Abstract
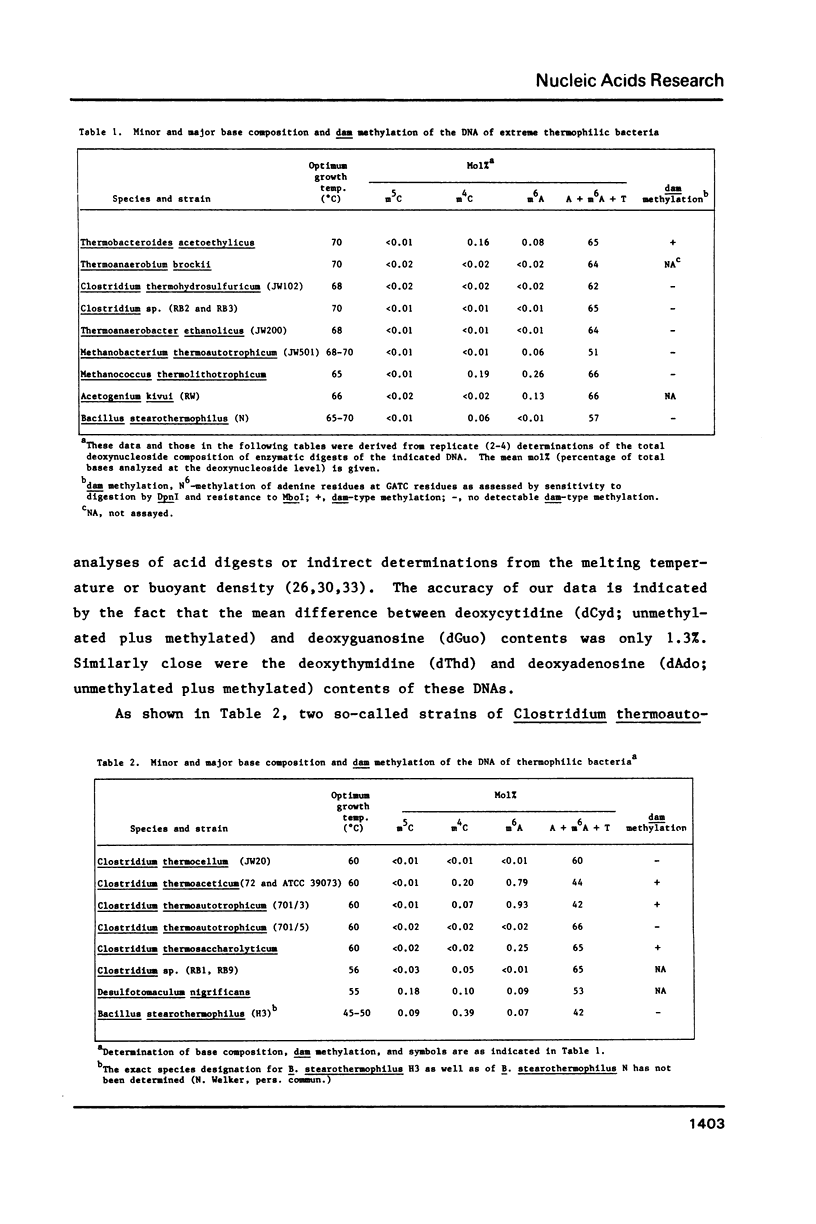
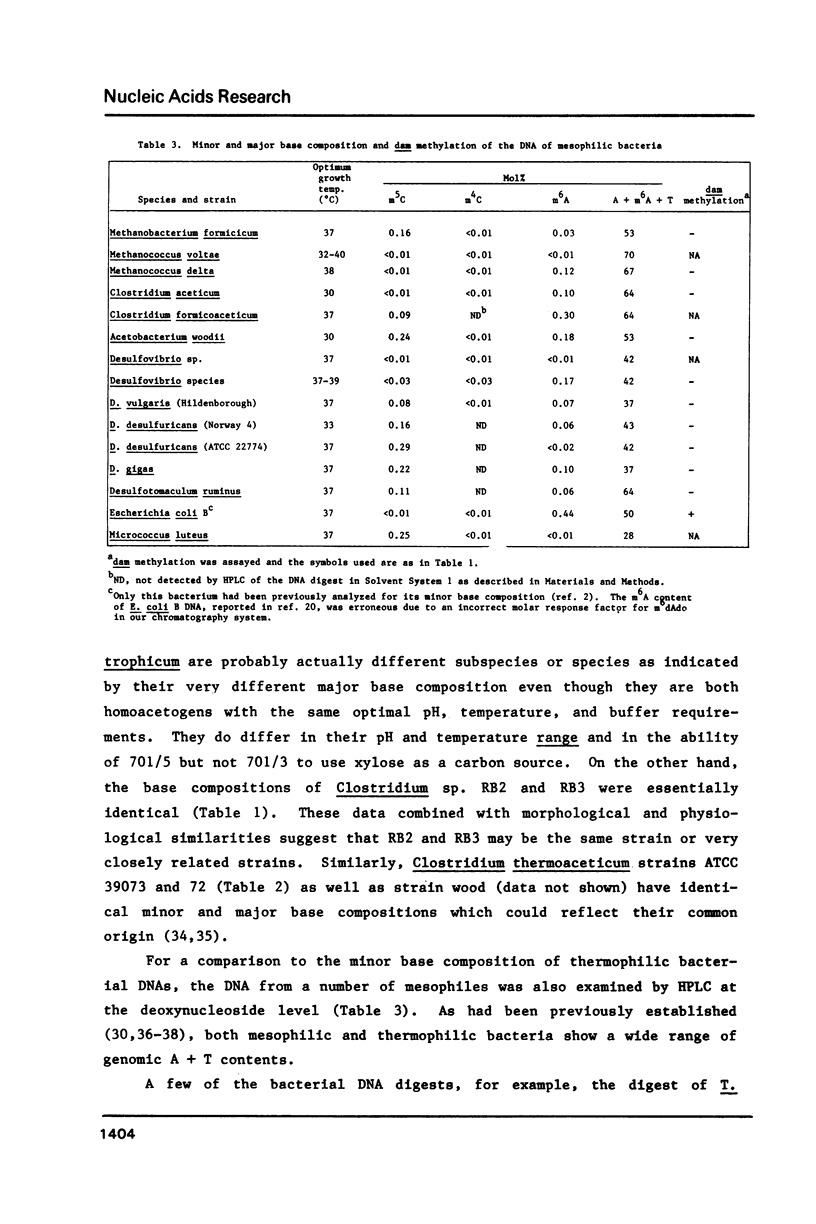
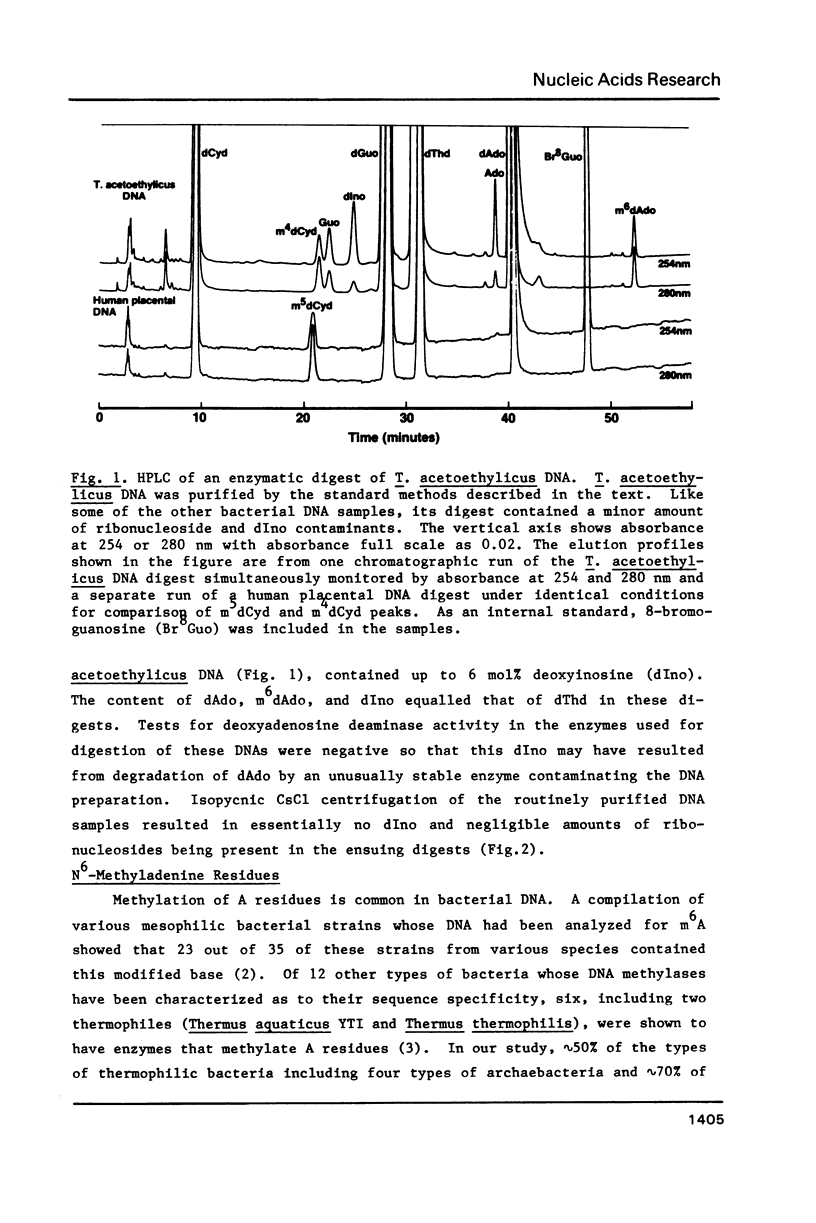
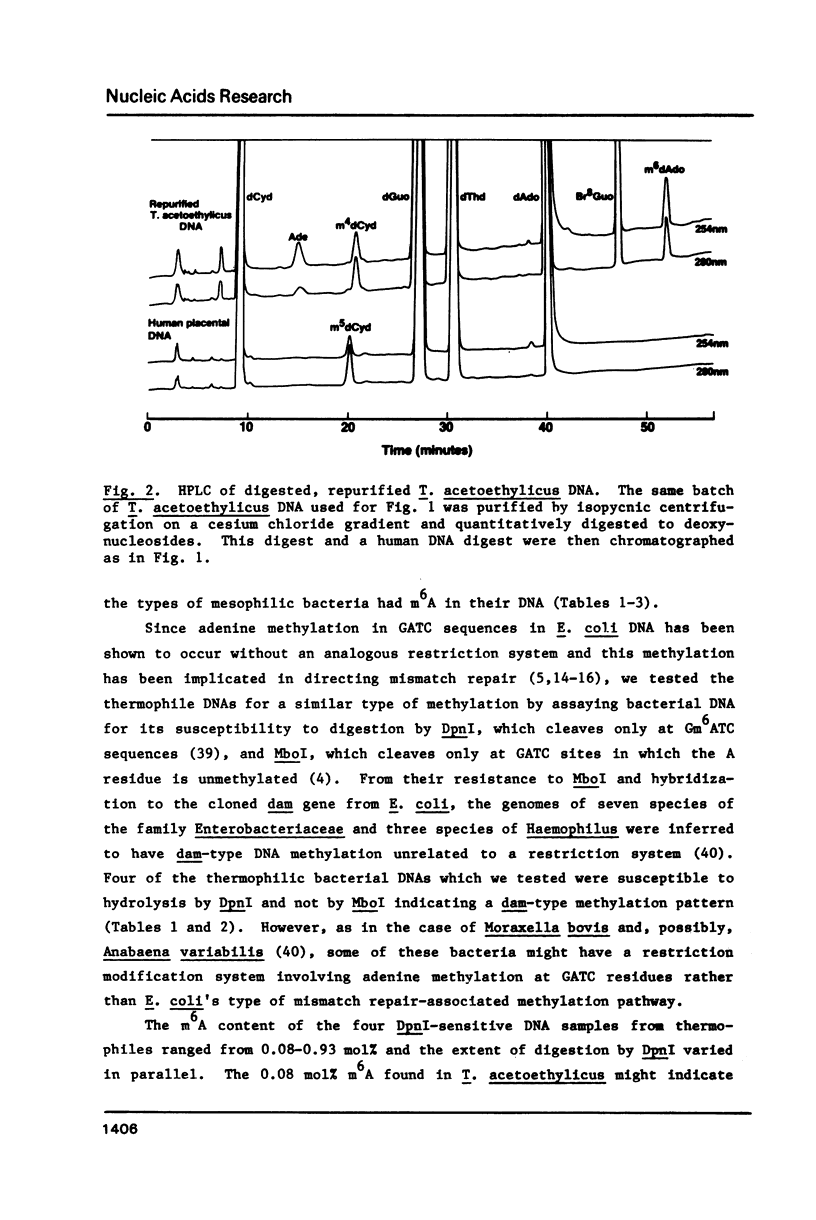
While determining the minor and major base composition of the DNA from 17 types of thermophilic bacteria by high performance liquid chromatography (HPLC) of enzymatic digests, we have discovered a novel base, N4-methylcytosine (m4C). Its structure was proven by comparison of the DNA-derived nucleoside to the analogous authentic compound by HPLC, UV spectroscopy, and mass spectroscopy. Eight of the bacterial DNAs contained m4C. Only two contained the common minor base, 5-methylcytosine (m5C), and neither of these was from an extreme thermophile. The other prevalent modified base of bacterial DNA, N6-methyladenine (m6A), was found in nine of the DNAs. Restriction analysis revealed that four of the DNAs had dam-type (Gm6ATC) methylation patterns. Due to the propensity of m5C residues to be deaminated by heat to thymine residues and to inefficient repair of the resulting mismatched base pairs, thermophiles with optimal growth temperatures of greater than or equal to 60 degrees C generally may avoid having m5C in their genomes. Instead, some of them have deamination-resistant m4C residues.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bale A., d'Alarcao M., Marinus M. G. Characterization of DNA adenine methylation mutants of Escherichia coli K12. Mutat Res. 1979 Feb;59(2):157–165. doi: 10.1016/0027-5107(79)90153-2. [DOI] [PubMed] [Google Scholar]
