Abstract
Distributions of stable isotopes have been used to infer an organism's trophic niche width, the ‘isotopic niche’, and examine resource partitioning. Spatial variation in the isotopic composition of prey may however confound the interpretation of isotopic signatures especially when foragers exploit resources across numerous locations. In this study the isotopic compositions from marine assemblages are modelled to determine the role of variation in the signature of prey items and the effect of dietary breadth and foraging strategies on predator signatures. Outputs from the models reveal that isotopic niche widths can be greater for populations of dietary specialists rather than for generalists, which contravenes what is generally accepted in the literature. When a range of different mixing models are applied to determine if the conversion from δ to p-space can be used to improve model accuracy, predator signature variation is increased rather than model precision. Furthermore the mixing models applied failed to correctly identify dietary specialists and/or to accurately estimate diet contributions that may identify resource partitioning. The results presented illustrate the need to collect sufficiently large sample sizes, in excess of what is collected under most current studies, across the complete distribution of a species and its prey, before attempts to use stable isotopes to make inferences about niche width can be made.
Introduction
Stable isotope analysis is often used by ecologists to identify trophic interactions [1]. This approach can be less problematic than others such as gut analysis, which may have logistical constraints and require regular and large sampling regimes [2], [3]. In the last decade, a number of authors have used stable isotopes to estimate trophic niche width [1], [3] and to examine resource partitioning [4], [5]. There has, however, been a growing realisation that interpreting patterns of stable isotope relies heavily on a comprehensive understanding of habitat use by predators, and the spatial patterns of isotopic variation among organisms at all trophic levels [6], [7], [8], [9], [10]. Post [3] has concluded, that without a suitable quantification of the isotopic composition of prey items, comparisons of consumers among and across habitats will be confounded by variations in prey signatures. The challenge for ecologists is to determine where isotopic variation exists and why.
An assumption of many studies aiming to estimate isotopic niche breadth, developed from the niche variation hypothesis proposed by Van Valen in 1965 [11], is that niche width correlates positively with diet breadth [12]. In this case, dietary specialists, i.e. those that utilise only a small number of food types at the population level, will have a narrow isotopic niche width, whereas dietary generalists, i.e. those that utilise a wide range of food resources at the population level, will have a broad isotopic niche width [11], [13], [14]. More recently this assumption has been challenged by studies which indicate that the converse can be true, i.e. the isotopic niche width of specialists can be broader than that for generalists, and that habitat use may complicate any conclusions that can be drawn from isotopic data [15], [16]. In addition, variation in isotopic signatures in δ-space (the dimensional space occupied by two or more isotopic signatures) may lead to incorrect estimates of the range of resources a population utilises. One suggestion to overcome this is to convert isotopic signatures from δ to p-space (relative proportions of prey items contributing to delta space signature) [17], [18]. The transformation to dietary proportions (p-space) is thought to resolve scaling discrepancies in δ-space, allowing direct comparison with a metric based measure of niche width [1], [19].
Flaherty and Ben-David [15] examined the effects of diet and habitat use on isotopic derived trophic niche width, in both δ-space and p-space, by modelling the isotopic composition of predators employing different feeding strategies. Their findings revealed that populations of dietary generalists display narrower isotopic niches than dietary specialists, suggesting that estimates from isotopic values of trophic niche may be confounded by habitat-derived differences (see also [12]). Our aim in this paper was to develop the models of Flaherty and Ben-David [15] by adding new degrees of ecological realism and statistical robustness by taking advantage of a rich new isotopic database, while also extending the models from a terrestrial to a marine context.
The isotopic data used for the modelling were derived from marine assemblages collected from artificial reefs (decommissioned oil drilling wellheads) on the North-West Shelf of Australia. These data offer several novel and significant features for such an investigation: a) the fauna sampled at each location was complete (i.e. isotopic signatures from the entire community were collected); b) the wellheads are replicated (the same) structures that differ in location and depth; and c) the wellheads had a range of species from a similar trophic level, representing a good system to investigate the effects of a predator that forages widely but without many additional differences between food patches. In addition, the wellheads were in deep-water locations (i.e. relatively unstudied) and the sample sizes for individual species were large (n = 4 – 195).
The objective of this study is to use similar models as those applied by Flaherty and Ben-David [15] to determine effects of habitat variability in prey on the isotopic outcomes of the predator. We will apply both multi-source and Bayesian-based models to determine if trophic interactions, such as trophic niche widths and resource partitioning can be accurately estimated. To confidently address this hypothesis we will improve the modelling approach and test the resulting outcomes with more rigorous statistical analysis. In addition, the isotopic outcomes of our model predator will be tested under more ecologically realistic assumptions that represent conditions a predator is likely to face in the real environment. The basis of this approach adopted the four basic foraging models (as outlined in the methods) tested by Flaherty and Ben-David [15], using all combinations of dietary and habitat specialists or generalists. For this study, habitat generalists refers to those collected from a range of wellhead (artificial reef) locations. Isotopic differences were simulated by: 1) using four common prey species of known isotopic signatures at each location; 2) incorporating the effects of distance between sites on the signatures of the predator feeding on the common prey species at each habitat/location; and 3) using the entire assemblage of prey sampled within a similar trophic level at each site.
Methods
Animals were collected in 2008 from the North West Shelf of Australia approximately 100km offshore from Dampier, Western Australia, from isolated wellhead structures (see Table 1). The wellheads were remotely severed and brought onboard a construction vessel as part of the decommissioning works, allowing organisms to be collected directly by hand from the structures (see [20] for full details). The wellheads had been in place for 12 – 16 years, such that they were colonised by extensive communities of deep reef species. δ13C and δ15N isotopes from muscle tissue were collected as a part of a trophic study of the wellhead communities. Where potential for carbonate tissue existed i.e. decapod exoskeleton, ground tissue samples were treated with 2N phosphoric acid. Isotope signatures of freeze dried tissue were measured from 0.5 mg material at Washington State University using an Isoprime isotope ratio mass spectrometer (IRMS) (for detailed methods see [21]. The data used included signatures from a range of fishes, crustaceans, molluscs, anthozoans, asteroids and ophiuroids, from 4 of the 5 wellhead sites (Yodel, Goodwyn, Echo, and Wanaea) as these were the most comprehensively sampled sites.
Table 1. Depths (metres) and distance (kilometres) between the sites.
| Wellhead | Goodwyn | Wanaea | Yodel | Cossack |
| Depth | 136 m | 84 m | 137 m | 82 m |
| Distance between well heads | ||||
| Wanaea | 45 km | |||
| Yodel | 31 km | 74 km | ||
| Cossack | 55 km | 10 km | 84 km | |
Using individual isotopic signatures, prey species of a similar trophic level (i.e. their δ15N signatures did not differ by more than 4‰) and common to all four of the selected sites (see Table 2) were identified. Generally, isotopic fractionation between trophic levels is assumed to be 3 – 4‰ [22].
