Abstract
Identification of marker genes expressed in specific cell types is essential for the genetic dissection of neural circuits. Here we report a new strategy for classifying heterogeneous populations of neurons into functionally distinct types and for identifying associated marker genes. Quantitative single-cell expression profiling of genes related to neurotransmitters and ion channels enables functional classification of neurons; transcript profiles for marker gene candidates identify molecular handles for manipulating each cell type. We apply this strategy to the mouse medial vestibular nucleus (MVN), which comprises several types of neurons subserving cerebellar-dependent learning in the vestibulo-ocular reflex. Ion channel gene expression differed both qualitatively and quantitatively across cell types and could distinguish subtle differences in intrinsic electrophysiology. Single-cell transcript profiling of MVN neurons established six functionally distinct cell types and associated marker genes. This strategy is applicable throughout the nervous system and could facilitate the use of molecular genetic tools to examine the behavioral roles of distinct neuronal populations.
Introduction
A major neuroscientific challenge is to determine how cellular signaling and plasticity mediate behavioral performance and learning. The ability to link neuronal firing directly with quantifiable behavioral performance and sensory-motor learning makes the vestibulo-ocular reflex (VOR) a particularly attractive model. The VOR stabilizes images on the retina during self-motion by transforming head motion signals from the inner ear into oculomotor commands. The primary circuit is simple: a “three neuron arc” comprising vestibular ganglion cells, medial vestibular nucleus (MVN) neurons, and ocular motoneurons mediates gaze stability in the horizontal plane via a pair of extraocular muscles. Excellent behavioral performance is maintained throughout life via cerebellar learning (du Lac et al., 1995; Blazquez et al., 2004; Broussard and Kassardjian, 2004; Schubert and Zee, 2010). Progress at the cellular mechanistic level has been limited, however, by the challenges of classifying and manipulating functionally distinct cell types which are spatially intermingled within central vestibular nuclei.
In vivo recordings have revealed several types of vestibular nucleus neurons with differential synaptic connections and/or firing responses during head and eye movements (Highstein and Ito, 1971; Keller and Kamath, 1975; Lisberger and Miles, 1980; McCrea et al., 1987; Scudder and Fuchs, 1992). Cerebellar learning was originally thought to be mediated by Purkinje cell synapses onto one of these cell types (Lisberger et al., 1994; Ramachandran and Lisberger, 2008). Although initial in vitro recordings classified MVN neurons into two types on the basis of action potential waveforms (Serafin et al., 1991; Johnston et al., 1994), subsequent studies demonstrated that intrinsic physiological properties are distributed continuously rather than bimodally (du Lac and Lisberger, 1995; Bagnall et al., 2007). Many studies have now demonstrated specific differences in intrinsic excitability across MVN neurons with different axonal projections (Sekirnjak and du Lac, 2006; Kolkman et al., 2011a,b) and/or transmitter expression (Takazawa et al., 2004; Bagnall et al., 2007; Shin et al., 2011). The observation that Purkinje cells differentially innervate the somata and dendrites of several types of MVN neurons (Sekirnjak et al., 2003; Shin et al., 2011) complicates analyses of cerebellar learning. Collectively, these findings indicate that the central VOR circuit comprises multiple functionally and molecularly distinct cell types. Given this heterogeneity, how can we target recordings and experimental manipulations to neurons playing distinct roles in behavior and learning?
In this study, we employ a new strategy for classifying heterogeneous neurons and for identifying “marker genes” which can be used in conjunction with modern molecular tools (Dymecki and Kim, 2007; Luo et al., 2008; Knöpfel et al., 2010; Deisseroth, 2011) to label, monitor, and manipulate distinct cell types. In our approach, the quantitative, single-cell expression profiling of genes related to neurotransmitters, ion channels, and candidate molecular markers enables mature neurons to be classified into molecularly distinct types. Anterograde and retrograde tracings provide insights into the functional role of each molecularly defined class of neurons. Application of this approach to the MVN resulted in the identification of six major neuronal classes and associated marker genes. This strategy for classifying neurons and identifying cell-type-specific marker genes is widely applicable to other brain regions and could facilitate the use of molecular genetic tools to examine behavioral roles of distinct neuronal populations.
Materials and Methods
Single-cell sampling and global cDNA amplification.
Experimental protocols for this study were approved by Salk Institute Animal Care and Use Committee. Coronal brainstem slices through the middle part of the rostrocaudal extent of the MVN (bregma from −5.80 to −6.24 mm) were prepared from male juvenile mice of YFP16, GIN, and GlyT2–13 lines in C57BL/6 background. Neurons were acutely dissociated from mouse brainstem slices. Following anesthesia with Nembutal and decapitation, slices (thickness, 250 μm) were prepared by vibratome in ice-cold modified artificial CSF (ACSF) containing the following (in mm): 125 NaCl, 1.25 KCl, 3 MgCl2, 25 NaHCO3, 1 CaCl2, 1.25 NaH2PO4, and 25 dextrose aerated with 95% CO2/5% O2. The central portion of the MVN (including both parvocellular and magnocellular regions) was punched out using a micropipette (660 μm, i.d.), enzymatically dissociated using papain (40 U/ml; Worthington) with cysteine (2 mm) in HEPES-buffered ACSF containing the following (in mm): 140 NaCl, 1.25 KCl, 1.25 NaH2PO4, 10 HEPES, 25 dextrose, 3 MgCl2, 1 CaCl2, 0.0001 tetrodotoxin, 0.02 6-cyano-7-mitroquinoxaline-2,3-dione, 0.05 d-APV at 37°C for 10 min, then triturated using a micropipette (350 μm, o.d.) in 1% BSA in HEPES-buffered ACSF. Dissociated cells were carefully washed one by one to avoid contamination of any tissue debris using glass capillaries (Hempel et al., 2007) and individually transferred to tubes containing cell lysis buffer (Kurimoto et al., 2007). In each sampling batch, we processed eight cells, one negative control (including HEPES-buffered ACSF in the dish used to wash cells), and one positive control (including 10 pg of mouse brain total RNA). Both controls were processed in the same way as the sample containing cells. The negative control was helpful in detecting potential contamination via cell suspension buffer. Each tube also contained spike-in RNAs (Lys, Dap, Phe, and Thr, amounting to 1000, 100, 20, and 5 copies, respectively). The following steps including cell lysis, reverse transcription (RT), exonuclease I treatment, poly(dA) addition, second-strand cDNA synthesis, and the 20 cycle PCR amplification were exactly as described previously (Kurimoto et al., 2007), except for exonuclease I treatment (1.5 U/μl exonuclease I, 37°C for 60 min, followed by 95°C for 30 min). Note that this method amplifies ∼700 bp of the 3′ end of cDNAs. To avoid change in transcriptional state and RNA degradation, tissues and cells were kept on ice whenever possible, and the procedure from decapitation to RT was finished in less than 2 h.
After the initial 20 cycle PCR, we ran an additional 10 cycle PCR to yield a sufficient amount of cDNA fragments for a large number of quantitative PCRs (qPCRs). One microliter of the 20 cycle PCR product was added to 200 μl of reaction mixture of 1× ExTaq buffer containing 0.25 mm each of dATP, dCTP, cGTP, and dTTP, 0.02 μg/μl primer V1(dT)24, 0.02 μg/μl primer V3(dT)24, and 0.05U/μl ExTaq Hot Start Version (Takara Bio), which was divided into four 50 μl scale reactions. The 10 cycle PCR was performed as follows: 95°C for 5 min, 10 cycles of 95°C for 30 s, 67°C for 1 min, and 72°C for 5 min (+ 6 s for each cycle), followed by 72°C for 10 min. The 10 cycle PCR products were mixed together after the reaction, purified using QIAquick PCR purification kit (Qiagen), and eluted into 400 μl of TE buffer. We used the 10 cycle PCR products for qPCR. No obvious distortion in transcript abundance representation was observed after the 10 cycle PCR (see Fig. 1D).
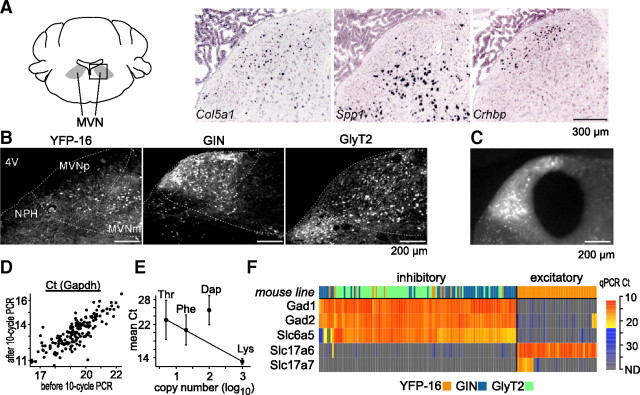
Figure 1.
Molecular anatomy of the MVN. A, Examples of marker gene candidates. Using the Allen Mouse Brain Atlas (Lein et al., 2007), genes expressed in subpopulations of MVN neurons were identified as marker candidates. In situ hybridization images of exemplar marker gene candidates Col5a1, Spp1, and Crhbp are shown. The images were taken from the boxed region in the schematic drawing of the mouse coronal brainstem section (left,∼−5.9 mm from bregma). The distinct distribution of hybridization signal across the three genes imply their potential use as marker genes. Scale bar, 300 μm. B, Representative MVN fluorescent images of YFP-16, GIN, and GlyT2 mouse lines. The single-cell cDNAs were prepared from fluorescent protein-expressing MVN neurons in the three lines. MVNp, parvocellular MVN; MVNm, magnocellular MVN; NPH, nucleus prepositus hypoglossi; 4V, fourth ventricle (darkened for contrast). Scale bar, 200 μm. C, A representative image of MVN after punching out its central portion for single-cell cDNA amplification. Scale bar, 200 μm. D, qPCR results for Gapdh before and after the 10 cycle amplification. Data from all of the single-cell cDNAs (n = 167) are plotted. The linear relationship indicates no systematic distortion in abundance representation of transcripts by the 10 cycle amplification. E, The abundance representation of the spike-in RNAs in the single-cell cDNAs (the 10 cycle PCR product). The mean Ct values in qPCR are plotted against the original copy number of the spike-in RNAs (log-scaled). The cases where Ct value was not determined (no transcript was detected) are excluded from averaging. Lys, Phe, and Thr were on a regression line in the order of copy number, indicating that abundance representation of the three spike-in RNAs was preserved. Dap is not on the same regression line, which is at least partly due to the lower amplification rate of Dap in qPCR (1.79) than the other three (Lys, 2.01; Phe, 1.97; Thr, 2.00). Y-intercept of the regression line is 26.2. Error bar indicates SD. F, The heat map of the single-cell cDNAs (columns) based on the transcript profiles of the transmitter-related genes (rows). Color in the first row indicates the mouse lines from which the single-cell cDNAs were prepared (YFP-16: n = 61, orange; GIN: n = 52, blue; GlyT2: n = 54, green). In the heat map showing qPCR results, Ct values lesser and greater than the mean Ct of spike-in RNA Thr (five copies per sample) are coded in red-yellow and blue-gray gradation, respectively. Excitatory (Slc17a6-positive) and inhibitory (Gad1-positive) cell groups are defined clearly by the transcript profiles. As expected, YFP-16 neurons are both excitatory and inhibitory, while GIN and GlyT2 cells are only inhibitory. Note the coexpression of Gad1, Gad2, and Slc6a5 in the inhibitory cell group.
Quality control of single-cell cDNA.