- Brooks J. E., Blumenthal R. M., Gingeras T. R. The isolation and characterization of the Escherichia coli DNA adenine methylase (dam) gene. Nucleic Acids Res. 1983 Feb 11;11(3):837–851. doi: 10.1093/nar/11.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks J. E., Roberts R. J. Modification profiles of bacterial genomes. Nucleic Acids Res. 1982 Feb 11;10(3):913–934. doi: 10.1093/nar/10.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H., Farabaugh P. J., Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978 Aug 24;274(5673):775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- De Ley J. Reexamination of the association between melting point, buoyant density, and chemical base composition of deoxyribonucleic acid. J Bacteriol. 1970 Mar;101(3):738–754. doi: 10.1128/jb.101.3.738-754.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnen S. T., Morris N. R. Deoxyribonucleic acid methylation and development in Caulobacter bacteroides. J Bacteriol. 1973 Oct;116(1):48–53. doi: 10.1128/jb.116.1.48-53.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dybvig K., Swinton D., Maniloff J., Hattman S. Cytosine methylation of the sequence GATC in a mycoplasma. J Bacteriol. 1982 Sep;151(3):1420–1424. doi: 10.1128/jb.151.3.1420-1424.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M., Gama-Sosa M. A., Huang L. H., Midgett R. M., Kuo K. C., McCune R. A., Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982 Apr 24;10(8):2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M., Wang R. Y. 5-Methylcytosine in eukaryotic DNA. Science. 1981 Jun 19;212(4501):1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- Fontaine F. E., Peterson W. H., McCoy E., Johnson M. J., Ritter G. J. A New Type of Glucose Fermentation by Clostridium thermoaceticum. J Bacteriol. 1942 Jun;43(6):701–715. doi: 10.1128/jb.43.6.701-715.