Table 2. Values of δ13C and δ15N (mean and standard deviation) for the prey species collected from each site.
| Global | Goodwyn | Yodel | Echo | <@emph type="sup">Wanaea<@/emph> | ||||||||||||
| Prey | n | C13δ | N15δ | n | C13δ | N15δ | n | C13δ | N15δ | n | C13δ | N15δ | n | C13δ | N15δ | |
| Pseudanthias rubrizonatus* | Reef Fish | 195 | −17.80 (0.63) | 11.25 (0.88) | 95 | −17.99 (0.69) | 11.0 (0.99) | 37 | −17.71 (0.49) | 11.62 (0.89) | 8 | −17.84 (0.54) | 11.54 (0.87) | 55 | −17.51 (0.50) | 11.40 (0.46) |
| Rhynchocinetes balssi* | Shrimp | 34 | −16.29 (0.41) | 11.34 (0.83) | 10 | −16.54 (0.20) | 11.89 (0.27) | 10 | −16.48 (0.29) | 11.85 (0.34) | 4 | −15.77 (0.58) | 11.58 (0.22) | 10 | −16.04 (0.31) | 10.17 (0.38) |
| Petrolisthes militaris* | Crab | 36 | −18.10 (1.39) | 10.77 (0.81) | 10 | −16.19 (0.25) | 11.37 (0.40) | 9 | −18.49 (0.57) | 10.60 (0.33) | 8 | −18.65 (0.42) | 11.27 (0.36) | 8 | −19.44 (1.06) | 9.62 (0.69) |
| Pilumnus scabriusculus* | Crab* | 31 | −16.82 (0.64) | 11.22 (0.87) | 10 | −17.15 (0.40) | 11.27 (0.44) | 9 | −16.68 (0.35) | 12.15 (0.42) | 7 | −16.13 (0.43) | 10.14 (0.41) | 5 | −17.41 (0.78) | 10.95 (0.76) |
| Maja spinigera | Spider Crab | 16 | −16.92 (0.61) | 11.60 (1.52) | 6 | −17.06 (0.48) | 11.58 (1.30) | 8 | −17.05 (0.63) | 12.28 (1.58) | ||||||
| Portunus nipponensis | Crab | 7 | −17.71 (1.54) | 11.30 (0.51) | 3 | −19.33 (0.25) | 10.86 (0.33) | 4 | −16.49 (0.34) | 11.62 (0.33) | ||||||
| Pylopaguropsis pustulosa | Hermit Crab | 31 | −17.87 (0.75) | 9.78 (1.05) | 9 | −17.97 (0.30) | 9.82 (0.32) | 8 | −18.41 (0.91) | 11.2 (0.40) | 14 | −17.51 (0.48) | 9.03 (0.87) | |||
| Munida rogeri | Squat Lobster | 19 | −17.29 (0.81) | 11.19 (0.48) | 10 | −16.80 (0.52) | 11.45 (0.45) | 9 | −17.83 (0.73) | 10.90 (0.34) | ||||||
| Lysmata amboinensis | Shrimp | 18 | −16.02 (0.76) | 11.72 (0.74) | 10 | −15.62 (0.37) | 12.03 (0.41) | 5 | −16.49 (1.04) | 11.66 (0.95) | 3 | −16.55 (0.58) | 10.80 (0.36) | |||
| Lysmata sp. | Shrimp | 11 | −17.05 (0.62) | 11.26 (0.64) | 8 | −16.76 (0.21) | 10.93 v(0.36) | 3 | −17.82 (0.74) | 12.14 (0.17) | ||||||
| Alpheus gracilipes | Shrimp | 3 | −16.53 (0.59) | 11.48 (0.83) | 3 | −16.53 (0.59) | 11.48 (0.83) | |||||||||
| Paranthus sp. | Anemone | 29 | −18.04 (1.71) | 12.03 (0.66) | 10 | −16.20 (0.36) | 11.87 (0.57) | 10 | −18.32 (0.34) | 12.36 (0.84) | 7 | −19.75 (1.50) | 11.74 (0.41) | |||
| Megabalanus tintinnabulum | Barnacle | 20 | −18.57 (0.40) | 10.02 (0.83) | 8 | −18.72 (0.18) | 9.89 (0.36) | 9 | −18.52 (0.32) | 9.89 (1.12) | 3 | −17.80 (0.40) | 10.72 (0.31) | |||
| Bait fish | Fish | 8 | −17.89 (0.23) | 11.57 (0.24) | 8 | −17.90 (0.23) | 11.60 (0.24) | |||||||||
| Pseudanthias shemii | Reef Fish | 13 | −18.25 (0.15) | 11.96 v(0.53) | 3 | −18.29 (0.22) | 12.29 (0.58) | 8 | −18.25 (0.15) | 11.73 (0.50) | ||||||
| Pseudanthias sp. | Reef Fish | 5 | −18.16 (0.63) | 12.50 (0.30) | 5 | −18.16 (0.63) | 12.50 (0.30) | |||||||||
| Gobiidae sp. | Fish | 3 | −18.13 (0.34) | 11.39 (0.93) | 3 | −18.13 (0.34) | 11.39 (0.93) | |||||||||
| Apogonidaesp. | Fish | 3 | −17.41 (0.71) | 11.10 (0.52) | 3 | −17.41 (0.71) | 11.10 (0.52) | |||||||||
| Turritellidae sp. | Gastropod | 4 | −15.68 (0.41) | 12.66 (0.99) | 4 | −15.68 (0.41) | 12.66 (0.99) | |||||||||
| Ranellidae sp. | Gastropod | 5 | −17.78 (0.09) | 10.38 (0.33) | 5 | −17.78 (0.09) | 10.38 (0.33) | |||||||||
| Ophiuridae sp. | Brittle Star | 3 | −16.91 (0.13) | 10.40 (0.86) | 3 | −16.91 (0.13) | 10.40 (0.86) | |||||||||
Global values were calculated from pooled site data. *Four common prey species.
Foraging models
Models were created using the MATLAB software package. The large pelagic fish Almaco Jack (Seriola rivoliana) was chosen as a model predator in the simulations. Almaco Jack are known to feed opportunistically on a wide range of prey [23] including both fish and invertebrates [24], [25], and foraging across distances of up to 50 km [26] with the capacity to migrate hundreds of kilometres [27]. All parts of this study used the basis of the same modelling approach as Flaherty and Ben-David (2010), but to enhance reliability, modelling was based on 100 000 replicates per model rather than the 250 (see Text S1 for a detailed description of the model).
In the first part of this study to mimic the original model [15], four focal species; the fish P. rubrizonatus and decapods R. balssi, P. militaris and P. scabriusculus (see also [28]) common to all sites were designated as prey (Table 2), for four different predator models, as follows:
DsHs – the predator is a dietary and habitat specialist (preys on specific items but has site fidelity) (Model 1);
DsHg – the predator is a dietary specialist and habitat generalist (preys on specific items and forages between sites) (Model 2);
DgHg – the predator is a dietary and habitat generalist (preys on everything and forages between sites) (Model 3);
DgHs – the predator is a dietary generalist and habitat specialist (preys on everything but has site fidelity) (Model 4).