The expression level of a housekeeping gene (Gapdh) was examined by qPCR with the 20 cycle PCR products. We rejected single-cell cDNAs showing an extraordinarily low Gapdh level, which were defined as those scoring qPCR threshold cycle (Ct) greater than mean + SD (4.3%; 9 of 208 cells). The rejected samples were assumed to have suffered severe RNA degradation or cDNA amplification failure. To exclude RNA contamination via the cell-suspending solution during the cell harvest, the sample batches in which Gapdh was detected in the negative control were all rejected (15.4%; 32 of 208 cells). Qualified single-cell cDNAs were further amplified by the 10 cycle PCR.
qPCR.
qPCR was performed using the 7900 Real-Time PCR System (Applied Biosystems) according to the manufacturer's instruction. Reaction mixture (10 μl per well) includes 1 μl of the 10 cycle PCR product, 200 nm of forward and reverse primers, and 100 nm of probe conjugated with 6-FAM at the 5′ end and Iowa Black FQ (Integrated DNA Technologies) or MGBNFQ (Applied Biosystems) at the 3′ end. For the quality control experiment (qPCR for Gapdh), 1 μl of 1/40 dilution of the 20 cycle PCR product was added as a template. Reaction preparation was assisted by the pipetting robot, epMotion (Eppendorf). The sequence of primers and probes are shown in Table 1; genes examined in this study are listed in Table 2. Each reaction plate included a duplicate or triplicate of the standard reaction which was qPCR for Gapdh with cDNA corresponding to 25 ng of mouse brain total RNA. The threshold to determine Ct was set so that Ct values of the standard reaction became identical across all the reaction plates. All reactions were done in duplicate or triplicate, and averaged Ct values were used for further analyses. If Ct values showed a large variation between duplicates (difference >1) or among triplicates (SD > 1), the same reaction was retried until consistent Ct values were obtained. Note that Ct values >30 generally showed larger variation because of the stochastic error associated with PCR and were not subjected to retest.
Table 1.
Sequences of primers and probes used in quantitative real-time qPCR
| Gene symbol | Amplicon size (bp) | Forward primer (5′…3′) | Probe (5′…3′) | Reverse primer (5′…3′) |
|---|---|---|---|---|
| Gapdh | 140 | gcatggccttccgtgttc | tacccccaatgtgtccgtcgtgga | gtagcccaagatgcccttca |
| Lys | 53 | tgcttcccgttacggttttga | cattccggaaatttg* | aacggcccggttcgat |
| Dap | 69 | cactggtggaagaagcgatct | tccgccggaatccata* | gcgccggaaggctttt |
| Phe | 79 | ggcccaaaacgaccttaatgg | ccggactgatcatcc* | gaaaatgcagacagcactctatgc |
| Thr | 57 | ccgatgccgtaaaagcaagtg | ctgtcagcaatatcc* | tggacatgatcgcagcaatga |
| Cacna1a | 140 | atctttcgttttcccggtgaa | ctgcacgccccacacacttctcag | cttgtagttgatctctgcaggcagta |
| Cacna1b | 140 | cttgcttcctgactttgggttt | tcttttcaagatgttgtctgtttccccctg | ccactgcttgacaggttcca |
| Cacna1c | 144 | gctatttgaaagcaagaacaaacca | cccacgagatatgcaaaagcaatgcaa | atgctgacacaactcacaaccat |
| Cacna1d | 142 | gcagagtgctgttctcctaaagg | tggactctgtcgtgctgggtgtctgt | cgatcacccattccagaacac |
| Cacna1e | 140 | gctgccacccacattatcaa | tgagtcaagcccatgagacgccc | tctgggtgggccagcat |
| Cacna1g | 140 | gcacctcgcatgccttctt | ctgaagagaacacagccctggagccc | cgagagccttggccacact |
| Cacna1 h | 140 | gtctcgttcctgtccatcatcac | cggcatccttccctctcacagtaccc | tgcatgggccagtactcctt |
| Cacna1i | 140 | cagagccagggctgaactg | cagaggccgccttgtccagca | aactaggagggaagttcgttcaca |
| Hcn1 | 140 | gtgccacaacgtgtcacctt | tccgacagatgtcctcgggagcc | tctgcatctgggtctgtatttagg |
| Hcn2 | 63 | gctgtgccccgcatgt | cggtggcctccgaggaagaatatgg | gggccgagtattgcacttga |
| Hcn4 | 140 | ctgagagtcttcggacctgtgtt | agccagctggacgccacagaacc | gacaagagacccctcactccttatc |
| Kcna1 | 140 | ttgccaattgcttcctcctt | taaggacacgtctcatctcatcacccctg | tgaaaactatgcgcatccttagg |
| Kcna2 | 79 | cccatgccaggcacaaag | tctgtgcttctcttcttctcctttgtggga | caccccatgtcatgaaattcag |
| Kcna3 | 140 | cgggagaaacaacagaaattcct | tgtcagacaccccttccagcgga | cacgggttgctatgggaact |
| Kcna4 | 156 | ttctggaggaaatgtaacaagttttg | tctatgccgaagcccgagagagacttc | cccattcttccaatgattgtca |
| Kcna5 | 142 | cccatcaaagtccggatcct | ctttccccgtttacccttcccaccc | aacggctcctgcgtcatg |
| Kcna6 | 140 | cacgaggccactctggaaaa | acttctctggaggatgagacccccgc | ccgtctcagtcactgcttgtg |
| Kcnb1 | 140 | ggaaggccccccttacaata | agcaccccaatcccatcagctcaga | tccttgaaagatggagcaatcc |
| Kcnc1 | 147 | ccaatgtgaccaagtttcataacaa | cctgcagtccagggccaccaga | cagttctcgccttgatgaaggt |
| Kcnc2 | 140 | atagcctaactataacccccctgtatc | cttttgtgaccatctcaaggtgcatgcc | ttcaagtctatccccagatcagttg |
| Kcnc3 | 248 | gcagcctgagcagcattgt | caccccccctggctctcccc | gcatcgtctcttttgcatattattg |
| Kcnc4 | 140 | cttcctgcccatccttggta | acgcctaggctgggcccactgt | tttattggtactcgagcacagcat |
| Kcnd1 | 140 | ctgacccctgggagagatagaa | atcgcctccttctctgctgtaactcctttg | ggagacagggaacctgagatga |
| Kcnd2 | 152 | agctgagtgacagggaaaagaact | ctgaccctcgggaagacctggctt | tgctctagaagcataaagtgacaaca |
| Kcnd3 | 140 | tgggagggatggtatatttttga | caatgttttccttagaggtcatgcgctcc | ggtggtaagcaaagcccaataa |
| Kcnf1 | 140 | gggttgggtgtggagttttg | tgtccaggctacaccccattgctgt | ggatttaacccaatcaccatgac |
| Kcng4 | 140 | cgcccctactttcctgactgt | tgccccagcatctactctaaagtgcatctt | aggagcatgtgtgtgcatctg |
| Kcnh1 | 140 | cacccccaccccaatga | tggagactgtccaccatccaagttgc | atgtttgtatgagaagcctcaggat |
| Kcnh2 | 144 | tgcccactgtggacatgtg | cagggcattacctgaccgctctcct | cagggtattgcaggaggagatg |
| Kcnma1 | 140 | cagaactgcaacatttcaaacataaa | cccaggtggtacaatcacatcctgcc | aagtctcttttcggcatctgtgt |
| Kcnn1 | 141 | gggatcctgatgtcaaagtaagga | tacagacgagcagagtcaccacggtca | cccatgcctgtgaggtgtct |
| Kcnn2 | 140 | aaaggatcgtcaccctggaa | tagcattcatgccctccctgggc | gacatgcttgtcgtagttctccat |
| Scn1a | 149 | gcttcagaatttcttttcctcaaaa | cagaagcagaccgtaggcacctctttttct | gcagttcatgaatacggtttgc |
| Scn1b | 119 | tgcatgatgggtgaagcaat | ccccaccctgcttttgctgcct | gcctatgtgcaaatcagcaaaa |
| Scn2a1 | 140 | ttgaatgcatgcgggaaaat | ctaccggtaagccacattacaatcaagcca | catctcgaggatagaaacacagaca |
| Scn2b | 140 | ataggaatggcttggtgcagtt | cagccttgctgtgcctcctgctg | tatgtacaatcgcatcagcaagct |
| Scn3a | 175 | cacgggctctgtcctttctaa | caggaaccaagtgggaaagttgtgcaa | gccattccctgcatgca |
| Scn3b | 140 | tataaatgagaaagccctggttctg | aattcactcagtcaatgca* | gcctaacctagtgctcgacaca |
| Scn4b | 140 | ggatgcccaaatgagggatt | tattttgtcccaggttggagtccctgctc | ccagaggactaaaccatgcataaga |
| Scn8a | 140 | cagcaggtcgagaaatgtctttc | tatctccttccttacccctccactcgttcc | ctcaagcatttctgccatttcc |
| Adcyap1 | 141 | tggtgaaactgggctctagtga | agtctaacttagtgttcacgtgg* | cagtttagtaaggctcttggcattt |
| Cck | 140 | tccttctggccgcatgtcc | cttaagaacctgcagagcctggacccc | tggcccactacgatgggtat |
| Coch | 71 | gcccttagaaactcaggaaagaaga | ttgtcatggattaacattg* | tccacttgaatgcatatttgaactc |
| Col5a1 | 140 | gaccccatgcccagagaga | acggagccgaatcacatgacctagcc | cggccaacggtccattaac |
| Crh | 140 | cgcccatctctctggatctc | agttagctcagcaagctcacagcaacagg | ttggccaagcgcaacatt |
| Crhbp | 151 | ggccggcttcagtgattg | ccaggccttgaacacgtgcacaca | caaaaggaagcaaaattggaactc |
| Grin2c | 140 | gttcctgctggactggattctg | cctctgccttcttctttgaaattctggcc | ggtcataagctggccatgga |
| Gsg1l | 140 | gggtcagattccagaccaatga | catgctcaggccccatcccagt | agacaaagttgacctggcagagt |
| Hspb8 | 142 | gggcaggtcttatatccccatat | tccttcaccacacaacacagtgcctttt | ggaccttagccagagaattaactga |
| Nrn1 | 140 | gccggtttcacctgctattg | cgatgcccctcgttcaggtcca | taacattgattacccccttgtgatc |
| Sema3a | 140 | agtttctatagatgctccattccagtt | tcaccatcatcctcaagtgcaagatttcc | gacctggcaaaaagcatgct |
| Spp1 | 107 | cagtcggatgtgatcgatagtca | tccaaagccagcctggaacatcaga | ttactcttagggtctaggactagcttgtc |
| Sst | 140 | ttctggaagacattcacatcctgtt | ttgttgtcctagccagacctctgatccctc | tcttcaatttctaatgcagggtcaa |
| Tacr3 | 141 | gtgtgcagataaacactatcaaaaatgaat | tcacaacaggattgccccaaaagcc | aaacatgcacacaatgcttcgt |
| Gad1 | 158 | tcatgttatggaaatcttgcttcag | agccttcgctcttctctttcccccc | cgagtcacagagattggtcatatactact |
| Gad2 | 140 | caactaagtcccaccctaagaaatct | tgtgatggcacacaaaatgcttcatctct | tccaagaccctgtagagtcaatacc |
| Slc17a6 | 141 | ttgcttgtggtagcagtgatttc | tgcgctgataagaatgcacatcccaat | aaatacccaaaactgacaggtaaaca |
| Slc17a7 | 140 | ccgggccttgaccttagc | ccaaaatggctctgctcaccccg | cctcgagccgctgaattaat |
| Slc6a5 | 140 | ctgtaaactgcctgcagaggaa | gcaggtgaccagctgccttacaggtg | cagcagcccaatgatcaaaa |
*Conjugated with MGBNQF (Applied Biosystems) instead of Iowa Black FQ (Integrated DNA Technologies) at 3′.
Table 2.
