Abstract
Microbes dominate most global biogeochemical cycles, and microbial metagenomics (studying the collective microbial genomes) provides invaluable new insights into microbial systems, independent of cultivation. Metagenomic approaches targeting specific genes, e.g. small subunit (ssu) ribosomal RNA (rRNA), can be used to investigate microbial community organization by efficiently showing which taxa of organisms are present, while shotgun approaches show all genes and can indicate what functions the organisms are capable of. But collecting and organizing comprehensive shotgun data is extremely challenging and costly, and, in theory, predicting functionalities from microbial identities alone would save immense effort. However, we don’t yet know to what extent such predictions are applicable.
Introduction
Microbes are critical to the functioning of all ecosystems on earth, not to mention most animals including ourselves [1], and often are the dominant players in most biogeochemical cycles (C, N, S, etc. [2]), so understanding the makeup and organization of microbial communities is crucial to understanding natural systems. Traditional studies that relied on cultivation missed the vast majority of organisms, with rare exceptions. But newer ways can assess microbial communities, based on studying collective community DNA. This article will discuss how this approach has evolved considerably, yielding several important discoveries, and now generates a veritable tsunami of sequence data. While such data contain immense amounts of useful information, we certainly do not need all of it to ascertain community organization. But are shortcuts suitable?
The development of metagenomics
In the 1980s, Norman Pace’s lab introduced the game-changing idea that microorganisms could be studied by the wholesale extraction of mixed microbial nucleic acid (DNA and RNA) from environmental samples and then analysis of the sequences, first RNA and then DNA [3,4]. Originally, when sequencing was time-consuming and costly, only certain phylogenetic marker genes were analysed, initially rRNA, and then when polymerase chain reaction (PCR) was invented it was used to selectively amplify rRNA genes, which were then cloned and sequenced to indicate which organisms were present [5]; ssu rRNA (16S and 18S) genes were used because they are universally present in cellular life and allow every organism to be placed on a single phylogenetic “tree of life” [6]. With such an approach, any organism, even uncultured and distantly related to anything previously studied, could be put into phylogenetic context. So, we could finally list what kinds of microbes occurred where, with the rRNA sequences providing “names”. This approach yielded remarkable and unexpected discoveries, such as the existence and high abundance of “non extremophile” marine archaea in a novel major division deeply related to thermoacidophiles [7,8]. Dozens of new major microbial divisions, at the phylum or perhaps even kingdom level, were discovered, greatly expanding our view of the microbial universe [6,9].
As sequencing got cheaper, more than just phylogenetic marker genes could be studied, allowing us, in theory, to predict the potential functions of the collective organisms in a sample. A new name was coined in 1998 when Handelsman used the term “metagenome” to describe the collective genomes of soil microflora [10], and now “metagenome” is used to describe the collective genomes of any sample (usually microbial). Handelsman’s lab and DeLong’s lab were among the first to examine large cloned fragments (>40 kb) of genomic DNA extracted from nature, with a goal of linking organisms and functions. Early on, Beja et al. reported a marine proteorhodopsin [11], among what we now know are incredibly widespread rhodopsins in many bacterial and archaeal lineages [11-13], apparently with an evolutionary origin in Euryarchaea [14]. Although many such rhodopsins appear to function as light-driven proton pumps, few organisms with the gene seem to gain a direct growth benefit from light, and the ecological functions of these rhodopsins are still enigmatic [13,15,16], a reminder that even well-studied genes may have unclear functions.
In contrast to metagenomics with large DNA fragments, “random shotgun sequencing” uses a different approach where the DNA is fragmented into pieces a few thousand bases long, cloned and sequenced (at least the ends), and assembled. Assembly is on the basis of overlapping identical sequences and the knowledge that the two ends of a single fragment are connected. This shotgun assembly approach was used by Venter et al. [12] for the Global Ocean Survey, yielding many discoveries [17,18]. One such assembled fragment pointed to the possibility that the marine archaea oxidize ammonia to nitrite, a key step in the global nitrogen cycle, a function previously thought confined to bacteria [12]. Metagenomics further clarified this unexpected archaeal function, with a fosmid-based study in soils [19] that showed an ammonia oxidation gene unambiguously connected to archaeal genes, and this functionality was confirmed by cultivation of an ammonia oxidizing archaeon, whose isolation was driven by metagenomic discoveries [20]. We now recognize that such archaea, unknown until 1992, are major players globally in the nitrogen cycle of waters and soils, with many implications for ecology and agriculture [21].