In part two of the study, the effect of distance between foraging sites on isotopic signatures of the Almaco Jack predator feeding on the four focal species was modelled. The aim was to model isotopic outcomes under conditions that are more likely to reflect a marine predator that is highly mobile and forages across large spatial scales. This was achieved by dictating the relative contribution of each habitat to reflect the effect of distance between foraging sites on habitat generalists (DsHg and DgHg; see Text S1 for a detailed description of the model).
In the third part, to further increase ecological realism, entire prey assemblages at each site were used to reflect site composition (see Table 2). Hence, for this part, only dietary generalists (DgHg and DgHs) were simulated. Unless otherwise denoted niche width is equal to the variance produced by the models.
Data analysis
To determine if the common invertebrates varied in isotopic signatures among sites, a Multiple Analysis of Variance (MANOVA) was performed. All statistical tests were performed using the SPSS statistical package. The dependent variables δ13C and δ15N were compared among the fixed factors site and prey species. Variances were compared separately for both δ13C and δ15N to determine the effects of habitat/location variability on isotopic composition in δ-space. An O'Brien's transformation 29] was applied to convert the variance data into a format suitable for Analysis of Variance (ANOVA), as follows:
Where ni is the number of observations of group i, yik is the k th observation of group i, 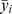 is the mean of the observations of group i, and
is the mean of the observations of group i, and 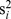 is the variance of the observations of group i.
is the variance of the observations of group i.
In order to avoid Type II error (i.e. falsely accept the null hypotheses) rarefaction curves were generated to determine the optimal sampling size of modelled variances [30]. For the modelled data it was found that optimal sample size ranged between 100 and 950 observations, hence a median of 475 observations was randomly selected from the 100 000 modelled observations for analysis of their means and variances.
To compare the four models, all combinations relevant to that model were pooled. For example, for the model DsHg this includes each of the prey species, which equates to four combinations). Analysis of Variance (ANOVA) was employed to test for differences between models, followed by Tukey's post hoc comparisons to identify where significant differences between models existed. However, the pooling of scenarios for models may confound some comparisons (i.e. where the effects of scenarios are opposite within each model, such differences due to pooling will not be apparent). Therefore, additional analysis using a one-way ANOVA with Tukey's post hoc comparisons between all possible scenarios of each model was performed. This same procedure was followed for all three parts of the study.
Mixing models were applied to the data to determine if converting δ-space to p-space (proportion space) as proposed by Newsome et al [19], could reduce variance to more accurately estimate trophic niche width, and to identify resource partitioning. To model the effects of prey variability across habitats that a “naive researcher” may encounter, 50 Almaco Jack were randomly sampled from the simulated populations. A sample size of 50 (predators) was deemed appropriate following initial runs which determined that a sample size of >15, as used by Flaherty and Ben-David [15], was required because the mixing space derived from the four reef species in this study was smaller.
Following the procedures of Flaherty and Ben-David [15], we constructed mixing spaces using the four focal prey species and selected Almaco Jack that fell only within this mixing space from simulated populations in both Part 1 and 2 for conversion to p-space. For models involving habitat generalists (DgHg and DsHg), global means of the four common prey species (sources) were used to distinguish the mixing space. However global means for habitat specialists were deemed inappropriate as they fell outside the mixing space, therefore the appropriate site means were used (Table 2).
In addition to the multi-source mixing model SISUS [31] applied by Flaherty and Ben-David [15], we also used the IsoSource [32], SIAR [33] and MixSIR [34] models to convert variances to p-space and estimate proportions of prey species contributions. Unless otherwise denoted, the model default settings were used and no trophic enrichment factors (TEF's) were defined other than program defaults, where appropriate. For the SISUS (Bayesian based) model [14], 10 000 samples were selected to be retained for analysis within the model, which generated mean proportions and variances for each of the mixtures (fractions of prey contributing to predator signature). For the IsoSource model (multiple source dual isotope mixing-model) [32], an increment of 1% and tolerance of 0.05 were selected for each possible mixture to generate mean proportions and variances. For the MixSIR (Bayesian based) model [34] 1 000 000 iterations were run and a posterior density ratio of <0.01 was ensured. For the SIAR (Bayesian based) model [33], 1 000 000 iterations with a burnin of 400 000 iterations (“very long” default setting in the package) were run, standard trophic enrichment factors (TEF's) of 3.54‰ (standard devation (SD) of 0.74) for δ15N and and 1.63‰ (SD = 0.63) for δ13C for trophic level were used, no elemental concentration corrections and/or priors were defined. Mean proportions and variances were calculated by randomly selecting a number, equal to the sample sizes of the mixtures for any one scenario. Mixture sample size was determined from the number of predator signatures that fell within the two dimensional mixing space (defined by the delta values of the prey).
For models containing dietary and habitat specialists, Pilumnus scabriusculus and the Yodel site were randomly selected for part 1 (for DsHg and DsHs), and Pseudanthias rubrizonatus in part 2 (DsHg). To determine the combined variances amongst proportions of each prey source in p-space, the Shannon-Wiener information measure (H) was used to estimate variances (niche width) [35]. These estimates were then compared with one way analysis of variance (ANOVA) followed by a Tukey's post hoc comparisons.
Results
Niche width estimates in δ-space
Part 1
The isotopic signatures of the common prey species varied among the sites (MANOVA, p<0.05; Table 2). Mean differences among sites were 0.5 ‰ (δ13C) and 0.6 ‰ (δ15N) for Pseudanthias rubrizonatus, 0.7 ‰ (δ13C) and 1.7 ‰ (δ15N) for Rhynchocinetes balssi, 3.2 ‰ (δ13C) and 1.8 ‰ (δ15N) for Petrolisthes militaris, and 1.3 ‰ (δ13C) and 2.1 ‰ (δ15N) for Pilumnus scabriusculus. Simulated models of Almaco Jack isotopic compositions from feeding on the common prey species (Part 1) found that their position within δ-space was variable (Fig 1, 2). In the majority of cases, higher variances indicated that dietary specialists (DsHg and DsHs) occupied greater bivariate space than dietary generalists (DgHs and DgHg). Pooled (i.e. the mean sum of all possible scenarios/combinations within each model) results for each model show that the isotopic niche can be greater for dietary specialists (DsHg and DsHs) with variances of 1.7 to 5.6 and 2 – 3 times greater for δ13C and δ15N, respectively, than dietary generalists (DgHs and DgHg) (Table 3). Comparison of O'Brien's variances among models with all possible scenarios pooled found significant differences for both δ13C (ANOVA, F3, 11875 = 8.27, p<0.001) and δ15N (ANOVA, F3, 11875 = 74.11, p<0.001). Post hoc comparisons revealed that for δ13C DsHg populations had significantly greater variances than DgHg, while DsHs variances were significantly greater than those of DgHs; however other comparisons e.g. DsHs and DgHs, were not significantly different (Table 4). For δ15N, significant differences were only found for comparisons of DsHg with all other models (Table 4).