List of the profiled genes
| Gene symbol | Gene name |
|---|---|
| Gapdh | Glyceraldehyde-3-phosphate dehydrogenase |
| Dap | Spike-in RNA derived from Bacillus subtilis |
| Lys | Spike-in RNA derived from Bacillus subtilis |
| Phe | Spike-in RNA derived from Bacillus subtilis |
| Thr | Spike-in RNA derived from Bacillus subtilis |
| Gad1 | Glutamic acid decarboxylase 1 (GAD67) |
| Gad2 | Glutamic acid decarboxylase 2 (GAD65) |
| Slc17a6 | Vesicular glutamate transporter 2 (VGluT2) |
| Slc17a7 | Vesicular glutamate transporter 1 (VGluT2) |
| Slc6a5 | Glycine transporter 2 (GlyT2) |
| Cacna1a | Voltage-dependent calcium channel, P/Q type, α 1A subunit (CaV2.1) |
| Cacna1b | Voltage-dependent calcium channel, N type, α 1B subunit (CaV2.2) |
| Cacna1c | Voltage-dependent calcium channel, L type, α 1C subunit (CaV1.2) |
| Cacna1d | Voltage-dependent calcium channel, L type, α 1D subunit (CaV1.3) |
| Cacna1e | Voltage-dependent calcium channel, R type, α 1E subunit (CaV2.3) |
| Cacna1g | Voltage-dependent calcium channel, T type, α 1G subunit (CaV3.1) |
| Cacna1 h | Voltage-dependent calcium channel, T type, α 1H subunit (CaV3.2) |
| Cacna1i | Voltage-dependent calcium channel, T type, α 1I subunit (CaV3.3) |
| Hcn1 | Hyperpolarization-activated, cyclic nucleotide-gated nonselective cation channel 1 |
| Hcn2 | Hyperpolarization-activated, cyclic nucleotide-gated nonselective cation channel 2 |
| Hcn4 | Hyperpolarization-activated, cyclic nucleotide-gated nonselective cation channel 4 |
| Kcna1 | Voltage-dependent potassium channel, shaker-related subfamily, member 1 (KV1.1) |
| Kcna2 | Potassium voltage-gated channel, shaker-related subfamily, member 2 (KV1.2) |
| Kcna3 | Potassium voltage-gated channel, shaker-related subfamily, member 3 (KV1.3) |
| Kcna4 | Potassium voltage-gated channel, shaker-related subfamily, member 4 (KV1.4) |
| Kcna5 | Potassium voltage-gated channel, shaker-related subfamily, member 5 (KV1.5) |
| Kcna6 | Potassium voltage-gated channel, shaker-related, subfamily, member 6 (KV1.6) |
| Kcnb1 | Potassium voltage gated channel, Shab-related subfamily, member 1 (KV2.1) |
| Kcnc1 | Potassium voltage gated channel, Shaw-related subfamily, member 1 (KV3.1) |
| Kcnc2 | Potassium voltage gated channel, Shaw-related subfamily, member 2 (KV3.2) |
| Kcnc3 | Potassium voltage gated channel, Shaw-related subfamily, member 3 (KV3.3) |
| Kcnc4 | Potassium voltage gated channel, Shaw-related subfamily, member 4 (KV3.4) |
| Kcnd1 | Potassium voltage-gated channel, Shal-related family, member 1 (KV4.1) |
| Kcnd2 | Potassium voltage-gated channel, Shal-related family, member 2 (KV4.2) |
| Kcnd3 | Potassium voltage-gated channel, Shal-related family, member 3 (KV4.3) |
| Kcnf1 | Potassium voltage-gated channel, subfamily F, member 1 (KV5.1) |
| Kcng4 | Potassium voltage-gated channel, subfamily G, member 4 (KV6.4) |
| Kcnh1 | Potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| Kcnh2 | Potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| Kcnma1 | Calcium-dependent potassium channel, large conductance subfamily M, α member 1 (BK) |
| Kcnn1 | Calcium-dependent potassium channel, intermediate/small conductance, subfamily N, member 1 (SK1) |
| Kcnn2 | Calcium-dependent potassium channel, intermediate/small conductance, subfamily N, member 2 (SK2) |
| Scn1a | Voltage-dependent sodium channel, type I, α subunit (NaV1.1) |
| Scn1b | Voltage-dependent sodium channel, type I, β subunit |
| Scn2a1 | Voltage-dependent sodium channel, type II, α subunit (NaV1.2) |
| Scn2b | Voltage-dependent sodium channel, type II, βsubunit |
| Scn3a | Voltage-dependent sodium channel, type III, α subunit (NaV1.3) |
| Scn3b | Voltage-dependent sodium channel, type III, β subunit |
| Scn4b | Voltage-dependent sodium channel, type IV, β subunit |
| Scn8a | Voltage-dependent sodium channel, type VIII, α subunit (NaV1.6) |
| Adcyap1 | Adenylate cyclase activating polypeptide 1 (PACAP) |
| Cck | Cholecystokinin |
| Coch | Cochlin |
| Col5a1 | Collagen, type V, α 1 |
| Crh | Corticotropin releasing hormone |
| Crhbp | Corticotropin-releasing hormone binding protein |
| Grin2c | Ionotropic glutamate receptor, NMDA2C (ϵ 3) |
| Gsg1l | GSG1-like |
| Hspb8 | Heat shock protein 8 |
| Nrn1 | Neuritin 1 (CPG15) |
| Sema3a | Semaphorin 3A |
| Spp1 | Secreted phosphoprotein 1 (osteopontine) |
| Sst | Somatostatin |
| Tacr3 | Tachykinin receptor 3 |
qPCR primer/probe design.
The target cDNA sequences for which primer/probe sets were designed were selected from 3′-end sequences flanking to the polyadenylation signal identified by polyadq (Tabaska and Zhang, 1999) or PolyA_DB 2 (Lee et al., 2007). In case multiple polyadenylation signals were found, the one flanked by the greatest number of 3′ expressed sequence tag (EST) annotations in UniGene (built 174, http://www.ncbi.nlm.nih.gov/unigene) was chosen. Notably, the qPCR primer/probe for Kcnma1 was designed for the transcript (A830039N20Rik) flanking to the designated 3′ end of Kcnma1 gene in the mouse genome, which was assigned as a part of the KCNMA1 gene in the human genome. This is because the primer/probe designed for the 3′ end of Kcnma1 designated in the mouse genome database (i.e., MGSCv37) detected no transcript in most cells, in striking contrast to the ubiquitously observed BK channel current in MVN neurons (Smith et al., 2002; Gittis and du Lac, 2007; van Welie and du Lac, 2011). We speculate that the recognition of the 3′ end of Kcnma1 in the mouse genome database is not correct, and that A830039N20Rik correctly corresponds to the actual 3′ end, as is the case with the human genome. Primers and probes were designed by Primer Express 3.0 (Applied Biosystems). The specificity of the primer/probe sequence was confirmed by BLAT homology search implemented in Ensembl transcript database (http://www.ensembl.org).
Slice preparation and electrophysiology.
The procedure for obtaining brainstem slices for electrophysiological recordings has been described previously (Bagnall et al., 2007; Shin et al., 2011). Mice were deeply anesthetized with Nembutal and decapitated. Coronal slices (thickness, 250 μm) were prepared by vibratome in ice-cold ACSF containing the following (in mm): 124 NaCl, 5 KCl, 1.3 MgSO4, 26 NaHCO3, 2.5 CaCl2, 1 NaH2PO4, and 11 dextrose aerated with 95% CO2/5% O2, then incubated at 34°C for 30 min, and held at RT for at least 30 min before recording. Kynurenic acid (2 mm), picrotoxin (100 μm), and strychnine (10 μm) were added to the ACSF to block glutamatergic, GABAergic, and glycinergic synaptic transmission during recording, respectively. Whole-cell patch-clamp recording from green fluorescent protein (GFP)-positive cells in the magnocellular region of the middle third of the MVN (in the rostrocaudal plane: −5.85 mm to −6.21 mm from bregma), was performed with glass electrodes (4–8 MΩ) filled with the internal recording solutions containing (in mm): 140 K-gluconate, 8 NaCl, 10 HEPES, 0.1 EGTA, 2 Mg-ATP, and 0.3 Na-GTP (pH 7.2, 280–290 mOsm). Data were analyzed as described by Bagnall et al. (2007).
In situ hybridization.
Double fluorescent in situ hybridization was performed using the protocol used in the Allen Mouse Brain Atlas (Lein et al., 2007), with some modification described below. Experiments were performed in 2- to 6-month-old mice. Two mRNAs were simultaneously detected by riboprobes labeled by digoxigenin (DIG), fluorescein (FLU), or biotin (BIO). The riboprobes were synthesized to cover almost the entire sequence of target mRNA, the specificity of which was assessed by comparing the results with those of the Allen Mouse Brain Atlas (Lein et al., 2007). Labeled riboprobes were sequentially processed with peroxidase-conjugated antibodies against DIG, FLU, or BIO, amplified by tyramide-BIO and tyramide-DIG (Hopman et al., 1998), then visualized by streptavidin conjugated with Alexa 488 (Invitrogen) and anti-DIG antibody conjugated with alkaline phosphatase followed by reaction with HNPP/Fast red TR (Roche Diagnostics), respectively. In situ hybridization with retrograde labeling by fluorogold was performed as described previously (Watakabe et al., 2010). Fluorogold 2.5% (Fluorochrome) in 0.9% NaCl was unilaterally injected into the flocculus by pressure through a craniotomy on the petrosal bone. After 7 d, animals were perfused by 4% paraformaldehyde (PFA) in PBS, and the brainstem was processed for in situ hybridization.
Retrograde tracer injection and immunostaining.
Ten percent biotinylated dextran amine (BDA) (3000 MW; Invitrogen) dissolved in 0.1 m PBS (pH 7.4) was stereotaxically injected into the abducens nucleus, the oculomotor nucleus (OMN), or the MVN of mice (2–6 months old) as described previously (Shin et al., 2011). Rabies virus expressing GFP instead of glycoprotein (Osakada et al., 2011) was unilaterally injected into the flocculus as was fluorogold. Five to seven days after injection, mice were transcardially perfused by 0.1 m PBS, followed by 4% PFA in 0.1 m PBS, under deep anesthesia by Nembutal. Coronal brainstem sections (30 μm) were prepared and subjected to immunostaining and BDA staining. Sections were first blocked and permeabilized by 3% normal donkey serum in 0.3% Triton X-100 in PBS for 1 h at room temperature, then incubated in primary antibody solution including anti-Spp1 (1:400, goat; R & D Systems #AF808) and/or anti-Pcp2 (1:200, rabbit; Abgent #AP6356a) for overnight at 4°C. Immunoreactivity was visualized by anti-goat-DyLight549, anti-goat-DyLight649, and/or anti-rabbit-Cy3 (for all the secondary antibodies, 1:400, raised in donkey; Jackson ImmunoResearch). Subsequently, if needed, BDA was visualized by streptavidin-Alexa Fluor 647 (1 μg/ml; Invitrogen). Distributions of immunoreactivity and retrogradely labeled cells were first mapped by fluorescent images (1297 × 1038 μm) taken with 10× objective lens, which was further confirmed by direct inspection by eye at higher magnification. GFP signal intensity in the GlyT2 line greatly varied across cells; we regarded any trace of GFP signal as GFP positive. Analysis was performed using the Cell Counter plug-in of ImageJ (http://rsbweb.nih.gov/ij/).
Data analysis and statistics.
Ct values are presented in the figures (Figs. 1, 2, 3) and subjected to the hierarchical clustering analysis without normalization. For dendrogram computation with the qPCR results (Figs. 2, 3), the distance (dissimilarity) between leafs (single-cell cDNAs) was defined as “1-Pearson's correlation coefficient of Ct values.” In the cases where the Ct value was greater than average Ct of spike-in RNA Thr (23.07) or not determined (ND), they were assigned Ct values of 40 so that gene expression fluctuation at low levels (<5 copies) did not influence the analyses. All of the dendrograms were calculated by Ward's hierarchical clustering method. In Figure 2A, mutual information (MI) was calculated as follows:
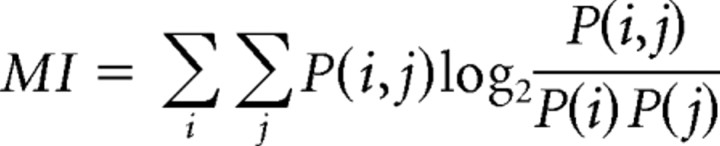 |
where i is the index of the primary dichotomy of the dendrogram (YFP-16-rich cluster/GIN and GlyT2-rich cluster; i = 1, 2), and j corresponds to rounded Ct value (j = 1, …, 40). Multiple pairwise comparison was done by the Wilcoxon rank sum test followed by Benjamini–Hochberg false discovery rate correction. All analyses were done by R (http://www.r-project.org/). Variances were reported in the text and the figures are SDs.