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Sosa M. A., Midgett R. M., Slagel V. A., Githens S., Kuo K. C., Gehrke C. W., Ehrlich M. Tissue-specific differences in DNA methylation in various mammals. Biochim Biophys Acta. 1983 Jun 24;740(2):212–219. doi: 10.1016/0167-4781(83)90079-9. [DOI] [PubMed] [Google Scholar]
- Gama-Sosa M. A., Slagel V. A., Trewyn R. W., Oxenhandler R., Kuo K. C., Gehrke C. W., Ehrlich M. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983 Oct 11;11(19):6883–6894. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke C. W., McCune R. A., Gama-Sosa M. A., Ehrlich M., Kuo K. C. Quantitative reversed-phase high-performance liquid chromatography of major and modified nucleosides in DNA. J Chromatogr. 1984 Sep 28;301(1):199–219. doi: 10.1016/s0021-9673(01)89189-5. [DOI] [PubMed] [Google Scholar]
- Glickman B. W., Radman M. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc Natl Acad Sci U S A. 1980 Feb;77(2):1063–1067. doi: 10.1073/pnas.77.2.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman B., van den Elsen P., Radman M. Induced mutagenesis in dam- mutants of Escherichia coli: a role for 6-methyladenine residues in mutation avoidance. Mol Gen Genet. 1978 Jul 25;163(3):307–312. doi: 10.1007/BF00271960. [DOI] [PubMed] [Google Scholar]
- Janulaitis A., Klimasauskas S., Petrusyte M., Butkus V. Cytosine modification in DNA by BcnI methylase yields N4-methylcytosine. FEBS Lett. 1983 Sep 5;161(1):131–134. doi: 10.1016/0014-5793(83)80745-5. [DOI] [PubMed] [Google Scholar]