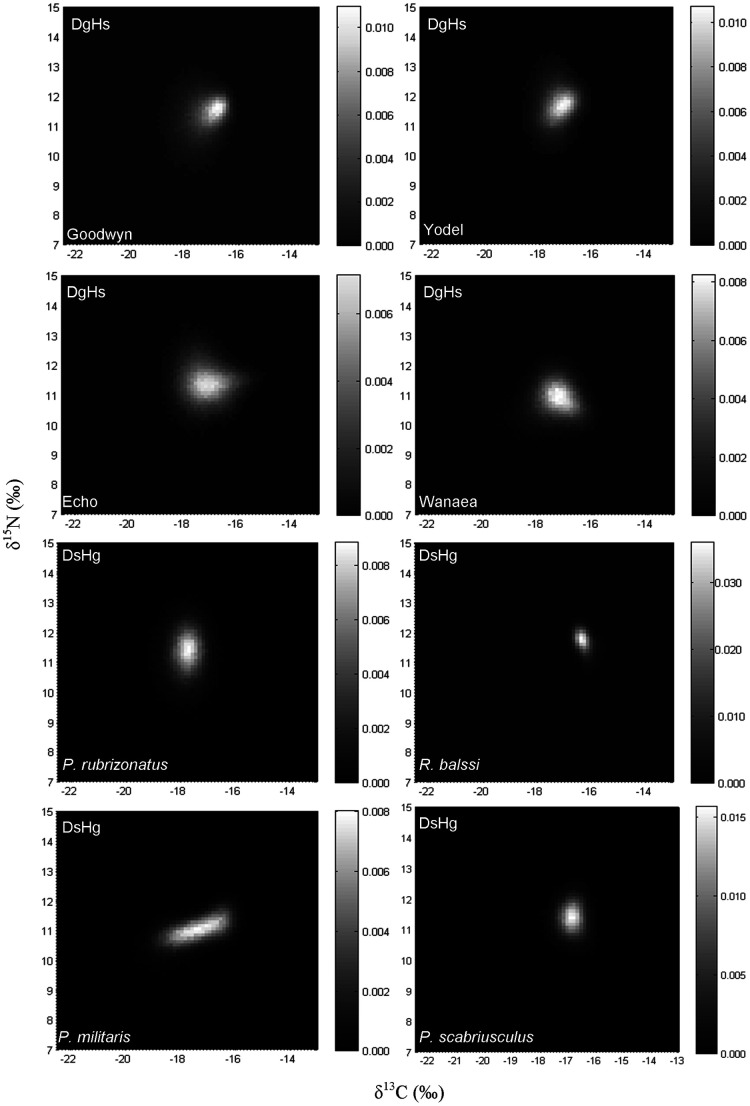
Figure 1. 2D histograms showing the distribution of the results obtained for 100 000 of the isotopic signatures from the modelled Almaco Jack in δ-space for the common species (Part 1) for dietary generalists and habitat specialists (DgHs) and dietary specialists and habitat generalists (DsHg).
Individual histogram greyscale bars indicate the relative frequency for each class.
Figure 2. 2D histograms showing the distribution of the results obtained for 100 000 of the isotopic signatures from the modelled Almaco Jack in δ-space for the common species (Part 1) for dietary generalists and habitat generalists (DgHg), habitat specialists that prey only on Petrolisthes militaris (DsHs) and habitat specialists that prey only on Pilumnus scabriusculus (DsHs).
Individual histogram greyscale bars indicate the relative frequency for each class.
Table 3. Mean and variance of δ13C and δ15N for models of simulated Almaco Jack populations.
| Model | Treatment | C13δ | N15δ | ||
| Part 1 | Mean | Variance | Mean | Variance | |
| DsHg (1) | P. rubrizonatus | −17.85 | 0.20 | 11.31 | 0.49 |
| DsHg (1) | R. balssi | −16.38 | 0.06 | 11.66 | 0.11 |
| DsHg (1) | P. militaris | −17.51 | 0.67 | 10.93 | 0.18 |
| DsHg (1) | P. scabriusculus | −16.90 | 0.14 | 11.32 | 0.21 |
| DsHg (1) | Pooled | −17.16 | 0.58 | 11.30 | 0.31 |
| DgHg (2) | Generalist | −17.40 | 0.24 | 11.27 | 0.26 |
| DsHs (3) | Goodwyn – P rubrizonatus | −17.96 | 0.45 | 11.00 | 0.97 |
| DsHs (3) | Goodwyn – R. balssi | −16.54 | 0.04 | 11.89 | 0.07 |
| DsHs (3) | Goodwyn – P. militaris | −16.19 | 0.07 | 11.36 | 0.17 |
| DsHs (3) | Goodwyn – P. scabriusculus | −17.17 | 0.14 | 11.27 | 0.20 |
| DsHs (3) | Yodel – P. rubrizonatus | −17.77 | 0.25 | 11.63 | 0.78 |
| DsHs (3) | Yodel – P. balssi | −16.47 | 0.09 | 11.85 | 0.12 |
| DsHs (3) | Yodel – P. militaris | −18.43 | 0.34 | 10.61 | 0.11 |
| DsHs (3) | Yodel – P. scabriusculus | −16.68 | 0.11 | 12.16 | 0.16 |
| DsHs (3) | Echo – P. rubrizonatus | −17.85 | 0.29 | 11.59 | 0.71 |
| DsHs (3) | Echo – R. balssi | −15.78 | 0.36 | 11.57 | 0.05 |
| DsHs (3) | Echo – P. militaris | −18.66 | 0.17 | 11.26 | 0.12 |
| DsHs (3) | Echo – P. scabriusculus | −16.12 | 0.16 | 10.11 | 0.18 |
| DsHs (3) | Wanaea – R. rubrizonatus | −17.50 | 0.25 | 11.39 | 0.23 |
| DsHs (3) | Wanaea – R. balssi | −16.04 | 0.09 | 10.13 | 0.14 |
| DsHs (3) | Wanaea – P. militaris | −19.47 | 1.20 | 9.58 | 0.49 |
| DsHs (3) | Wanaea – P. scabriusculus | −17.34 | 0.58 | 10.96 | 0.59 |
| DsHs (3) | Pooled | −17.25 | 1.35 | 11.15 | 0.80 |
| DgHs (4) | Goodwyn | −17.29 | 0.29 | 11.29 | 0.41 |
| DgHs (4) | Yodel | −17.36 | 0.23 | 11.55 | 0.32 |
| DgHs (4) | Echo | −17.17 | 0.45 | 11.33 | 0.32 |
| DgHs (4) | Wanaea | −17.34 | 0.35 | 10.80 | 0.28 |
| DgHs (4) | Pooled | −17.29 | 0.34 | 11.24 | 0.41 |
Those models in bold elucidate mean values for each population based on diet (specialist vs. generalist) and habitat (Wellhead). Where a model consists of numerous variations (different specialisations) a ‘Pooled’ value is provided as an accumulative mean value for the model. Models for Part 1 used the four common prey species. Models for Part 2 used the common prey species and incorporating distance between sites. Models for Part 3 used the entire prey assemblage.