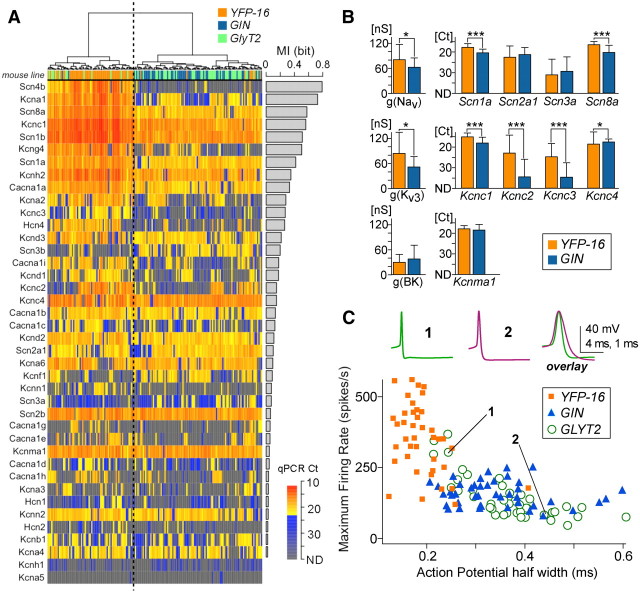
Figure 2.
Ion channel transcript profiles correspond with electrophysiological properties of MVN neurons. A, Dendrogram and heat map of single-cell cDNAs from each neuron (columns) based on the ion channel transcript profiles (rows). The color coding of the mouse lines and the color scale of heat map are the same as in Figure 1F. Ion channel transcript profiles divide the single-cell cDNAs into two groups, bisected by the vertical dotted line; one predominantly comprises YFP-16 cells, and the other largely comprises GIN and GlyT2 cells. The bar graph at right indicates MI of the ion channel genes, reflecting their contribution to the primary dichotomy of the dendrogram. Genes in the heat map are sorted by MI; genes ranking high in MI are expressed at higher levels in the YFP-16-rich branch and include ion channels essential for sustained firing at high rates. Ct values represented as gray-blue indicate very low expression levels and were not used in the dendrogram calculations to exclude the influence of stochastic error in gene expression regulation. B, Comparison of NaV, KV3, and BK channel conductances recorded in acutely dissociated MVN neurons, g(NaV), g (KV3), and g(BK), respectively, with corresponding transcript levels (qPCR Ct values). The transcript profiles are congruent with electrophysiological data and reveal the ion channel subtypes underlying the electrophysiologically measured conductances. Significant differences in conductances and transcript levels between YFP-16 and GIN neurons are indicated by *p < 0.05 and ***p < 0.0001. The conductance data were reanalyzed from Gittis and du Lac (2007) (for NaV: YFP-16, n = 39, GIN, n = 35; for KV3: YFP-16, n = 20, GIN, n = 19; for BK: YFP-16, n = 10, GIN, n = 5). C, Action potential waveforms from 2 GlyT2 neurons are shown above a plot of maximum firing rate evoked during 1 s of intracellular depolarization versus action potential width (measured at half-height) for neurons recorded from three mouse lines. As predicted from ion channel gene expression profiles, GlyT2 neurons range widely in electrophysiological properties. YFP-16: n = 40, orange squares; GIN: n = 43, blue triangles; GlyT2, n = 41, green open circles. YFP-16 and GIN data are replotted from Bagnall et al. (2007). Scale for action potential waveform: 40 mV and 4 ms for cells 1 and 2 and 40 mV and 1 ms for the overlay.
Figure 3.
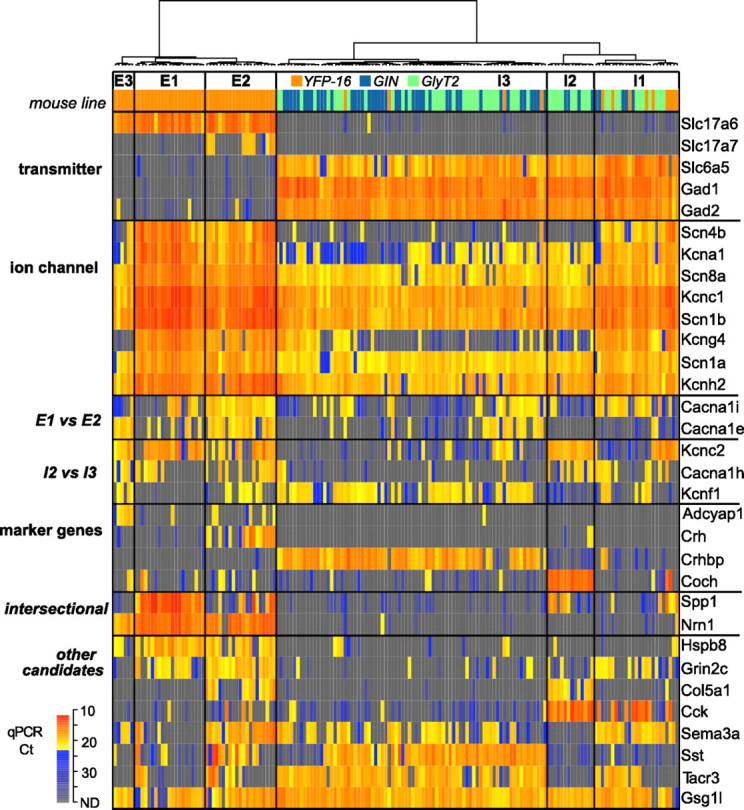
Identification of marker genes associated with the neurochemically and electrophysiologically homogeneous cell types. The dendrogram of the single-cell cDNAs (columns) was computed using the transcript profiles of the transmitter-related genes, the ion channel genes, and the marker gene candidates selected using Allen Mouse Brain Atlas (Lein et al., 2007). The color code rows under the dendrogram indicate mouse lines as in Figure 1F and Figure 2A. In the dendrogram, the single-cell cDNAs are divided into six cell types (E1–3, I1–3) within which expression profiles of the transmitter-related and ion channel genes are generally homogeneous. The heat map shows the transcript profiles (qPCR Ct values) of the transmitter-related genes (n = 5), the ion channel genes that ranked high MI in Figure 2A (n = 8) or differentially expressed between E1 and E2 (E1 vs E2, n = 2) or I2 and I3 (I2 vs I3, n = 3), and the marker gene candidates. Those associated with the defined cell types are labeled as “marker genes” (n = 4), those serving as marker in combination are labeled as “intersectional” (n = 2), and the others are labeled as “other candidates” (n = 8).
Results
Our strategy for identifying cell types and associated marker genes was to perform single-cell expression profiling of specific sets of genes and to classify neurons based on quantitative gene expression patterns. We chose three distinct classes of genes for analysis: transmitter-related genes, ion channel genes, and candidate marker genes identified in the Allen Mouse Brain Atlas (Lein et al., 2007). Motivated by observations that functionally distinct cell types are partially segregated in different regions of the MVN (Scudder and Fuchs, 1992; Bagnall et al., 2007), we selected genes as marker candidates if they exhibited spatially distinct patterns of hybridization signal (Fig. 1A). Transcripts were analyzed in neurons from the MVN, a brainstem nucleus containing several cell types which subserve the horizontal VOR.
Preparation of single-cell cDNAs from MVN neurons
MVN neurons were harvested from acute brainstem slices obtained from three strains of transgenic mice (YPF-16, Feng et al., 2000; GIN, Oliva et al., 2000; GlyT2, Zeilhofer et al., 2005) which express fluorescent protein in spatially distributed subsets of neurons (Fig. 1B). Fluorescently labeled neurons were isolated with an enzymatic and mechanical dissociation method which preserves neuronal physiology (Gittis and du Lac, 2007). Juvenile mice, age 24–32 d (27.2 ± 2.2), were used to attain a higher yield of healthy neurons; at this stage, performance and plasticity of the horizontal VOR are mature (Faulstich et al., 2004). Neurons sampled primarily from the central portion of the vestibular complex (Fig. 1C) were identified with a fluorescent stereoscope, washed, and individually harvested into PCR tubes by a manual sorting method (Hempel et al., 2007). After RT, single-cell cDNAs were globally amplified using a PCR-based method (Kurimoto et al., 2007), in which the 3′ rapid amplification of cDNA ends method (Tietjen et al., 2003) was modified to attain high amplification of cDNAs with minimal distortion of the transcript abundance distribution (see Materials and Methods). Using this method, a single neuron can yield as much as 50 μg of cDNA (fragments of ∼700 bp from 3′ end), enabling repetition of sufficient numbers of qPCR to ensure reliable, extensive transcript profiling. In total, we obtained cDNAs from 167 individual neurons (YPF-16: n = 61; GIN: n = 52; GlyT2: n = 54) which passed stringent quality control tests (see Materials and Methods).
To evaluate the detection power of the single-cell transcript profiling, the abundance of spike-in control RNAs added to each single-cell sample was examined by qPCR. Lys (1000 copies), Dap (100 copies), and Phe (20 copies) were detected in virtually all the samples (Lys, 167/167; Dap, 164/167; Phe, 164/167), whereas Thr (five copies) was detected in ∼60% of the samples (Thr, 102/167). This indicates that transcripts of >20 copies are nearly always represented in the single-cell transcript profiling. The qPCR results of the spike-in RNAs generally followed the initial copy numbers, supporting the notion that transcript abundance representation in a single cell was maintained after amplification (Fig. 1E).
Expression profiling of neurotransmitter-related genes
To confirm that the methods for dissociation and molecular amplification could reliably evaluate gene expression and to determine neurotransmitter profiles across neurons, single-cell cDNAs were probed with qPCR for five genes related to neurotransmitter synthesis or transport. Genes for vesicular glutamate transporters (VGluT) 1 and/or 2 (Slc17a7 and/or Slc17a6) and glycine transporter 2 (Slc6a5) were used to identify glutamatergic and glycinergic neurons, respectively. GABAergic neurons were identified as those expressing genes for glutamic acid decarboxylase (Gad1 and/or Gad2).
Neurotransmitter-related gene expression in 167 MVN neurons from three lines of transgenic mice is shown as a heat map in Figure 1F. Hierarchical clustering analyses divided the population into two distinct groups, here defined as “excitatory” and “inhibitory” on the basis of mutually exclusive expression of Slc17a6 and Gad1, respectively. All glutamatergic neurons expressed Slc17a6, and a small subset of glutamatergic neurons coexpressed Slc17a7. In contrast, all inhibitory neurons expressed both Gad1 and Gad2. Surprisingly, Slc6a5 was coexpressed in most inhibitory neurons, suggesting that GABA and glycine may be coreleased (Walberg et al., 1990; Wentzel et al., 1993; Tanaka and Ezure, 2004; Lu et al., 2008). Excitatory neurons were obtained exclusively from YFP-16 mice while inhibitory neurons were obtained from all three lines of mice, as is consistent with previous reports indicating that glutamatergic or glycinergic neurons are labeled in the YFP-16 line while GABAergic and glycinergic neurons are labeled in the GIN line (Bagnall et al., 2007).