Metagenomics can also be used to ascertain essentially complete genomes of uncultivated organisms – stitching them together bioinformatically from fragments. Initially, this was done from low diversity samples like acid mine drainage where only a few taxa dominated, making the job easier [22]. Next generation sequencing of metagenomes, which requires no cloning steps, has now enabled such work in very complex environments like cow rumen, where 268 gigabases of DNA sequences were used to assemble 15 microbial genomes [23], and 58 gigabases of mate-paired short-read sequences allowed assembly of several near-complete genomes from uncultivated, relatively minor constituents of complex marine samples [14].
These metagenomic studies have greatly expanded our knowledge of what organisms occur in the “wild” and what collections of functions they possess, but how do they contribute to our understanding of microbial community organization? And more to the point, is metagenomics a suitable, efficient, and cost-effective approach to routinely assess microbial community organization? Metagenomic studies are generating terabases of raw sequence, which are hard to transmit between labs, let alone readily compare across studies or even easily comprehend. Obviously, we don’t want (and usually can’t afford) to analyse gigabases of sequence just to assess which organisms are in one sample, when we might need to analyse hundreds or thousands of samples in one study. For such questions, another version of metagenomic analysis is more suitable, a logical extension of the original PCR approach used initially to yield individual rRNA clones, where the ssu rRNA genes are first amplified then the products are sequenced directly. In this “tag-sequencing” approach, next generation sequencing effectively supercharges the data collection of phylogenetic marker genes like 16S rRNA, generating a million or more rRNA sequences in a single run, with numbers and lengths of sequences depending on the sequencing platform used, e.g. 454 [24-27] or Illumina [28]. Barcoding allows hundreds of samples to be combined in one run, yielding easily tens of thousands of tag sequences per sample at reasonable cost per sample. Therefore, even rare organisms are readily detected and compared across samples or globally [25]. And the data can be readily compared, all being based on a single gene that has been incredibly well-characterized phylogenetically [9], ideally when the same primers are used. Another advantage of this approach over the shotgun approach is that when one is interested in the bacteria and archaea, but they can’t be separated well from large amounts of animal/plant/or protistan biomass (hence the shotgun sequences would be dominated by eukaryotic DNA in the bulk extract), targeted bacterial/archaeal PCR primers amplify only the DNA of interest, although chloroplasts do amplify as cyanobacteria.
Which metagenomic approach is best for community organization?
From a metagenomic sample, tag sequencing efficiently provides the distribution of phylogenentic/taxonomic types with considerable sensitivity and depth of coverage. Shotgun sequencing provides genome-wide information about all potential functionalities, and has yielded remarkable results in many systems [29-34]. But shotgun results are “diluted” and most informative about the more abundant members of the community, providing much less information about rarer organisms. So, which is more valuable, tag or shotgun, for evaluating community organization? If all you want are identities, the tag sequencing is a clear choice in terms of “bang for the buck”. But if you care about functional types, will tag sequences do? How well can we predict functions from taxonomy – e.g. how closely correlated are phylogenetic marker genes to the ones that define functions? If they are well correlated then identities alone may suffice. The question is important because microbes can have remarkably plastic genome content, even in a single species. For example, the genomes of two Escherichia coli strains can differ by as much as a third, and sometimes two organisms with extremely close 16S rRNA sequences have significant differences in their major functions [35]. If such variation were the norm and happens randomly then predicting functions from identity alone would be almost hopeless. Yet it does not seem hopeless; at least for some habitats, there is evidence that particular phylogenetic types have predictable distributions in time and space, and such predictability suggests that particular functions correlate consistently with particular identities (from phylogenetic markers). One set of examples come from two different long-term ocean plankton time series off California and England, where microbial communities exhibit annually repeating patterns of community composition, whether measured by community fingerprinting [36], or 16S tag sequencing [37]. Another example is the highly structured and predictable global distributions of closely related varieties of the abundant marine cyanobacterium Prochlorococcus, suggesting niche partitioning [38]. A further example is the consistent co-occurrence patterns of microbes, as identified by 16S rRNA sequences, across multiple habitats [39]. These robust patterns would not exist if niche-defining functions were not well correlated to marker-gene based identities. Also consistent with such correlations, a large study of gut microbiota of 18 humans and 33 mammals, as related to diet, showed strong concordance between patterns of 16S rRNA and functional gene distributions [1].