Table 4. Tukey's post hoc results comparing variances between models using the four common prey species (Part 1) for δ13C and δ15N.
| Isotope | Comparison | DsHg | DgHg | DsHs |
| δ13C | DgHg | * | ||
| δ13C | DsHs | NS | *** | |
| δ13C | DgHs | NS | NS | * |
| δ15N | DgHg | *** | ||
| δ15N | DsHs | *** | NS | |
| δ15N | DgHs | *** | NS | NS |
NS: no significant difference; asterisks indicate significant differences at *p<0.05, ** p<0.01 and *** p<0.001. A).
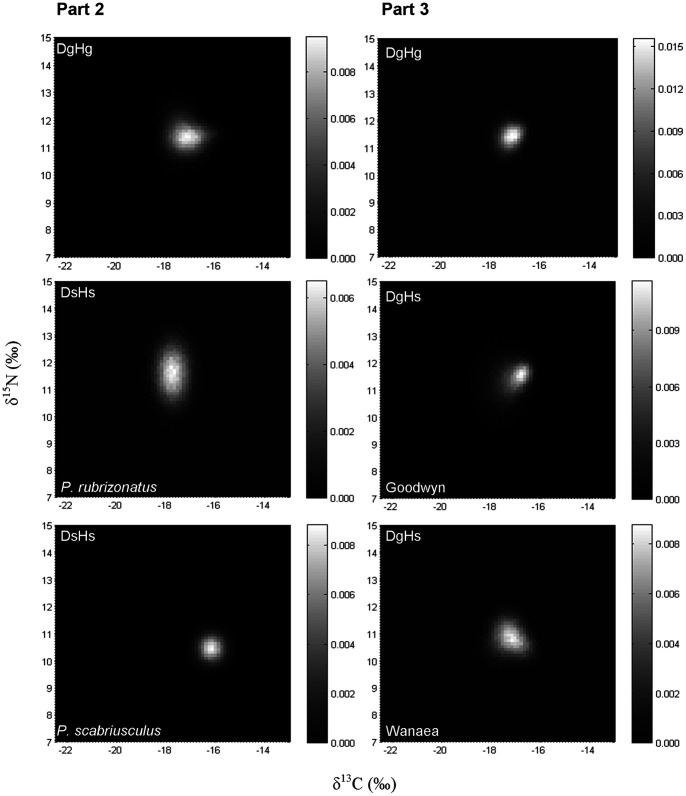
A closer inspection of the modelled data for Part 1 revealed that the niche width displayed by the predator varied both among and within models (Fig 1, 2) (for additional plots see Figure S1). Further comparison among the modelled outcomes found that isotopic niche width varied between both sites and prey species for the simulated populations of Almaco Jack (Table 3). The data show that differences in isotopic variances of the predator are prey species specific. For DsHg, δ13C variances ranged from being 2.8 times greater to 4 times less than those of DgHg, while for δ15N, DsHg variances ranged from 1.9 times greater to 2.4 times less than DgHg. In a similar manner the data reveal that for all models, differences in variances are prey source and/or habitat specific (see Table 3).
Part 2
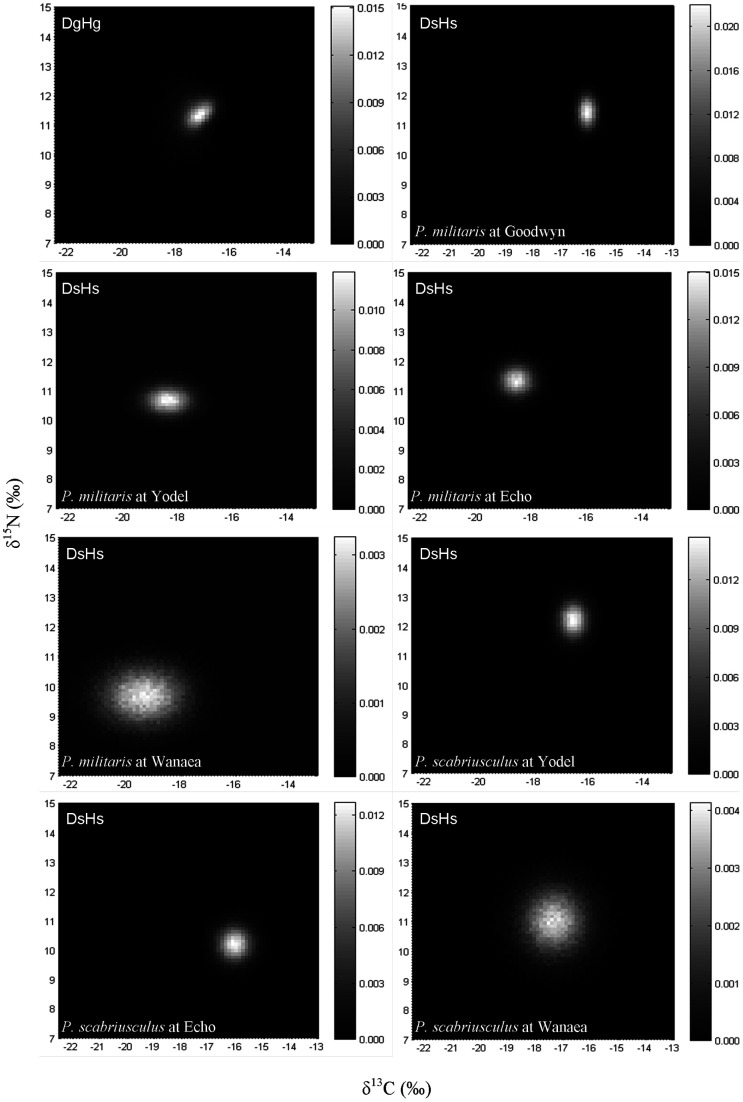
The isotopic composition of habitat generalists was found to further vary when the distance that the predator travels between foraging sites was added to the model (See DgHg Fig 3) (for additional plots see Figure S2). In δ-space the differences in variances of dietary specialists (DsHg) was variable between prey sources, ranging from being the same to 1.9 times greater than dietary generalists (DgHg) for δ13C, and 6 times less to 2.3 times greater than dietary generalists (DgHg) for δ15N (Table 3). Comparison of variances between models with scenarios pooled was significant for δ13C (ANOVA, F1, 2375 = 106.636, p<0.001) and δ15N (ANOVA, F1, 2375 = 6.083, p<0.05), while comparisons among all scenarios within each of the two models (1 and 4) were significant for both δ13C (ANOVA, F4, 2375 = 60.719, p<0.001) and δ15N (ANOVA, F4, 2375 = 208.679, p<0.001). All scenarios of dietary specialists (DsHg) were found to be different to dietary generalists (DgHg) for both δ13C and δ15N, while some comparisons between the different dietary specialists (DsHg) were also different (see Table 5).
Figure 3. 2D histograms showing the distribution of the results obtained for 100 000 of the isotopic signatures from the modelled Almaco Jack in δ-space incorporating distance between sites for the common prey species (Part 2), and the entire prey assemblages (Part 3).