Expression profiling of ion channel genes
Previous reports demonstrate that functionally distinct MVN neurons differ in their expression of ionic currents and intrinsic excitability. Relative to interneurons targeted in GIN mice, projection neurons targeted in YFP-16 mice have larger repolarizing currents and narrower action potentials which enable faster firing (Bagnall et al., 2007; Gittis and du Lac, 2007; Gittis et al., 2010). To determine whether ion channel genes reflect these differences, we performed quantitative analyses on 36 transcripts encoding α subunits of voltage-gated or calcium-dependent ion channels and on 4 sodium channel β subunits.
Ion channel expression patterns divided the population of 167 MVN neurons into two major groups, each comprising both excitatory and inhibitory neurons, with one group dominated by YFP-16 neurons and the other by GIN and GlyT2 neurons (Fig. 2A). Qualitative and quantitative differences in gene expression across cells were evident. The contribution of each gene to this dichotomy was quantified as MI (see Materials and Methods). Genes related to the initiation and repolarization of action potentials were primarily responsible for the classification into the two groups. Genes with the highest MI values included Na channel α and β subunits (Scn8a, Scn1a, Scn4b, and Scn1b) and repolarizing K channel subunits (Kcna1, Kcnc1). Surprisingly, a “silent” KV α subunit (Kcng4) also ranked high in MI, revealing a potentially important role for Kv6.4 channels in regulating excitability.
Recordings of ionic currents from dissociated MVN neurons demonstrated that Na and KV3, but not BK, currents are significantly larger in YFP-16 than GIN neurons (Gittis and du Lac, 2007). A comparison of whole-cell conductances from that study and transcript levels of the related genes is shown in Figure 2B. Larger Na channel conductance in YFP-16 than GIN neurons is paralleled by significantly higher expression of Na channel genes Scn1a and Scn8a but no difference in Scn2a1 or Scn3a transcript levels. Larger KV3 conductances in YFP-16 versus GIN neurons are matched by significantly higher expression of associated Kcnc1, Kcnc2, and Kcnc3 transcripts. BK channel conductances and corresponding Kcnma1 gene expression were equivalent in YFP-16 and GIN neurons. The parallels between ionic currents measured electrophysiologically and gene expression levels indicates that the relative strength of ionic conductances can be inferred from transcript levels.
Action potential speed and the ability to sustain high firing rates are determined primarily by the amplitude and kinetics of Na and KV3 channels in YFP-16 and GIN neurons (Gittis and du Lac, 2007; Gittis et al., 2010). Expression levels of genes associated with action potentials vary widely in GlyT2 neurons (Fig. 2A), predicting similar variations in action potentials and firing properties. To test this prediction, GlyT2 neurons were targeted for whole-cell recordings in the MVN in acute brainstem slices. As expected from ion channel expression profiling, GlyT2 neurons ranged widely in intrinsic physiology (Fig. 2C). A subset of GlyT2 neurons with narrow action potentials and relatively high maximum firing rates overlapped with the distribution of YFP-16 neurons, while the majority overlapped with GIN neurons (Fig. 2C). The congruence between results from transcript profiling and physiological experiments indicates that ion channel expression profiles can distinguish neurons which exhibit graded differences in intrinsic physiology.
Postinhibitory rebound firing varies widely within vestibular nucleus neurons (Sekirnjak and du Lac, 2002); YFP-16 neurons exhibit significantly more pronounced rebound firing than do GIN neurons (Bagnall et al., 2007). Rebound firing in vestibular and cerebellar nucleus neurons relies predominantly on cationic H channels and T-type Ca channels (Aizenman and Linden, 1999; Sekirnjak and du Lac, 2002; Molineux et al., 2008). Consistent with these findings, transcripts associated with these channels differed significantly between YFP-16 and GIN neurons (Fig. 2A). H channel genes (Hcn1, Hcn2, and Hcn4) were expressed solely or in combination with each other in 88% of YFP-16 neurons but only 57% of GIN neurons (p < 0.001 Fisher exact test; n = 52 and 61, respectively). In neurons expressing H channel genes, transcript levels were significantly higher in YFP-16 than GIN neurons (p < 0.0001; sum of threshold cycle (Ct) values, 90 ± 16 vs 109 ± 12; note that the smaller the Ct is, the more transcript there is). At least one T-type calcium channel gene (Cacna1 g, Cacna1 h, Cacna1i) was expressed in most neurons, but combined T-type channel transcript levels were significantly higher in YFP-16 than GIN neurons (p < 0.0001; sum of Ct values, 90 ± 17 vs 122 ± 11). Together, these results demonstrate that transcript profiling of ion channel genes can reliably differentiate between physiologically distinct cell types.
Neuronal classification incorporating marker gene candidates
Marker gene candidates were identified in the Allen Mouse Brain Atlas (Lein et al., 2007) via systematic screening of the Anatomical Gene Expression Atlas (Ng et al., 2009) and manual queries. Hierarchical clustering analyses incorporating quantitative expression patterns of 14 marker gene candidates together with the 5 neurotransmitter and 40 ion channel-related genes classified neurons into functionally and molecularly distinct groups (Fig. 3). Six major cell types emerged from this classification; three excitatory and three inhibitory (designated E1–3 and I1–3, respectively).
A subset of marker gene candidates identified in the Allen Mouse Brain Atlas (Lein et al., 2007) were expressed predominantly in one cell type, including Adcyap1 (E3), Crh (E2), Crhpb (I3), and Coch (I2). The gene for VGlutT1 (Slc17a7) was also expressed exclusively in E2. Nrn1 was expressed in virtually all glutamatergic neurons, while Spp1 was expressed predominantly in E1 as well as in subsets of E2, I1, and I2. As such, E1 could be defined by the intersection of Spp1 and Nrn1 or Slc17a6. Several marker gene candidates, including neuropeptide genes Sst and Cck, were expressed in multiple cell types.
Two excitatory cell types and one inhibitory cell type (E1, E2, and I1) expressed patterns of ion channel transcripts associated with narrow action potentials and faster firing neurons (e.g., high levels of Scn8a and Kcnc1). Calcium channel transcripts distinguished these cell types; Cacna1e was coexpressed with Cacna1i in E2 but not in E1 or I1 (Fig. 3, E1 vs E2). Inhibitory cell types expressing low levels of narrow action potential-related genes indicative of slower firing neurons could be distinguished by differential expression of Kcnc2 (I2), Cacna1 h (I2), and Kcnf1 (I3) (Fig. 3, I2 vs I3). These results support the principle that molecular classification based on quantitative expression of genes related to neurotransmitters and ion channels can define functionally distinct cell types.
Validation and anatomical distribution of marker genes via in situ hybridization
To confirm marker genes identified with single-cell transcript profiling, we examined their coexpression with neurotransmitter-related genes (excitatory: Slc17a7, Slc17a6; inhibitory: Gad1) using double fluorescent in situ hybridization (DFISH). As predicted, marker genes for excitatory groups E2 (Crh) and E3 (Adycap1) were coexpressed with Slc17a7 (Fig. 4A; 83.0% of Crh-positive neurons, n = 141) and Slc17a6 (Fig. 4B; 81.1% of Adcyap1-positive neurons, n = 74), respectively. Note that Slc17a7 is another marker for E2. Similarly, marker genes for inhibitory groups I2 and I3 (Coch and Crhbp) were expressed exclusively in Gad1-positive neurons (Fig. 4C,D, 98.7% of Coch-positive neurons, n = 75; 97.5% of Crhbp-positive neurons, n = 118).
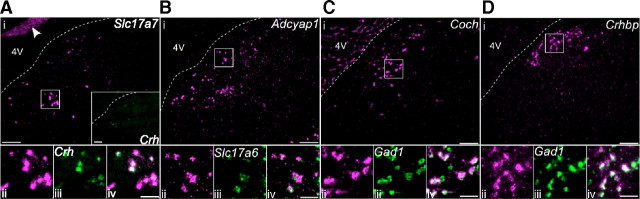
Figure 4.
Validation of identified marker genes with double fluorescent in situ hybridization. In each part, an in situ hybridization image of a coronal MVN section is shown in the top row (i), and magnified images of the white-boxed area are shown in the bottom row (ii–iv). A, Slc17a7 (magenta) and Crh (inset, green). The in situ hybridization signals of Slc17a7 and Crh were sparsely distributed over the MVN, and greatly overlapped each other. The arrowhead indicates the granule cell layer in the cerebellum. B, Adcyap1 (magenta) and Slc17a6 (green). Adcyap1-positive cells were scattered in the MVN, especially near the fourth ventricle. In the majority of cases, they colabeled with Slc17a6. C, Coch (magenta) and Gad1 (green). Coch-positive cells were nearly always colabeled by Gad1. The hybridization signal was also observed in the choroid plexus in the fourth ventricle. D, Crhbp (magenta) and Gad1 (green). Crhbp-positive cells were also colabeled by Gad1 (green, iii and iv), forming a dense cluster near the fourth ventricle and showing distinct distribution from that of Coch-positive cells. Scale bar: 150 μm for the top row images (i) and the inset images in A; 50 μm for the bottom images (ii, iii, iv).
The anatomical distribution of marker genes provides further evidence that they are expressed in distinct cell types. Adycap1-positive cells (E3) were distributed near the fourth ventricle (Fig. 4B). In contrast, Crh- and Slc17a7-positive neurons (E2) were sparsely distributed in the central portion of the MVN (Fig. 4A) and formed a dense cluster in the supragenual nucleus (data not shown). Of the inhibitory cell markers, Crhbp (I3) was densely expressed near the fourth ventricle (Fig. 4D), dorsal to Coch-positive neurons (I2, Fig. 4C). No cells coexpressed Crhbp and Coch (n = 167 expressing either but not both), confirming that these genes are expressed in distinct populations of neurons.
Functional identity of cell types classified by gene expression profiles
The MVN comprises neurons that differentially project to the cerebellum, ocular motoneurons or locally within the bilateral vestibular system (Highstein and Holstein, 2006). To determine the axonal projections of molecularly defined cell classes, we combined retrograde tracer labeling with in situ hybridization or immunohistochemistry. Injection of fluorogold or glycoprotein-deleted rabies virus expressing enhanced GFP (Osakada et al., 2011) into the cerebellar flocculus labeled neurons in the central portion of the MVN and in the supragenual nucleus which expressed Slc17a7 and/or Crh (Fig. 5A; 21/25 and18/28 fluorogold-positive cells, respectively, n = 2 animals), but not Spp1 (3/47, 0/51, 3/79, and 4/56 rabies virus infected neurons, n = 4 animals). This result indicates that the E2 population includes precerebellar mossy fiber neurons.
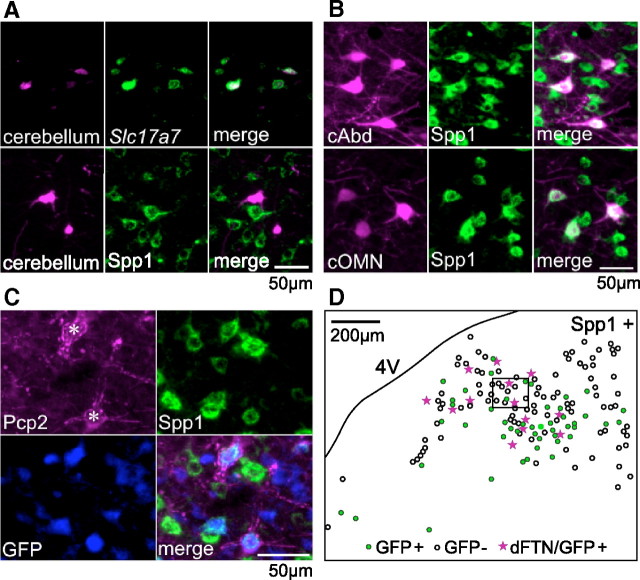
Figure 5.