- Korch C., Hagblom P., Normark S. Sequence-specific DNA modification in Neisseria gonorrhoeae. J Bacteriol. 1983 Sep;155(3):1324–1332. doi: 10.1128/jb.155.3.1324-1332.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo K. C., McCune R. A., Gehrke C. W., Midgett R., Ehrlich M. Quantitative reversed-phase high performance liquid chromatographic determination of major and modified deoxyribonucleosides in DNA. Nucleic Acids Res. 1980 Oct 24;8(20):4763–4776. doi: 10.1093/nar/8.20.4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. DNA glycosylases, endonucleases for apurinic/apyrimidinic sites, and base excision-repair. Prog Nucleic Acid Res Mol Biol. 1979;22:135–192. doi: 10.1016/s0079-6603(08)60800-4. [DOI] [PubMed] [Google Scholar]
- Ljungdahl L. G. Physiology of thermophilic bacteria. Adv Microb Physiol. 1979;19:149–243. doi: 10.1016/s0065-2911(08)60199-x. [DOI] [PubMed] [Google Scholar]
- Lu A. L., Clark S., Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MYLROIE R. L., HUNGATE R. E. Experiments on the methane bacteria in sludge. Can J Microbiol. 1954 Aug;1(1):55–64. doi: 10.1139/m55-008. [DOI] [PubMed] [Google Scholar]
- McClelland M. The effect of site specific methylation on restriction endonuclease cleavage (update). Nucleic Acids Res. 1983 Jan 11;11(1):r169–r173. doi: 10.1093/nar/11.1.235-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K., Marmur J. Purification and properties of deoxyribonucleic acid methylase from Bacillus subtilis. Biochemistry. 1966 Feb;5(2):761–773. doi: 10.1021/bi00866a051. [DOI] [PubMed] [Google Scholar]
- Postgate J. R., Kent H. M., Robson R. L., Chesshyre J. A. The genomes of Desulfovibrio gigas and D. vulgaris. J Gen Microbiol. 1984 Jul;130(7):1597–1601. doi: 10.1099/00221287-130-7-1597. [DOI] [PubMed] [Google Scholar]