Models include those for dietary generalists and habitat generalists (DgHg), dietary specialists and habitat specialists (DsHs) (Part 2 only) and dietary generalists and habitat specialists (DgHs) (Part 3 only). Individual histogram greyscale bars indicate the relative frequency for each class.
Table 5. Tukey's post hoc results comparing variances between models using the common prey species and incorporating distance between sites (Part 2).
| Isotope | Comparison | DsHg P. rubrizonatus | DsHg R. balssi | DsHg P. militaris | DsHg P. scabriusculus |
| δ13C | DsHg – R. balssi | NS | |||
| δ13C | DsHg – P. militaris | *** | *** | ||
| δ13C | DsHg – P. scabriusculus | *** | *** | NS | |
| δ13C | DgHg | *** | ** | *** | *** |
| δ15N | DsHg – R. balssi | *** | |||
| δ15N | DsHg – P. militaris | *** | NS | ||
| δ15N | DsHg – P. scabriusculus | *** | NS | NS | |
| δ15N | DgHg | *** | *** | ** | * |
NS: no significant difference; asterisks indicate significant differences at *p<0.05, **p<0.01 and ***p<0.001.
Part 3
Differences in δ-space were also variable when comparing models of dietary generalists (DgHg and DgHs) utilising the entire prey assemblages at each site (Fig 3; for additional plots see Figure S3). Variances of habitat specialists (DgHs) ranged from being 1.4 to 2.9 times greater than habitat generalists (DgHg) for δ13C, and 1.4 to 2.2 times greater than habitat generalists (DgHg) for δ15N (Table 3). Comparisons of pooled variances (i.e. those derived from the isotopic signatures) of simulated populations feeding on the entire prey assemblage were significant for both δ13C and δ15N isotopes (δ13C: ANOVA, F1, 2375 = 106.636, p<0.001; δ15N ANOVA, F1, 2375 = 66.189, p<0.001). Differences were also found for δ13C and δ15N variances among all scenarios within each of the two models compared (DgHg and DgHs) (δ13C: ANOVA, F4, 2375 = 73.911, p<0.001; δ15N: ANOVA, F4, 2375 = 25.942, p<0.001). All comparisons of individual scenarios within DgHg were different to DgHs (with the exception of habitat specialists at Yodel for δ13C and Wanaea for δ15N), while only some comparisons between the different habitat specialists were different (see Table 6).
Table 6. Tukey's post hoc results comparing variances between models using the entire prey assemblage (Part 3).
| Isotope | Comparison | DgHs Goodwyn | DgHs Yodel | DgHs Echo | DgHs Wanaea |
| δ13C | DgHs Yodel | *** | |||
| δ13C | DgHs Echo | *** | *** | ||
| δ13C | DgHs Wanaea | NS | *** | *** | |
| δ13C | DgHg | *** | NS | *** | *** |
| δ15N | DgHs Yodel | NS | |||
| δ15N | DgHs Echo | ** | NS | ||
| δ15N | DgHs Wanaea | *** | ** | NS | |
| δ15N | DgHg | *** | *** | *** | NS |
NS: no significant difference; asterisks indicate significant differences at *p<0.05, **p<0.01 and ***p<0.001.
Niche width estimates in p-space and prey source proportions
In Part 1, variances indicate that isotopic niche width in p-space was greater for the dietary specialists (DsHg and DsHs), than the dietary generalists (DgHg and DgHs) (Table 7), however only differences using the MIXSIR and SIAR models were found to be significantly different (SISUS: F3, 43 = 1.588, p = 0.208; IsoSource: F3, 43 = 2.082, p = 0.118; MIXSIR: F3, 43 = 5.013, p<0.05; SIAR: F3, 43 = 68.153, p<0.001). Post hoc comparisons for the MIXSIR model indicated that only dietary and habitat generalists (DgHg) were different from dietary generalist, habitat specialists (DgHs). In comparison, post-hoc analysis for the SIAR model revealed that dietary generalists and habitat specialists (DgHs) were different to all other categories, which were not different from each other (Table 8).
Table 7. Comparison of source proportion estimates for each of the common prey species and Shannon-Wiener information measure (H) means and variances in p-space, between mixing models (SISUS, IsoSource and MixSIR) for Part 1 and 2.
| Models | Prey sources | p-space | |||||
| Part 1 | Mixing model | Pseudanthias rubrizonatus | Rhynchocinetes balssi | Petrolisthes militaris | Pilumnus scabriusculus | Hmean | Hvarianc |
| DsHg | SISUS | 0.30 (0.06) | 0.44 (0.08) | 0.11 (0.02) | 0.15 (0.01) | −0.98 | 0.10 |
| n = 8 | IsoSource | 0.35 (0.11) | 0.33 (0.08) | 0.12 (0.02) | 0.19 (0.01) | −0.97 | 0.16 |
| MixSIR | 0.29 (0.15) | 0.09 (0.02) | 0.26 (0.05) | 0.36 (0.09) | −0.79 | 0.09 | |
| SIAR | 0.21 (0.01) | 0.10 (0.01) | 0.51 (0.03) | 0.16 (0.01) | −1.08 | 0.04 | |
| Mean | 0.29 (0.08) | 0.24 (0.19) | 0.25 (0.03) | 0.22 (0.01) | −0.96 | 0.01 | |
| DgHg | SISUS | 0.36 (0.05) | 0.15 (0.06) | 0.28 (0.09) | 0.20 (0.09) | −1.11 | 0.07 |
| n = 7 | IsoSource | 0.31 (0.06) | 0.13 (0.01) | 0.24 (0.08) | 0.19 (0.01) | −1.14 | 0.06 |
| MixSIR | 0.11 (0.03) | 0.22 (0.03) | 0.45 (0.06) | 0.22 (0.05) | −0.95 | 0.01 | |
| SIAR | 0.23 (0.04) | 0.10 (0.01) | 0.45 (0.02) | 0.22 (0.01) | −1.14 | 0.01 | |
| Mean | 0.25 (0.05) | 0.15 (0.03) | 0.36 (0.06) | 0.21 (0.04) | −1.09 | 0.04 | |
| DsHs | SISUS | 0.09 (0.01) | 0.49 (0.09) | 0.10 (<0.01) | 0.33 (0.10) | −0.88 | 0.11 |
| n = 8 | IsoSource | 0.10 (<0.01) | 0.47 (0.08) | 0.09 (<0.01) | 0.33 (0.08) | −0.95 | 0.07 |
| MixSIR | 0.19 (0.09) | 0.17 (0.02) | 0.55 (0.05) | 0.08 (0.01) | −0.88 | 0.06 | |
| SIAR | 0.06 (<0.01) | 0.02 (<0.01) | 0.90 (<0.01) | 0.03 (<0.01) | −0.99 | 0.02 | |
| Mean | 0.11 (0.03) | 0.29 (0.05) | 0.41 (0.01) | 0.19 (0.05) | −0.93 | 0.07 | |
| DgHs | SISUS | 0.23 (0.04) | 0.28 (0.04) | 0.31 (0.04) | 0.18 (0.01) | −1.09 | 0.04 |
| n = 20 | IsoSource | 0.25 (0.03) | 0.26 (0.04) | 0.30 (0.04) | 0.20 (0.01) | −1.15 | 0.01 |
| MixSIR | 0.09 (0.01) | 0.05 (<0.01) | 0.79 (0.01) | 0.07 (<0.01) | −0.66 | 0.02 | |
| SIAR | 0.21 (0.02) | 0.12 (0.01) | 0.60 (0.01) | 0.07 (<0.01) | −0.40 | 0.02 | |
| Mean | 0.20 (0.03) | 0.18 (0.02) | 0.50 (0.02) | 0.13 (<0.01) | −0.83 | 0.02 | |
Includes mean proportion estimates, and Shannon-Wiener means and variances for all three models, elucidated in bold.