Projection target and Purkinje cell innervation of molecularly defined cell types. A, Retrograde labeling of precerebellar neurons followed by in situ hybridization (top) or immunostaining against Spp1 (bottom) demonstrates that precerebellar neurons largely express Slc17a7, but not Spp1, indicating that E2 corresponds to precerebellar neurons. For retrograde labeling, FG and glycoprotein-deleted rabies virus are used for in situ hybridization and immunostaining, respectively. B, Excitatory premotor neurons retrogradely labeled by BDA. The top and bottom row show the cells labeled by injection into the contralateral abducens (cAbd) and the contralateral OMN (cOMN), respectively. The left, middle, and right columns show BDA signal, Spp1 immunoreactivity, and merged images, respectively. The majority of the retrogradely labeled neurons are immunostained against Spp1, indicating that E1 corresponds to premotor neurons. C, A representative image of MVN neurons densely innervated by Purkinje cell terminals (*). Pcp2 immunoreactivity visualizes Purkinje cell terminals. Spp1+/GFP + cells in the GlyT2 line mostly include cells densely innervated by Purkinje cell terminals. D, A representative distribution map of Spp1 immunoreactivity positive cells (Spp1+). Filled green dots and open circles indicate the presence and absence of GFP signal in the GlyT2 line, which presumably correspond to inhibitory and excitatory cells, respectively. Pink stars indicate cells densely innervated by Purkinje cell terminals (dFTN; Shin et al., 2011), which also express GFP. Note that Spp1-positive inhibitory cells in the ventrolateral MVN are likely to belong to I1, which include some dFTNs. dFTNs near the fourth ventricle might belong to I2, because they are in the territory of cells expressing Coch (Fig. 4C, I2 marker). The image shown in C corresponds to the boxed area.
To identify premotor cell types, we injected BDA into motor nuclei subserving horizontal eye movements: the abducens and OMN. Experiments were performed in the GlyT2 mouse line, in which GFP-negative neurons are predominantly glutamatergic (Zeilhofer et al., 2005); DFISH confirmed that virtually all MVN neurons expressing Slc6a5 (gene for glycine transporter 2) were GFP positive in the GlyT2 line (data not shown). Neurons retrogradely labeled from the contralateral abducens or OMN were devoid of GFP expression in the GlyT2 line, as expected from previous studies demonstrating that MVN neurons projecting contralaterally to ocular motoneurons are glutamatergic (Wentzel et al., 1995; Shin et al., 2011). Neurons projecting contralaterally to the abducens and OMN were immunopositive for Spp1 (Fig. 5B; for each of two animals, abducens: 64/77 and 88/96 neurons; OMN: 50/52 and 54/69 neurons). These results demonstrate that the E1 population comprises premotor neurons that control movements of the contralateral eye.
Neurons in the MVN that project axons to the ipsilateral abducens nucleus are predominantly glycinergic (Spencer et al., 1989; Shin et al., 2011). A subset of these neurons receive dense somatic innervation from the cerebellar flocculus (Sekirnjak et al., 2003; Shin et al., 2011). We identified such flocculus target neurons in GlyT2 mice with antibodies against the Purkinje cell marker Pcp2 (aka L7) and used immunostaining to determine whether they coexpress Spp1. The majority of glycinergic neurons that were densely surrounded by Pcp2 immunoreactivity were also immunostained for Spp1 (Fig. 5C,D; 40/43 and 69/79 neurons in two animals). These results suggest that flocculus target neurons that mediate cerebellar influence of ipsilateral eye movements are enriched in Spp1-positive neurons included in I1 and/or I2 (Fig. 3).
A summary of major cell types, axonal projections, and marker genes identified in this study is shown in Figure 6. The E1 population comprises glutamatergic neurons that project to contralateral motor nuclei and are identified by coexpression of Spp1 and Slc17a6 (or Nrn1). The E2 population includes glutamatergic precerebellar mossy fiber neurons which can be identified by expression of Crh or Slc17a7. The projections of E3, glutamatergic neurons which express the marker gene Adycap1, have yet to be identified; ion channel transcripts suggest that these neurons have slower firing properties than those in E1 and E2. The I1 population of inhibitory neurons expresses genes related to fast firing; these neurons can be identified by coexpression of Scn4b and inhibitory marker such as Slc6a5, and include cerebellar target neurons projecting to the ipsilateral abducens motor nucleus. I2 and I3 comprise anatomically distinct populations of inhibitory neurons distinguished by marker genes Coch (I2) and Crhbp (I3). The I2 population largely comprises GlyT2 neurons, some of which are likely to be cerebellar target neurons and project ipsilaterally to the abducens (Shin et al., 2011). Previous reports demonstrated that GIN neurons project axons locally or to the contralateral vestibular nucleus (Bagnall et al., 2007; McElvain et al., 2010). The I3 population, which is dominated by GIN neurons and includes transcriptionally similar GlyT2 neurons, is likely to comprise local and commissurally projecting interneurons. Although additional subdivision of these cell types may be prompted by further analyses, each of the previously known classes of vestibular nucleus neurons are represented in our molecularly defined classification scheme. Thus, single-cell transcript profiling was able to successfully classify a heterogeneous population of intermingled brainstem neurons into six anatomically and functionally distinct cell types.
Figure 6.
Summary of marker genes and projection targets of the six cell types identified with our molecular classification strategy. E1–3 are excitatory neurons; I1–3 are inhibitory neurons.
Discussion
This study presents a new strategy for classifying mature neurons into functionally distinct types and for identifying differentially expressed marker genes via quantitative single-cell transcript analyses. By applying this strategy to a heterogeneous population of MVN neurons, we identified six major cell types together with associated marker genes. In situ hybridization and immunohistochemical analyses in conjunction with retrograde and anterograde tracing confirmed that cell types expressing different marker genes occupy distinct positions within the vestibulo-motor circuits. This approach provides an efficient means of identifying differentially expressed genes which can be used to label and manipulate distinct cell types for functional analyses of neural circuitry and behavior.
Strategic and technical considerations
Several criteria differ across functionally distinct classes of neurons, including dendritic morphology, axonal projections, innervation patterns, intrinsic excitability, and neurotransmitter types. In designing our strategy, we reasoned that genes directly related to excitability and transmitter types have the best chance of distinguishing mature cell types with clear functional differences. To identify marker gene candidates, we used the Allen Mouse Brain Atlas (Lein et al., 2007) and selected 14 genes that appeared to be expressed in subsets of MVN neurons. Although most of these candidates were expressed in several cell types, four were expressed almost exclusively in single populations and thus served as useful markers genes. Additional differentially expressed genes which may further subdivide the major classes defined here could be identified via genome-wide analyses of pooled cDNAs from neurons of each type.
Quantitative analyses of ion channel transcripts serve as a powerful means to classify neurons, even in populations with subtle differences in electrophysiological profiles (Fig. 2). Our findings demonstrate that small variations in action potential width between cell types are evident in expression levels of several ion channel transcripts. In addition, variations in postinhibitory rebound firing within and between cell types is matched by variations in genes encoding channels activated at hyperpolarized membrane potentials (H and T-type Ca channel). Most brain regions comprise cell types which exhibit larger differences in intrinsic excitability than do vestibular nucleus neurons; our strategy should be valuable for identifying subtle differences among cell types even in the absence of prior electrophysiological information.
Although we used juvenile animals (P24–32) to prepare single-cell cDNAs, several lines of evidence indicate that MVN neurons are mature by that stage. First, action potential waveforms and intrinsic excitability attains developmental maturity before P20 (Murphy and du Lac, 2001). Second, performance and plasticity of the horizontal VOR are mature by P21–26 (Faulstich et al., 2004). Finally, in situ hybridization data from 2- to 6-month-old mice matched the gene expression profiles obtained with qPCR in neurons harvested from juveniles.
Single-cell transcript analyses performed with qPCR have several potential sources of variation and error. First, the stochastic nature of gene expression regulation (Elowitz et al., 2002) can result in variability in transcript numbers. This may account for atypical patterns of sparsely expressed transcripts (e.g., Slc17a6 in inhibitory neurons, shown as blue-gray in Fig. 1F). Second, inappropriate design of primers and probes can yield false negatives. We selectively amplified 3′-end sequences of the cDNAs, and, for primer/probe design, used the distribution of 3′ ESTs to identify the most frequently transcribed 3′-end sequence; however, annotation of 3′ ESTs to genes in the database (i.e., UniGene) is not always perfect. In one case (Kcnma1; see Materials and Methods) we redesigned primers and probes that were initially discrepant with previous electrophysiological or anatomical data. Third, any method involving several time-critical steps and biological tissue can be compromised by accumulation of errors. To minimize such errors, we used a robotic pipetting system for qPCR preparation, and performed rigorous quality control experiments, rejecting all samples that might be compromised by contamination or transcript degradation. Mouse lines in which neurons express fluorescent proteins are not necessary for our strategy but can be useful both for distinguishing neurons from debris during cell sorting and for enriching samples with cell types of interest.
Classification of vestibular nucleus neurons into six distinct types
Conjoint expression of transmitter-related, ion channel, and marker candidate genes defined six molecularly distinct classes of vestibular nucleus neurons. Three cell types (E1–3) expressed the glutamate transporter VGluT2 and were defined as excitatory. The remaining three cell types (I1–3) coexpressed genes encoding synthetic enzymes for GABA (Gad65 and Gad67) and for the glycine transporter GlyT2 and were thus defined as inhibitory. Although distinct types of vestibular nucleus neurons are thought to mediate GABAergic versus glycinergic inhibition (Precht et al., 1973; Spencer et al., 1989; Furuya et al., 1991; Camp et al., 2006; Bagnall et al., 2007), the coexpression of these transmitters suggests the possibility of corelease, as has been shown to speed the kinetics of synaptic responses in the auditory brainstem neurons (Lu et al., 2008).
Ion channel transcript levels defined two predominant types of MVN neurons (Fig. 2A), but genes responsible for initiating and repolarizing the action potential (Na and Kv3 channel α subunits) exhibited continuously graded differences across neurons in each type. These results bridge previously discrepant findings (Serafin et al., 1991; Johnston et al., 1994; du Lac and Lisberger, 1995; Straka et al., 2005; Bagnall et al., 2007) by demonstrating both bimodal and continuous patterns of ion channel gene expression. Differences in transmitter expression and axonal projections across neurons within each of the two cell types defined with gene expression pattern underscores the importance of a classification scheme which incorporates multiple functional properties.
Several types of vestibular nucleus neurons have been distinguished on the basis of firing responses during behavior, axonal projections, and/or synaptic inputs (Highstein and Holstein, 2006; Cullen, 2012). Two cell types are thought to directly control the VOR: excitatory “position-vestibular-pause (PVP)” neurons provide strong drive to motoneurons (Scudder and Fuchs, 1992) and flocculus target neurons (FTNs; Lisberger and Pavelko, 1988), subsequently identified as glycinergic (Shin et al., 2011), mediate cerebellar learning. The firing properties of these cell types differ from each other (Scudder and Fuchs, 1992; Lisberger et al. 1994; Ramachandran and Lisberger, 2008), from vestibular nucleus neurons projecting mossy fiber axons to the cerebellar flocculus (Zhang et al., 1995; Cheron et al., 1996), and from other vestibular nucleus neurons (Scudder and Fuchs, 1992, Cullen, 2012). Additional physiological (Precht et al., 1973; Furuya et al., 1991; Malinvaud et al., 2010) and anatomical (Barmack et al., 1993; De Zeeuw et al., 1993; Balaban and Beryozkin, 1994; Holstein et al., 1999a,b; Bagnall et al., 2007; McElvain et al., 2010) studies indicate the presence of several inhibitory cell types with axons projecting locally, commissurally, to the inferior olive, or to ocular motoneurons.