- Pukkila P. J., Peterson J., Herman G., Modrich P., Meselson M. Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics. 1983 Aug;104(4):571–582. doi: 10.1093/genetics/104.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raibaud A., Gaillard C., Longacre S., Hibner U., Buck G., Bernardi G., Eisen H. Genomic environment of variant surface antigen genes of Trypanosoma equiperdum. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4306–4310. doi: 10.1073/pnas.80.14.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Razin S. Methylated bases in mycoplasmal DNA. Nucleic Acids Res. 1980 Mar 25;8(6):1383–1390. doi: 10.1093/nar/8.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 1984;12 (Suppl):r167–r204. doi: 10.1093/nar/12.suppl.r167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHILDKRAUT C. L., MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its buoyant density in CsCl. J Mol Biol. 1962 Jun;4:430–443. doi: 10.1016/s0022-2836(62)80100-4. [DOI] [PubMed] [Google Scholar]
- Schein A., Berdahl B. J., Low M., Borek E. Deficiency of the DNA of Micrococcus radiodurans in methyladenine and methylcytosine. Biochim Biophys Acta. 1972 Jul 20;272(3):481–485. doi: 10.1016/0005-2787(72)90400-5. [DOI] [PubMed] [Google Scholar]
- Somogyi P. A., Maso Bel M., Földes I. Methylated nucleic acid bases in Mycobacterium and mycobacteriophage DNA. Acta Microbiol Acad Sci Hung. 1982;29(3):181–185. [PubMed] [Google Scholar]
- Sprinzl M., Gauss D. H. Compilation of tRNA sequences. Nucleic Acids Res. 1984;12 (Suppl):r1–57. [PMC free article] [PubMed] [Google Scholar]
- Vovis G. F., Lacks S. Complementary action of restriction enzymes endo R-DpnI and Endo R-DpnII on bacteriophage f1 DNA. J Mol Biol. 1977 Sep 25;115(3):525–538. doi: 10.1016/0022-2836(77)90169-3. [DOI] [PubMed] [Google Scholar]