Table 8. Comparison of variances in δ-space (for both δ13C and δ15N) with p-space (Shannon-Wiener information measure) for models using the common prey species (Part 1) and models using the common prey species and incorporated distance between sites (Part 2).
| Part | Model | δ13C | δ15N | P |
| DsHg (P. scabriusculus only) | ||||
| Part 1 | DgHg | 1.7 | 1.2 | 2.4 |
| Part 1 | DsHs (P. scabriusculus and Yodel only) | 1.3 | 1.3 | 1.5 |
| Part 1 | DgHs (Yodel only) | 1.7 | 1.2 | 6.0 |
| DgHg | ||||
| Part 1 | DsHs (P. scabriusculus and Yodel only) | 2.2 | 1.6 | 1.6 |
| Part 1 | DgHs (P. scabriusculus only) | 1.0 | 1.2 | 2.5 |
| DsHs (P. scabriusculus and Yodel only) | ||||
| Part 1 | DgHs (Yodel only) | 2.1 | 2.0 | 4.0 |
| DsHg (P. rubrizonatus only) | ||||
| Part 2 | DgHg | 1.1 | 2.3 | 2.0 |
In Part 2 (where foraging distance was included in the models) the dietary specialist (DsHg) was found to have narrower isotopic niche than the dietary generalist (DgHg). Three of the four models found these differences to be significant, SISUS (F1, 12 = 6.220, p<0.05), IsoSource (F1, 12 = 6.794, p<0.05) and MIXSIR (F1, 12 = 6.794, p<0.05). These results should be interpreted with care as the sample size was small (Table 7). Results from both Parts 1 and 2 show that model variances decreased on conversion from δ-space. However, the differences in variances between models remained similar or increased (Table 8).
Discussion
The results confirm that isotopic variability amongst habitats can confound estimates of isotopic niche in both δ-space and p-space. The modelling of isotopic compositions of simulated populations of Almaco Jack foraging between artificial reefs conforms with the terrestrial modelling by Flaherty and Ben-David [15]. In the present study, improved modelling techniques and more ecologically realistic conditions were applied to test the effects of isotopic variability between habitats on trophic niche width. In addition, data were converted from δ-space to p-space, as suggested by Newsome et al. [19] using a range of different mixing models to reduce scaling discrepancies. The modelling suggests that the isotopic variability of prey may confound any predictions of trophic niche, irrespective of an organism's trophic strategy (specialist vs. generalist) and/or the source of isotopic variation (spatial vs. compositional differences). In addition, the use of mixing models to convert δ-space variance to p–space variance offers little or no assistance. Interestingly, and in contrast to what is commonly accepted, although estimated isotopic niche breadth is a function of the variance of prey items (in this study global values of common prey species varied by 1.9‰ for δ13C and 0.5‰ for δ15N) and the spatial dispersion of that variance, dietary specialists appear to have a broader isotopic range than dietary generalists.
Analysis of the data revealed that prey variability in stable isotope signatures among habitats must be accounted for if we are to make realistic predictions about niche width. These results confirm that the natural variability that occurs across spatial scales of the study area will influence isotopic signatures, especially those of δ13C [16], [31], confounding comparisons of isotopic variances between many populations [36]. Natural variations in isotopic signatures will be evident amongst most basal resource pools. This is especially evident in the marine environment. For example phytoplankton are known to show trends of δ13C enrichment with decreasing latitude towards the equator [37], indicating fluctuations in the physiochemical environment may lead to variability. What remains clear, is that to interpret the variance amongst isotopic signatures of predators, isotopic variability of prey needs careful consideration [16], [38] and for many studies, adequate sampling across relevant spatial and temporal scales needs to be a prerequisite [39]. Despite this, a number of studies have attempted to estimate isotopic niche width as a measure of trophic niche [31], [40], [41], [42], [43], [44]. Where spatial variation in isotopic composition of prey can be dismissed, comparisons of trophic niche widths may be possible e.g. as in Willson et al. [40] who used a small, isolated study site to investigate aquatic snakes. Unfortunately for the majority of habitats and study species, it is clear that a detailed knowledge of species-specific feeding behaviour and the ecology of the community are required before variability in prey isotopic composition can be identified and accounted for [1], [15], [45]. The use of multiple methods may aid the accuracy of estimation of trophic niche width using stable isotopes, and as such, a number of studies have successfully utilised the information from stable isotopes combined with gut analysis to make informed estimates of trophic niches [42], [46], [47].
The ‘niche variation hypothesis’ proposed by Van Valen [11] predicts that “populations with wider niches are more variable than populations with narrower niches” [48]. Correspondingly, Bearhop et al. [12] predicted that populations consuming a wider range of prey and those that forage in a range of geographical areas could display wider isotopic variation than those that have a narrow range of prey and limited foraging capacity. In accordance with Bearhop et al's [12] predictions, Olsson et al. [41] examined the isotopic niche widths of invasive and native crayfish in Swedish streams. The greater niche width of the introduced species reflected a wider use of habitat and/or prey sources. However at the population level, the two species did not differ in niche widths, indicating that isotopic variability between habitats was confounding any differences [41]. Accordingly, our models have identified the confounding influence of habitat use on predictions of trophic niche width. Furthermore, comparisons of populations of dietary generalists feeding on the common four prey sources indicate that isotopic variation among habitat specialists was similar or greater than the equivalent habitat generalists (Table 3). Niche width may increase by either the entire population shifting to use all available resources or by an increase in inter-individual specialisation within a population (see [49]). Simulations of populations of dietary generalists here suggest that populations confined to one site may display greater isotopic variance within their population due to individual specialisation. This individual niche variation among conspecific individuals has been suggested as being widespread [49], indicating that the variation in isotopic niche within a population may further confound any estimates of a populations trophic niche width. For example, predators within the same population with different dietary specialisations can account for greater trophic variability at the population level than the same population composed of generalists.