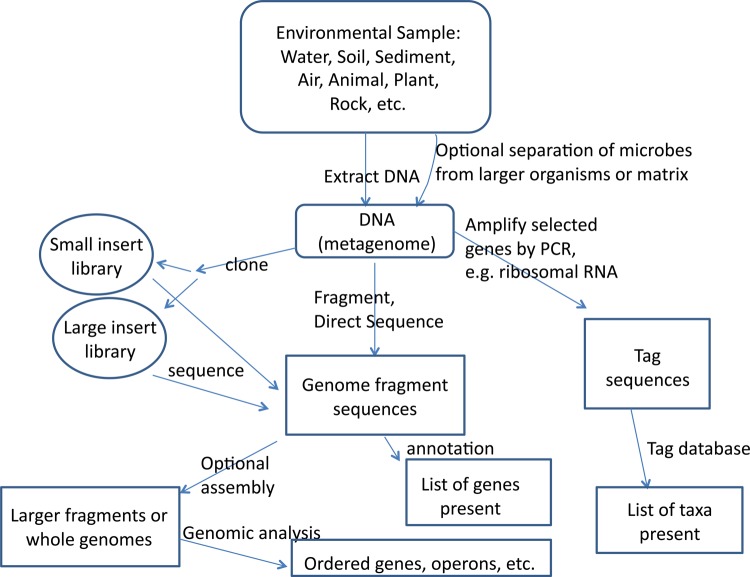
Figure 1. Schematic of shotgun and targeted metagenomic analysis.
The metagenome is the collective genomic DNA of organisms (usually microorganisms) in a given sample. After sampling, and sometimes attempts to separate microorganisms from larger organisms, DNA is extracted from the total biomass. For overall genomic analysis, older studies cloned various size fragments and sequenced them. Next generation sequencing shotgun sequencing studies now generally fragment the DNA and sequence the fragments directly, assembling some of them into overlapping ‘contigs’, larger ‘scaffolds’, or even whole genomes. In contrast, tag sequencing uses polymerase chain reaction to amplify specific genes of interest, most often 16S rRNA, and the amplified fragments are sequenced directly.
Future prospects
It remains to be seen how consistently identity from tag sequences correlates to functionality in non-marine environments, like soils and animal or plant microbiomes. Marine planktonic bacteria, which tend to be free-living and survive on low levels of nutrients, have streamlined genomes compared with most studied bacteria [40], which are probably more stable than genomes of other organisms like potential pathogens [41]. So rRNA tag sequencing alone is unsuitable to clearly identify pathogens. The phylogenetic resolution of the selected tag sequences also matters, and we need widely collected shotgun data and curated database systems [42,43] ,as well as sequenced genomes from infrequently studied organisms [44], to link functionalities to identities more broadly. Efforts like the Earth Microbiome Project [45] (http://www.earthmicrobiome.org) are working to integrate such information from samples collected globally to assess worldwide patterns of microbial diversity.
Acknowledgments
This work was supported by the NSF Microbial Observatories, Biological Oceanography, and Dimensions in Biodiversity programs, grants 0703159, 1031743, and 1136818. I thank Mitch Sogin, Rob Knight, Jack Gilbert, Janet Jansson, John Heidelberg, and Ian Hewson, for helpful discussions.
Abbreviations
- PCR
polymerase chain reaction
- rRNA
ribosomal RNA
- ssu
small subunit
Competing interests
The author declares that he has no competing interests.