- Wachsman J. T., Irwin V. Methylated bases of Bacillus megaterium KM nucleic acids: comparison with Escherichia coli. J Bacteriol. 1970 Nov;104(2):814–818. doi: 10.1128/jb.104.2.814-818.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R., Jr, Meselson M. Repair tracts in mismatched DNA heteroduplexes. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4135–4139. doi: 10.1073/pnas.73.11.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. Y., Kuo K. C., Gehrke C. W., Huang L. H., Ehrlich M. Heat- and alkali-induced deamination of 5-methylcytosine and cytosine residues in DNA. Biochim Biophys Acta. 1982 Jun 30;697(3):371–377. doi: 10.1016/0167-4781(82)90101-4. [DOI] [PubMed] [Google Scholar]
- Warner H. R. Base excision repair in the thermophile Thermus sp. strain X-1. J Bacteriol. 1983 Jun;154(3):1451–1454. doi: 10.1128/jb.154.3.1451-1454.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman W. B., Ankwanda E., Wolfe R. S. Nutrition and carbon metabolism of Methanococcus voltae. J Bacteriol. 1982 Mar;149(3):852–863. doi: 10.1128/jb.149.3.852-863.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegel J., Ljungdahl L. G., Rawson J. R. Isolation from soil and properties of the extreme thermophile Clostridium thermohydrosulfuricum. J Bacteriol. 1979 Sep;139(3):800–810. doi: 10.1128/jb.139.3.800-810.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeikus J. G., Wolfe R. S. Methanobacterium thermoautotrophicus sp. n., an anaerobic, autotrophic, extreme thermophile. J Bacteriol. 1972 Feb;109(2):707–715. doi: 10.1128/jb.109.2.707-713.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]


