google Scholar]
- 34.Edwards AO, Ritter R III, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424 [DOI] [PubMed] [Google Scholar]
- 35.Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haines JL, Hauser MA, Schmidt S, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421 [DOI] [PubMed] [Google Scholar]
- 37.Rivera A, Fisher SA, Fritsche LG, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14:3227–3236 [DOI] [PubMed] [Google Scholar]
- 38.Dewan A, Liu M, Hartman S, et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314:989–992 [DOI] [PubMed] [Google Scholar]
- 39.Gold B, Merriam JE, Zernant J, et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat genet. 2006;38:458–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maller J, George S, Purcell S, et al. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat Genet. 2006;38:1055–1059 [DOI] [PubMed] [Google Scholar]
- 41.Thakkinstian A, Han P, McEvoy M, et al. Systematic review and meta-analysis of the association between complement factor H Y402H polymorphisms and age-related macular degeneration. Human Mol Genet. 2006;15:2784–2790 [DOI] [PubMed] [Google Scholar]
- 42.Maller JB, Fagerness JA, Reynolds RC, Neale BM, Daly MJ, Seddon JM. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nat Genet. 2007;39:1200–1201 [DOI] [PubMed] [Google Scholar]
- 43.Yates JR, Sepp T, Matharu BK, et al. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357:553–561 [DOI] [PubMed] [Google Scholar]
- 44.Fagerness JA, Maller JB, Neale BM, Reynolds RC, Daly MJ, Seddon JM. Variation near complement factor I is associated with risk of advanced AMD. Eur J Hum Genet. 2009;17:100–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neale BM, Fagerness J, Reynolds R, et al. Genome-wide association study of advanced age-related macular degeneration identifies a role of the hepatic lipase gene (LIPC). Proc Nat Acad Sci U S A. 2010;107:7395–7400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen W, Stambolian D, Edwards AO, et al. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc Nat Acad Sci U S A. 2010;107:7401–7406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu Y, Bhangale TR, Fagerness J, et al. Common variants near FRK/COL10A1 and VEGFA are associated with advanced age-related macular degeneration. Hum Mol Genet. 2011;20:3699–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu Y, Reynolds R, Fagerness J, Rosner B, Daly MJ, Seddon JM. Association of variants in the LIPC and ABCA1 genes with intermediate and large drusen and advanced age-related macular degeneration. Invest Ophthalmol Vis Sci. 2011;52:4663–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pangburn MK. Host recognition and target differentiation by factor H, a regulator of the alternative pathway of complement. Immunopharmacology. 2000;49:149–157 [DOI] [PubMed] [Google Scholar]
- 50.Fraczek LA, Martin BK. Transcriptional control of genes for soluble complement cascade regulatory proteins. Mol Immunol. 2010;48:9–13 [DOI] [PubMed] [Google Scholar]
- 51.Rall SC Jr, Weisgraber KH, Innerarity TL, Mahley RW. Structural basis for receptor binding heterogeneity of apolipoprotein E from type III hyperlipoproteinemic subjects. Proc Nat Acad Sci U S A. 1982;79:4696–4700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiltshire S, de Bakker PI, Daly MJ. The value of gene-based selection of tag SNPs in genome-wide association studies. Eur J Hum Genet. 2006;14:1209–1214 [DOI] [PubMed] [Google Scholar]
- 53.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doerge RW, Churchill GA. Permutation tests for multiple loci affecting a quantitative character. Genetics. 1996;142:285–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vernon KA, Cook HT. Complement in glomerular disease. Adv Chronic Kidney Dis. 2012;19:84–92 [DOI] [PubMed] [Google Scholar]
- 56.Sobrin L, Ripke S, Yu Y, et al. Heritability and genome-wide association study to assess genetic differences between advanced age-related macular degeneration subtypes. [published online ahead of print June 15, 2012] Ophthalmology. 2012. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leveziel N, Tilleul J, Puche N, et al. Genetic factors associated with age-related macular degeneration. Ophthalmologica. 2011;226:87–102 [DOI] [PubMed] [Google Scholar]
- 58.Tamura Y, Konomi H, Sawada H, Takashima S, Nakajima A. Tissue distribution of type VIII collagen in human adult and fetal eyes. Invest Ophthalmol Vis Sci. 1991;32:2636–2644 [PubMed] [Google Scholar]
- 59.Ikuno Y, Sayanagi K, Soga K, et al. Lacquer crack formation and choroidal neovascularization in pathologic myopia. Retina. 2008;28:1124–1131 [DOI] [PubMed] [Google Scholar]
- 60.Peter I, Huggins GS, Ordovas JM, Haan M, Seddon JM. Evaluation of new and established age-related macular degeneration susceptibility genes in the Women's Health Initiative Sight Exam (WHI-SE) Study. Am J Ophthalmol. 2011;152:1005–1013.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thakkinstian A, Bowe S, McEvoy M, Smith W, Attia J. Association between apolipoprotein E polymorphisms and age-related macular degeneration: a HuGE review and meta-analysis. Am J Epidemiol. 2006;164:813–822 [DOI] [PubMed] [Google Scholar]
- 62.Tikellis G, Sun C, Gorin MB, et al. Apolipoprotein e gene and age-related maculopathy in older individuals: the cardiovascular health study. Arch Ophthalmol. 2007;125:68–73 [DOI] [PubMed] [Google Scholar]
- 63.Loane E, McKay GJ, Nolan JM, Beatty S. Apolipoprotein E genotype is associated with macular pigment optical density. Invest Ophthalmol Vis Sci. 2010;51:2636–2643 [DOI] [PubMed] [Google Scholar]
- 64.McKay GJ, Silvestri G, Chakravarthy U, et al. Variations in apolipoprotein E frequency with age in a pooled analysis of a large group of older people. Am J Epidemiol. 2011;173:1357–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malek G, Johnson LV, Mace BE, et al. Apolipoprotein E allele-dependent pathogenesis: a model for age-related retinal degeneration. Proc Nat Acad Sci U S A. 2005;102:11900–11905 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