The electronic version of this article is the complete one and can be found at: http://f1000.com/reports/b/4/15
References
- 1.Muegge BD, Kuczynski J, Knights D, Clemente JC, Gonzalez A, Fontana L, Henrissat B, Knight R, Gordon JI. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–4. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Jed Fuhrman 17 Jul 2012
- 2.Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive Earth's biogeochemical cycles. Science. 2008;320:1034–9. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Jed Fuhrman 17 Jul 2012
- 3.Olsen GJ, Lane DL, Giovannoni SJ, Pace NR. Microbial ecology and evolution: A ribosomal RNA approach. Ann. Rev. Microbiol. 1986;40:337–65. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Jed Fuhrman 17 Jul 2012
- 4.Pace NR, Stahl DA, Lane DL, Olsen GJ. The analysis of natural microbial populations by rrna sequences. Adv. Microbiol. Ecol. 1986;9:1–55. [Google Scholar]
- 5.Giovannoni SJ, Britschgi TB, Moyer CL, Field KG. Genetic diversity in Sargasso Sea bacterioplankton [see comments] Nature. 1990;345:60–3. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 6.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–40. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 7.Fuhrman JA, mccallum K, Davis AA. Novel major archaebacterial group from marine plankton. Nature. 1992;356:148–9. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- 8.Fuhrman J. Oceans of Crenarchaeota: a personal history describing this paradigm shift. Microbe. 2011;6:531–7. [Google Scholar]
- 9.Tringe SG, Hugenholtz P. A renaissance for the pioneering 16S rrna gene. Current Opinion in Microbiology. 2008;11:442–6. doi: 10.1016/j.mib.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 10.Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chemistry & Biology. 1998;5:R245–9. doi: 10.1016/S1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- 11.Beja O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, Jovanovich SB, Gates CM, Feldman RA, Spudich JL, Spudich EN, delong EF. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea [see comments] Science. 2000;289:1902–6. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Jed Fuhrman 17 Jul 2012
- 12.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu DY, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]; F1000 Factor 20Evaluated by Shiladitya DasSarma 09 Mar 2004, Rino Rappuoli 09 Mar 2004, Douglas Capone 21 Apr 2004, Gerard Muyzer 23 Apr 2004, Ian M Head 16 Aug 2004
- 13.Fuhrman JA, Schwalbach MS, Stingl U. Proteorhodopsins: an array of physiological roles? Nature Rev Microbiol. 2008;6:488–94. doi: 10.1038/nrmicro1893. [DOI] [PubMed] [Google Scholar]
- 14.Iverson V, Morris RM, Frazar CD, Berthiaume CT, Morales RL, Armbrust EV. Untangling Genomes from Metagenomes: Revealing an Uncultured Class of Marine Euryarchaeota. Science. 2012;335:587–90. doi: 10.1126/science.1212665. [DOI] [PubMed] [Google Scholar]; F1000 Factor 9Evaluated by Oded Beja 26 Apr 2012, Jed Fuhrman 17 Jul 2012
- 15.Gomez-Consarnau L, Akram N, Lindell K, Pedersen A, Neutze R, Milton DL, Gonzalez JM, Pinhassi J. Proteorhodopsin phototrophy promotes survival of marine bacteria during starvation. Plos Biol. 2010;8:e1000358. doi: 10.1371/journal.pbio.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomez-Consarnau L, Gonzalez JM, Coll-Llado M, Gourdon P, Pascher T, Neutze R, Pedros-Alio C, Pinhassi J. Light stimulates growth of proteorhodopsin-containing marine Flavobacteria. Nature. 2007;445:210–3. doi: 10.1038/nature05381. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Jed Fuhrman 17 Jul 2012