Two of the excitatory cell types defined by our transcript analyses have a clear relationship with vestibular neuronal classes previously identified in vivo. The E1 population, comprising large neurons which project contralaterally to abducens or OMNs, corresponds with PVP interneurons which provide the predominant excitatory drive to motoneurons. (Scudder and Fuchs, 1992; Ramachandran and Lisberger, 2008). The E2 population, which includes precerebellar neurons, corresponds with mossy fiber neurons projecting to the cerebellar flocculus (Zhang et al., 1995; Cheron et al., 1996). The remaining excitatory neurons (E3) have no known function in the VOR but might correspond with vestibulo-sympathetic neurons projecting to autonomic circuits in the medulla (Holstein et al., 2011). The marker gene Adycap1 provides a molecular handle for circuit and behavioral analyses of the role of E3 neurons.
Inhibitory cell types identified in our molecular classification scheme include premotor neurons densely targeted by Purkinje cell terminals (I1) and spatially segregated populations of neurons (I2 and I3) expressing distinct marker genes (Coch and Crhbp, respectively). The I1 population includes FTNs which were identified in vivo as vestibular nucleus neurons receiving powerful, monosynaptic cerebellar inhibition (Lisberger and Pavelko, 1988; Ramachandran and Lisberger, 2008). Glycinergic neurons with sparse somatic innervation by Purkinje cells also project axons to the abducens nucleus (Shin et al., 2011) and are likely to be included in the I2 population which express the premotor marker Spp1. The I3 population predominantly comprises GIN neurons known to restrict axonal projections to the bilateral vestibular complex (McElvain et al., 2010) and are likely to be a predominant source of feedforward inhibition (Biesdorf et al., 2008).
Future applications
The strategy for classifying neurons and identifying associated marker genes presented here can be applied throughout the nervous system. The extent of neuronal diversity in the cerebellar nuclei (Bagnall et al., 2009; Uusisaari and Knöpfel, 2012), which are anatomical and functionally similar to vestibular nuclei, could be readily addressed with single-cell transcript profiling. Heterogeneity in intrinsic excitability across vestibular ganglion cells (Kalluri et al., 2010; Eatock and Songer, 2011) and anatomical and functional differences between ocular motoneurons (Büttner-Ennever et al., 2001; Davis-López de Carrizosa et al., 2011) suggest that corresponding molecular differences could be identified in VOR sensory inputs and motor outputs. This strategy, which requires no pre-existing knowledge about neuronal diversity, should be particularly effective for fundamental brainstem circuits in which experimental traction was previously limited by an inability to unambiguously identify different cell types.
Cerebellar and vestibular circuits exhibit remarkable plasticity throughout life, and several studies indicate that mechanisms of plasticity differ across neurons (Straka et al., 2005; Dutia, 2010; McElvain et al., 2010; Pettorossi et al., 2011). Future studies exploiting the molecular markers identified here to generate viruses and/or mouse lines for cell-type-specific manipulations will make it possible to analyze cerebellar learning and vestibulo-motor plasticity at the level of gene expression in functionally identified classes of neurons.
Footnotes
This work was supported by the Howard Hughes Medical Institute, and National Institute of Health Grant EY-11027. We thank F. Osakada and E.M. Callaway for the recombinant rabies virus, K. Kurimoto for technical advice, S. Fenstermacher and M. Fuentes for technical assistance, and S. Sahara and P. Slesinger for valuable discussions.
References
- Aizenman CD, Linden DJ. Regulation of the rebound depolarization and spontaneous firing patterns of deep nuclear neurons in slices of rat cerebellum. J Neurophysiol. 1999;82:1697–1709. doi: 10.1152/jn.1999.82.4.1697. [DOI] [PubMed] [Google Scholar]
- Bagnall MW, Stevens RJ, du Lac S. Transgenic mouse lines subdivide medial vestibular nucleus neurons into discrete, neurochemically distinct populations. J Neurosci. 2007;27:2318–2330. doi: 10.1523/JNEUROSCI.4322-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnall MW, Zingg B, Sakatos A, Moghadam SH, Zeilhofer HU, du Lac S du. Glycinergic projection neurons of the cerebellum. J Neurosci. 2009;29:10104–10110. doi: 10.1523/JNEUROSCI.2087-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban CD, Beryozkin G. Organization of vestibular nucleus projections to the caudal dorsal cap of kooy in rabbits. Neuroscience. 1994;62:1217–1236. doi: 10.1016/0306-4522(94)90354-9. [DOI] [PubMed] [Google Scholar]
- Barmack NH, Fagerson M, Errico P. Cholinergic projection to the dorsal cap of the inferior olive of the rat, rabbit, and monkey. J Comp Neurol. 1993;328:263–281. doi: 10.1002/cne.903280208. [DOI] [PubMed] [Google Scholar]
- Biesdorf S, Malinvaud D, Reichenberger I, Pfanzelt S, Straka H. Differential inhibitory control of semicircular canal nerve afferent-evoked inputs in second-order vestibular neurons by glycinergic and GABAergic circuits. J Neurophysiol. 2008;99:1758–1769. doi: 10.1152/jn.01207.2007. [DOI] [PubMed] [Google Scholar]
- Blazquez PM, Hirata Y, Highstein SM. The vestibulo-ocular reflex as a model system for motor learning: what is the role of the cerebellum? Cerebellum. 2004;3:188–192. doi: 10.1080/14734220410018120. [DOI] [PubMed] [Google Scholar]
- Broussard DM, Kassardjian CD. Learning in a simple motor system. Learn Mem. 2004;11:127–136. doi: 10.1101/lm.65804. [DOI] [PubMed] [Google Scholar]
- Büttner-Ennever JA, Horn AK, Scherberger H, D'Ascanio P. Motoneurons of twitch and nontwitch extraocular muscle fibers in the abducens, trochlear, and oculomotor nuclei of monkeys. J Comp Neurol. 2001;438:318–335. doi: 10.1002/cne.1318. [DOI] [PubMed] [Google Scholar]
- Camp AJ, Callister RJ, Brichta AM. Inhibitory synaptic transmission differs in mouse type A and B medial vestibular nucleus neurons in vitro. J Neurophysiol. 2006;95:3208–3218. doi: 10.1152/jn.01001.2005. [DOI] [PubMed] [Google Scholar]
- Cheron G, Escudero M, Godaux E. Discharge properties of brain stem neurons projecting to the flocculus in the alert cat. I. Medical vestibular nucleus. J Neurophysiol. 1996;76:1759–1774. doi: 10.1152/jn.1996.76.3.1759. [DOI] [PubMed] [Google Scholar]
- Cullen KE. The vestibular system: multimodal integration and encoding of self-motion for motor control. Trends Neurosci. 2012;35:185–196. doi: 10.1016/j.tins.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis-López de Carrizosa MA, Morado-Díaz CJ, Miller JM, de la Cruz RR, Pastor AM. Dual encoding of muscle tension and eye position by abducens motoneurons. J Neurosci. 2011;31:2271–2279. doi: 10.1523/JNEUROSCI.5416-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K. Optogenetics. Nat Methods. 2011;8:26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Zeeuw CI, Wentzel P, Mugnaini E. Fine structure of the dorsal cap of the inferior olive and its GABAergic and non-GABAergic input from the nucleus prepositus hypoglossi in rat and rabbit. J Comp Neurol. 1993;327:63–82. doi: 10.1002/cne.903270106. [DOI] [PubMed] [Google Scholar]
- du Lac S, Lisberger SG. Cellular processing of temporal information in medial vestibular nucleus neurons. J Neurosci. 1995;15:8000–8010. doi: 10.1523/JNEUROSCI.15-12-08000.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Lac S, Raymond JL, Sejnowski TJ, Lisberger SG. Learning and memory in the vestibulo-ocular reflex. Annu Rev Neurosci. 1995;18:409–441. doi: 10.1146/annurev.ne.18.030195.002205. [DOI] [PubMed] [Google Scholar]
- Dutia MB. Mechanisms of vestibular compensation: recent advances. Curr Opin Otolaryngol Head Neck Surg. 2010;18:420–424. doi: 10.1097/MOO.0b013e32833de71f. [DOI] [PubMed] [Google Scholar]
- Dymecki SM, Kim JC. Molecular neuroanatomy's “Three Gs”: a primer. Neuron. 2007;54:17–34. doi: 10.1016/j.neuron.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eatock RA, Songer JE. Vestibular hair cells and afferents: two channels for head motion signals. Annu Rev Neurosci. 2011;34:501–534. doi: 10.1146/annurev-neuro-061010-113710. [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- Faulstich BM, Onori KA, du Lac S. Comparison of plasticity and development of mouse optokinetic and vestibulo-ocular reflexes suggests differential gain control mechanisms. Vis Res. 2004;44:3419–3427. doi: 10.1016/j.visres.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Furuya N, Yabe T, Koizumi T. Neurotransmitters regulating vestibular commissural inhibition in the cat. Acta Otolaryngol [Suppl] 1991;481:205–208. doi: 10.3109/00016489109131381. [DOI] [PubMed] [Google Scholar]
- Gittis AH, du Lac S. Firing properties of GABAergic versus non-GABAergic vestibular nucleus neurons conferred by a differential balance of potassium currents. J Neurophysiol. 2007;97:3986–3996. doi: 10.1152/jn.00141.2007. [DOI] [PubMed] [Google Scholar]
- Gittis AH, Moghadam SH, du Lac S. Mechanisms of sustained high firing rates in two classes of vestibular nucleus neurons: differential contributions of resurgent Na, Kv3, and BK currents. J Neurophysiol. 2010;104:1625–1634. doi: 10.1152/jn.00378.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel CM, Sugino K, Nelson SB. A manual method for the purification of fluorescently labeled neurons from the mammalian brain. Nat Protoc. 2007;2:2924–2929. doi: 10.1038/nprot.2007.416. [DOI] [PubMed] [Google Scholar]
- Highstein SM, Holstein GR. Neuroanatomy of the oculomotor system. Amsterdam: Elsevier; 2006. The anatomy of the vestibular nuclei; pp. 157–203. E-book available at http://www.sciencedirect.com/science/article/B7CV6-4H9939D-9/2/88be9cb6056d40c5a5cad4e3ce8469ee. [DOI] [PubMed] [Google Scholar]
- Highstein SM, Ito M. Differential localization within the vestibular nuclear complex of the inhibitory and excitatory cells innervating 3d nucleus oculomotor neurons in rabbit. Brain Res. 1971;29:358–362. doi: 10.1016/0006-8993(71)90043-6. [DOI] [PubMed] [Google Scholar]
- Holstein GR, Martinelli GP, Cohen B. Ultrastructural features of non-commissural GABAergic neurons in the medial vestibular nucleus of the monkey. Neuroscience. 1999a;93:183–193. doi: 10.1016/s0306-4522(99)00140-2. [DOI] [PubMed] [Google Scholar]
- Holstein GR, Martinelli GP, Cohen B. The ultrastructure of GABA-immunoreactive vestibular commissural neurons related to velocity storage in the monkey. Neuroscience. 1999b;93:171–181. doi: 10.1016/s0306-4522(99)00141-4. [DOI] [PubMed] [Google Scholar]
- Holstein GR, Friedrich VL, Jr, Kang T, Kukielka E, Martinelli GP. Direct projections from the caudal vestibular nuclei to the ventrolateral medulla in the rat. Neuroscience. 2011;175:104–117. doi: 10.1016/j.neuroscience.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopman AH, Ramaekers FC, Speel EJ. Rapid synthesis of biotin-, digoxigenin-, trinitrophenyl-, and fluorochrome-labeled tyramides and their application for in situ hybridization using CARD amplification. J Histochem Cytochem. 1998;46:771–777. doi: 10.1177/002215549804600611. [DOI] [PubMed] [Google Scholar]