- 17.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, Beeson K, Tran B, Smith H, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, Li K, Kravitz S, Heidelberg JF, Utterback T, Rogers YH, Falcon LI, Souza V, Bonilla-Rosso G, Eguiarte LE, Karl DM, Sathyendranath S, Platt T, Bermingham E, Gallardo V, Tamayo-Castillo G, Ferrari MR, Strausberg RL, Nealson K, Friedman R, Frazier M, Venter JC. The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. Plos Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williamson SJ, Rusch DB, Yooseph S, Halpern AL, Heidelberg KB, Glass JI, Andrews-Pfannkoch C, Fadrosh D, Miller CS, Sutton G, Frazier M, Venter JC. The Sorcerer II Global Ocean Sampling Expedition: Metagenomic Characterization of Viruses within Aquatic Microbial Samples. Plos One. 2008;3:e1456. doi: 10.1371/journal.pone.0001456. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Jed Fuhrman 17 Jul 2012
- 19.Schleper C, Jurgens G, Jonuscheit M. Genomic studies of uncultivated archaea. Nature Reviews Microbiology. 2005;3:479–88. doi: 10.1038/nrmicro1159. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Jed Fuhrman 17 Jul 2012
- 20.Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–6. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]; F1000 Factor 15Evaluated by Michael Wagner 04 Oct 2005, Ian M Head 02 Nov 2005, Douglas Capone 14 Nov 2005, Jed Fuhrman 17 Jul 2012
- 21.Francis CA, Beman MJ, Kuypers MMM. New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archeal ammonia oxidation. ISME Journal. 2007;1:19–27. doi: 10.1038/ismej.2007.8. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Jed Fuhrman 17 Jul 2012
- 22.Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]; F1000 Factor 14Evaluated by Julian Parkhill 05 Mar 2004, Joern Piel 16 Mar 2004, Michael Wagner 02 Apr 2004, Donald Ward 09 Jun 2004, Ian M Head 16 Aug 2004
- 23.Hess M, Sczyrba A, Egan R, Kim TW, Chokhawala H, Schroth G, Luo SJ, Clark DS, Chen F, Zhang T, Mackie RI, Pennacchio LA, Tringe SG, Visel A, Woyke T, Wang Z, Rubin EM. Metagenomic Discovery of Biomass-Degrading Genes and Genomes from Cow Rumen. Science. 2011;331:463–7. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]; F1000 Factor 11Evaluated by Chris Somerville 16 Apr 2012, Jed Fuhrman 17 Jul 2012
- 24.Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12115–20. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zinger L, Amaral-Zettler LA, Fuhrman JA, Horner-Devine MC, Huse SM, Mark Welch DB, Martiny JBH, Neal PR, Sogin M, Boetius A, Ramette A. Global patterns of bacterial beta-diversity in seafloor and seawater ecosystems. Plos ONE. 2011;6:e24570. doi: 10.1371/journal.pone.0024570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agogue H, Lamy D, Neal PR, Sogin ML, Herndl GJ. Water mass-specificity of bacterial communities in the North Atlantic revealed by massively parallel sequencing. Mol Ecol. 2011;20:258–74. doi: 10.1111/j.1365-294X.2010.04932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pommier T, Neal PR, Gasol JM, Coll M, Acinas SG, Pedros-Alio C. Spatial patterns of bacterial richness and evenness in the NW Mediterranean Sea explored by pyrosequencing of the 16S rrna. Aquatic Microbial Ecology. 2010;61:212–24. doi: 10.3354/ame01484. [DOI] [Google Scholar]
- 28.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rrna diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4516–22. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C, Chan AM, Haynes M, Kelley S, Liu H, Mahaffy JM, Mueller JE, Nulton J, Olson R, Parsons R, Rayhawk S, Suttle CA, Rohwer F. The marine viromes of four oceanic regions. Plos Biology. 2006;4:2121–31. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM, Furlan M, Desnues C, Haynes M, Li LL, mcdaniel L, Moran MA, Nelson KE, Nilsson C, Olson R, Paul J, Brito BR, Ruan YJ, Swan BK, Stevens R, Valentine DL, Thurber RV, Wegley L, White BA, Rohwer F. Functional metagenomic profiling of nine biomes. Nature. 2008;452:629–U8. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- 31.Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, Bork P, Hugenholtz P, Rubin EM. Comparative metagenomics of microbial communities. Science. 2005;308:554–7. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]; F1000 Factor 16Evaluated by Daniel Vaulot 25 Apr 2005, Jeffrey G Lawrence 27 Apr 2005, John Jaenike 28 Apr 2005, Robert B Jackson 04 May 2005, Gerard Muyzer 25 May 2005, Christos Ouzounis 01 Aug 2005