- Johnston AR, MacLeod NK, Dutia MB. Ionic conductances contributing to spike repolarization and after-potentials in rat medial vestibular nucleus neurones. J Physiol. 1994;481(Pt 1):61–77. doi: 10.1113/jphysiol.1994.sp020419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Xue J, Eatock RA. Ion channels set spike timing regularity of mammalian vestibular afferent neurons. J Neurophysiol. 2010;104:2034–2051. doi: 10.1152/jn.00396.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller EL, Kamath BY. Characteristics of head rotation and eye movement-related neurons in alert monkey vestibular nucleus. Brain Res. 1975;100:182–187. doi: 10.1016/0006-8993(75)90257-7. [DOI] [PubMed] [Google Scholar]
- Knöpfel T, Lin MZ, Levskaya A, Tian L, Lin JY, Boyden ES. Toward the second generation of optogenetic tools. J Neurosci. 2010;30:14998–15004. doi: 10.1523/JNEUROSCI.4190-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolkman KE, McElvain LE, du Lac S. Diverse precerebellar neurons share similar intrinsic excitability. J Neurosci. 2011a;31:16665–16674. doi: 10.1523/JNEUROSCI.3314-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolkman KE, Moghadam SH, du Lac S. Intrinsic physiology of identified neurons in the prepositus hypoglossi and medial vestibular nuclei. J Vestib Res. 2011b;21:33–47. doi: 10.3233/VES-2011-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Saitou M. Global single-cell cDNA amplification to provide a template for representative high-density oligonucleotide microarray analysis. Nat Protoc. 2007;2:739–752. doi: 10.1038/nprot.2007.79. [DOI] [PubMed] [Google Scholar]
- Lee JY, Yeh I, Park JY, Tian B. PolyA_DB 2: mRNA polyadenylation sites in vertebrate genes. Nucleic Acids Res. 2007;35:D165–D168. doi: 10.1093/nar/gkl870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lisberger SG, Miles FA. Role of primate medial vestibular nucleus in long-term adaptive plasticity of vestibuloocular reflex. J Neurophysiol. 1980;43:1725–1745. doi: 10.1152/jn.1980.43.6.1725. [DOI] [PubMed] [Google Scholar]
- Lisberger SG, Pavelko TA. Brain stem neurons in modified pathways for motor learning in the primate vestibulo-ocular reflex. Science. 1988;242:771–773. doi: 10.1126/science.3142040. [DOI] [PubMed] [Google Scholar]
- Lisberger SG, Pavelko TA, Broussard DM. Neural basis for motor learning in the vestibuloocular reflex of primates. I. Changes in the responses of brain stem neurons. J Neurophysiol. 1994;72:928–953. doi: 10.1152/jn.1994.72.2.928. [DOI] [PubMed] [Google Scholar]
- Lu T, Rubio ME, Trussell LO. Glycinergic transmission shaped by the corelease of GABA in a mammalian auditory synapse. Neuron. 2008;57:524–535. doi: 10.1016/j.neuron.2007.12.010. [DOI] [PubMed] [Google Scholar]
- Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinvaud D, Vassias I, Reichenberger I, Rössert C, Straka H. Functional organization of vestibular commissural connections in frog. J Neurosci. 2010;30:3310–3325. doi: 10.1523/JNEUROSCI.5318-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrea RA, Strassman A, May E, Highstein SM. Anatomical and physiological characteristics of vestibular neurons mediating the horizontal vestibulo-ocular reflex of the squirrel monkey. J Comp Neurol. 1987;264:547–570. doi: 10.1002/cne.902640408. [DOI] [PubMed] [Google Scholar]
- McElvain LE, Bagnall MW, Sakatos A, du Lac S. Bidirectional plasticity gated by hyperpolarization controls the gain of postsynaptic firing responses at central vestibular nerve synapses. Neuron. 2010;68:763–775. doi: 10.1016/j.neuron.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molineux ML, Mehaffey WH, Tadayonnejad R, Anderson D, Tennent AF, Turner RW. Ionic factors governing rebound burst phenotype in rat deep cerebellar neurons. J Neurophysiol. 2008;100:2684–2701. doi: 10.1152/jn.90427.2008. [DOI] [PubMed] [Google Scholar]
- Murphy GJ, du Lac S. Postnatal development of spike generation in rat medial vestibular nucleus neurons. J Neurophysiol. 2001;85:1899–1906. doi: 10.1152/jn.2001.85.5.1899. [DOI] [PubMed] [Google Scholar]
- Ng L, Bernard A, Lau C, Overly CC, Dong HW, Kuan C, Pathak S, Sunkin SM, Dang C, Bohland JW, Bokil H, Mitra PP, Puelles L, Hohmann J, Anderson DJ, Lein ES, Jones AR, Hawrylycz M. An anatomic gene expression atlas of the adult mouse brain. Nat Neurosci. 2009;12:356–362. doi: 10.1038/nn.2281. [DOI] [PubMed] [Google Scholar]
- Oliva AA, Jr, Jiang M, Lam T, Smith KL, Swann JW. Novel hippocampal interneuronal subtypes identified using transgenic mice that express green fluorescent protein in GABAergic interneurons. J Neurosci. 2000;20:3354–3368. doi: 10.1523/JNEUROSCI.20-09-03354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakada F, Mori T, Cetin AH, Marshel JH, Virgen B, Callaway EM. New rabies virus variants for monitoring and manipulating activity and gene expression in defined neural circuits. Neuron. 2011;71:617–631. doi: 10.1016/j.neuron.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettorossi VE, Dieni CV, Scarduzio M, Grassi S. Long-term potentiation of synaptic response and intrinsic excitability in neurons of the rat medial vestibular nuclei. Neuroscience. 2011;187:1–14. doi: 10.1016/j.neuroscience.2011.04.040. [DOI] [PubMed] [Google Scholar]
- Precht W, Schwindt PC, Baker R. Removal of vestibular commissural inhibition by antagonists of GABA and glycine. Brain Res. 1973;62:222–226. doi: 10.1016/0006-8993(73)90631-8. [DOI] [PubMed] [Google Scholar]
- Ramachandran R, Lisberger SG. Neural substrate of modified and unmodified pathways for learning in monkey vestibuloocular reflex. J Neurophysiol. 2008;100:1868–1878. doi: 10.1152/jn.90498.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert MC, Zee DS. Saccade and vestibular ocular motor adaptation. Restor Neurol Neurosci. 2010;28:9–18. doi: 10.3233/RNN-2010-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scudder CA, Fuchs AF. Physiological and behavioral identification of vestibular nucleus neurons mediating the horizontal vestibuloocular reflex in trained rhesus monkeys. J Neurophysiol. 1992;68:244–264. doi: 10.1152/jn.1992.68.1.244. [DOI] [PubMed] [Google Scholar]
- Sekirnjak C, du Lac S. Intrinsic firing dynamics of vestibular nucleus neurons. J Neurosci. 2002;22:2083–2095. doi: 10.1523/JNEUROSCI.22-06-02083.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirnjak C, du Lac S. Physiological and anatomical properties of mouse medial vestibular nucleus neurons projecting to the oculomotor nucleus. J Neurophysiol. 2006;95:3012–3023. doi: 10.1152/jn.00796.2005. [DOI] [PubMed] [Google Scholar]
- Sekirnjak C, Vissel B, Bollinger J, Faulstich M, du Lac S. Purkinje cell synapses target physiologically unique brainstem neurons. J Neurosci. 2003;23:6392–6398. doi: 10.1523/JNEUROSCI.23-15-06392.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafin M, de Waele C, Khateb A, Vidal PP, Mühlethaler M. Medial vestibular nucleus in the guinea-pig. I. Intrinsic membrane properties in brainstem slices. Exp Brain Res. 1991;84:417–425. doi: 10.1007/BF00231464. [DOI] [PubMed] [Google Scholar]
- Shin M, Moghadam SH, Sekirnjak C, Bagnall MW, Kolkman KE, Jacobs R, Faulstich M, du Lac S. Multiple types of cerebellar target neurons and their circuitry in the vestibulo-ocular reflex. J Neurosci. 2011;31:10776–10786. doi: 10.1523/JNEUROSCI.0768-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MR, Nelson AB, Du Lac S. Regulation of firing response gain by calcium-dependent mechanisms in vestibular nucleus neurons. J Neurophysiol. 2002;87:2031–2042. doi: 10.1152/jn.00821.2001. [DOI] [PubMed] [Google Scholar]
- Spencer RF, Wenthold RJ, Baker R. Evidence for glycine as an inhibitory neurotransmitter of vestibular, reticular, and prepositus hypoglossi neurons that project to the cat abducens nucleus. J Neurosci. 1989;9:2718–2736. doi: 10.1523/JNEUROSCI.09-08-02718.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straka H, Vibert N, Vidal PP, Moore LE, Dutia MB. Intrinsic membrane properties of vertebrate vestibular neurons: Function, development and plasticity. Prog Neurobiol. 2005;76:349–392. doi: 10.1016/j.pneurobio.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Tabaska JE, Zhang MQ. Detection of polyadenylation signals in human DNA sequences. Gene. 1999;231:77–86. doi: 10.1016/s0378-1119(99)00104-3. [DOI] [PubMed] [Google Scholar]
- Takazawa T, Saito Y, Tsuzuki K, Ozawa S. Membrane and firing properties of glutamatergic and GABAergic neurons in the rat medial vestibular nucleus. J Neurophysiol. 2004;92:3106–3120. doi: 10.1152/jn.00494.2004. [DOI] [PubMed] [Google Scholar]
- Tanaka I, Ezure K. Overall distribution of GLYT2 mRNA-containing versus GAD67 mRNA-containing neurons and colocalization of both mRNAs in midbrain, pons, and cerebellum in rats. Neurosci Res. 2004;49:165–178. doi: 10.1016/j.neures.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Tietjen I, Rihel JM, Cao Y, Koentges G, Zakhary L, Dulac C. Single-cell transcriptional analysis of neuronal progenitors. Neuron. 2003;38:161–175. doi: 10.1016/s0896-6273(03)00229-0. [DOI] [PubMed] [Google Scholar]
- Uusisaari MY, Knöpfel T. Diversity of neuronal elements and circuitry in the cerebellar nuclei. Cerebellum. 2012 doi: 10.1007/s12311-011-0350-6. [DOI] [PubMed] [Google Scholar]
- van Welie I, du Lac S. Bidirectional control of BK channel open probability by CAMKII and PKC in medial vestibular nucleus neurons. J Neurophysiol. 2011;105:1651–1659. doi: 10.1152/jn.00058.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walberg F, Ottersen OP, Rinvik E. GABA, glycine, aspartate, glutamate and taurine in the vestibular nuclei: an immunocytochemical investigation in the cat. Exp Brain Res. 1990;79:547–563. doi: 10.1007/BF00229324. [DOI] [PubMed] [Google Scholar]
- Watakabe A, Komatsu Y, Ohsawa S, Yamamori T. Fluorescent in situ hybridization technique for cell type identification and characterization in the central nervous system. Methods. 2010;52:367–374. doi: 10.1016/j.ymeth.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Wentzel PR, De Zeeuw CI, Holstege JC, Gerrits NM. Colocalization of GABA and glycine in the rabbit oculomotor nucleus. Neurosci Lett. 1993;164:25–29. doi: 10.1016/0304-3940(93)90848-f. [DOI] [PubMed] [Google Scholar]
- Wentzel PR, De Zeeuw CI, Holstege JC, Gerrits NM. Inhibitory synaptic inputs to the oculomotor nucleus from vestibulo-ocular-related nuclei in the rabbit. Neuroscience. 1995;65:161–174. doi: 10.1016/0306-4522(94)00471-g. [DOI] [PubMed] [Google Scholar]
- Zeilhofer HU, Studler B, Arabadzisz D, Schweizer C, Ahmadi S, Layh B, Bösl MR, Fritschy JM. Glycinergic neurons expressing enhanced green fluorescent protein in bacterial artificial chromosome transgenic mice. J Comp Neurol. 2005;482:123–141. doi: 10.1002/cne.20349. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Partsalis AM, Highstein SM. Properties of superior vestibular nucleus flocculus target neurons in the squirrel monkey. I. General properties in comparison with flocculus projecting neurons. J Neurophysiol. 1995;73:2261–2278. doi: 10.1152/jn.1995.73.6.2261. [DOI] [PubMed] [Google Scholar]