- 32.Gilbert JA, Dupont CL. Microbial Metagenomics: Beyond the Genome. Annual Review of Marine Science, Vol 3. 2011;3:347–71. doi: 10.1146/annurev-marine-120709-142811. [DOI] [PubMed] [Google Scholar]
- 33.Von Mering C, Hugenholtz P, Raes J, Tringe SG, Doerks T, Jensen LJ, Ward N, Bork P. Quantitative phylogenetic assessment of microbial communities in diverse environments. Science. 2007;315:1126–30. doi: 10.1126/science.1133420. [DOI] [PubMed] [Google Scholar]; F1000 Factor 12Evaluated by Russell Hill 14 Mar 2007, Ian M Head 27 Mar 2007
- 34.De Bruijn FJ, editor. Handbook of molecular microbial ecology II. Metagenomics in different habitats. Series. Hoboken, New Jersey: Wiley-Blackwell; 2011. [DOI] [Google Scholar]
- 35.Jaspers E, Overmann J. Ecological significance of microdiversity: Identical 16S rrna gene sequences can be found in bacteria with highly divergent genomes and ecophysiologies. Applied and Environmental Microbiology. 2004;70:4831–9. doi: 10.1128/AEM.70.8.4831-4839.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, Naeem S. Annually reoccurring bacterial communities are predictable from ocean conditions. Proc Natl Acad Sci U S A. 2006;103:13104–9. doi: 10.1073/pnas.0602399103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilbert JA, Steele J, Caporaso JG, Steinbrück L, Reeder J, Temperton B, Huse S, Joint I, mchardy AC, Knight R, Somerfield P, Fuhrman JA, Field D. Defining seasonal marine microbial community dynamics. ISME Journal. 2011;6:298–308. doi: 10.1038/ismej.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson ZI, Zinser ER, Coe A, mcnulty NP, Woodward EM, Chisholm SW. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science. 2006;311:1737–40. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Jed Fuhrman 17 Jul 2012
- 39.Chaffron C, Rehrauer H, Pernthaler J, von Mering C. A global network of coexisting microbes from environmental and whole-genome sequence data. Genome Res. 2010;20:947–59. doi: 10.1101/gr.104521.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biers EJ, Sun SL, Howard EC. Prokaryotic Genomes and Diversity in Surface Ocean Waters: Interrogating the Global Ocean Sampling Metagenome. Applied and Environmental Microbiology. 2009;75:2221–9. doi: 10.1128/AEM.02118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ochman H, Davalos LM. The nature and dynamics of bacterial genomes. Science. 2006;311:1730–3. doi: 10.1126/science.1119966. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Jed Fuhrman 17 Jul 2012
- 42.Markowitz VM, Chen IMA, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang JH, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Research. 2012;40:D115–22. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Markowitz VM, Chen IMA, Chu K, Szeto E, Palaniappan K, Grechkin Y, Ratner A, Jacob B, Pati A, Huntemann M, Liolios K, Pagani I, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. IMG/M: the integrated metagenome data management and comparative analysis system. Nucleic Acids Research. 2012;40:D123–9. doi: 10.1093/nar/gkr975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu DY, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, Hooper SD, Pati A, Lykidis A, Spring S, Anderson IJ, d'haeseleer P, Zemla A, Singer M, Lapidus A, Nolan M, Copeland A, Han C, Chen F, Cheng JF, Lucas S, Kerfeld C, Lang E, Gronow S, Chain P, Bruce D, Rubin EM, Kyrpides NC, Klenk HP, Eisen JA. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–60. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gilbert J, O’Dor R, King N, Vogel T. The importance of metagenomic surveys to microbial ecology: or why Darwin would have been a metagenomic scientist. Microbial Informatics and Experimentation. 2011;5:1. doi: 10.1186/2042-5783-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Jed Fuhrman 17 Jul 2012