Fundamentally, anything which prevents or causes an organism to sample only a portion of the complete distribution of prey signatures where variation exists could result in incomplete and inaccurate estimates of niche width. Our data indicates that as the variance in prey items increases, the greater there is for the potential of inaccuracy (dependant on the spatial distribution of the signatures). The influence of distance between resources on the foraging behaviour of animals has been well established [50], [51], and such effects may be driven by macronutrient regulation [52], [53] and prey availability [54]. Data from simulated populations of Almaco Jack accounting for distance between foraging locations revealed that isotopic values were variable and prey species-dependent. Many communities are vastly more complex than a four prey model [49] and large predators are likely to feed on a greater diversity of prey [55]. Inclusion of all prey species of a similar trophic level to the model, to further increase ecological realism, showed that habitat generalists displayed narrower niches than habitat specialists. Dietary specialists will typically exhibit a broader trophic niche than dietary generalists because they lack the influence of different prey items that are variable in their isotopic signature. That is across many sites where variation in prey signatures exists, the range between means will be less for predators that eat multiple prey items (dietary generalist) than for those that only eat specific prey (dietary specialists).
This problematic nature of estimates of niche width using variance in δ-space has been addressed by Newsome et al. [19], who proposed the use of mixing models to transform data into p-space (dietary proportions). The transformation provides a value comparable to other common variables used in studies of ecological niches, and corrects for magnitude differences amongst isotopic composition of prey [19]. In the present study the mixing models reduced the variances observed in p-space (Table 6) compared with those observed in δ-space (Table 3), however, they failed to reduce the differences in variances observed amongst models of the Alcamo Jack populations. In both parts of the study (1 & 2) where variances were compared in both δ-space and p-space, it was clear that this transformation maintained and in many instances increased the observed differences in isotopic variances between the simulated models (Table 7). We therefore concur with the findings of Flaherty and Ben-David [15] who raised concerns with the use of such transformation. Furthermore, many mixing models used to estimate proportional values are reliant on amounts of a priori information, in such cases isotopic mixing models are sometimes less informative than non-isotopic information in its raw form i.e. stomach content data (see [1] for discussion).
Flaherty and Ben-David [15] modelled the attempts of a “naive researcher” who ignores habitat use of the study species when using isotopic data to estimate the trophic niche. In a similar manner, we used mixing spaces [32] to reproduce these simulations within a marine ecosystem. In comparison, mixing spaces for habitat specialists (DgHs and DsHs) were defined using source values from each site. If habitat variability in isotopic signatures is an important source of variation [15], [16], [56], it seems only appropriate that we define mixing space accordingly. Like Flaherty and Ben-David [15], we too encountered many isotopic values that fell outside of the mixing space. Because simulations are based on the isotopic signatures of the global or site mean of the prey species, when populations of specialist predators are observed a large proportion of the calculated isotopic values will fall outside their mixing space, independent of mixing space width. As variability in δ13C and δ15N of the primary producers in food webs exist among habitats [57], [58], [59], comparisons of δ13C and δ15N among habitats will be confounded by isotopic variability of the prey source [3].
Mixing models that provide estimates of prey item proportions within diets are becoming popular to determine partitioning of dietary resources. Such models have been refined [32], [34], [60], [61] and debated [62], [63] over recent years. Very recent examples of their use include Kristensen et al. [64], who applied mixing models to δ13C and δ15N isotopes to determine resource partitioning amongst leaf-eating mangrove crabs, and Flaherty et al. [65] used similar models to determine the contribution of different prey items to overall diet of flying squirrels. We tested and compared numerous models to determine if the partitioning of a resource by populations could be identified. It can be seen that in the majority of cases SISUS and IsoSource made very similar estimates, but different to those from the MixSIR and SIAR models (Table 6). The mixing models all predicted that Almaco Jack fed in a relatively generalist manner on all four prey species, with the exception of the SIAR model for DsHs in Part 1. This includes models generated in part 2 for dietary specialists (DsHg and DsHs), which were simulated to feed exclusively on P. scabriusculus. Of concern was that on closer inspection of the proportions estimated, it was evident that no mixing model was able to accurately estimate proportions of the dietary specialists, possibly with the exception of SIAR for DsHs, irrespective of isotopic variation of habitats (Table 6). For part 2, SIAR failed to allocate the majority of the diet to the specialist prey item, P. rubrizonatus.
Transformation of the data to dietary proportions failed to distinguish the correct partitioning of prey sources for dietary specialists. In Part 1 mean estimates among mixing models for predators specialising on P. scabriusculus determined that this prey source, only counted for approximately a ¼ of their diet irrespective of the habitat model. In part 2, mean estimates amongst mixing models for predators specialising on P. rubrizonatus revealed that P. rubrizonatus accounted for only 31% of their diet, while other “uneaten” individual prey species contributing up to 49% of the diet (Table 6). Because no mixing model was able to accurately estimate proportions of the dietary specialists, irrespective of isotopic variation of habitats (Table 5), our data therefore show that inaccuracies amongst estimates provided by linear mixing models may go well beyond problems associated with habitat variability.
Like Flaherty and Ben-David [15], we too provide simplistic approaches to what are in reality, much more complex systems [49] that are likely to substantially underestimate the true extent of isotopic variability. We have attempted to include greater ecological complexity by including foraging distance and by using entire assemblages across a trophic level as prey sources. With these additions our models show that isotopic variability amongst habitats will confound estimations of trophic niche derived from measures of isotopic niche width in both δ-space [12] and p-space [19]. While the variability of prey isotopes is lower than may be encountered in some ecological systems but still likely reflective of many, it remains clear that isotopic niche is not a reliable indicator of trophic niche. Of greater concern was the failure of mixing models to correctly identify dietary specialisations and potential resource partitioning. Additionally, our simulations bring into question the accuracy of mixing models in identifying contribution sources, irrespective of whether isotopic variability amongst habitats exists. Our findings emphasise that progress in isotopic studies in animal ecology will require a greater understanding of the functional traits and behaviour of organisms.
Supporting Information
Data output from simulations of the isotopic signatures for Part 1 from the modelled Almaco Jack in δ-space that were both dietary and habitat specialists (DsHs) for the common.
(TIF)
Data output from simulations of the isotopic signatures for Part 1 from the modelled Almaco Jack in δ-space that were both dietary and habitat specialists (DsHs) for the common species.
(TIF)
Data output from simulations of the isotopic signatures from the modelled Almaco Jack in δ-space. A) Habitat generalists specialising on the common species (DsHg) accounting for distance between sites – Part 2. B) – Habitat specialists feeding on the entire prey assemblages (DgHs) – Part 3.
(TIF)
Detailed model description.
(DOCX)
Acknowledgments
We gratefully acknowledge Roger Springthorpe, Shane Ahyong and Stephen Keable of the Australian Museum, and Michela Mitchell from the Museum of Victoria, for assistance with specimen identification. We are grateful to SEA SERPENT personnel D. Skropeta, N. Pastro and A. Fowler who assisted in the collection of specimens and V. Valenzuela Davie for maintaining the database of these specimens. We thank the Captain, business partners and crew of the C/V Havila Harmony and Woodside Energy who made this research possible by providing access and logistical support. The quality and content of this manuscript was significantly improved, by the comments of two anonymous reviewers, to whom we are grateful. The paper is contribution #8 from the South East Asian Branch of the SERPENT (Scientific and Environmental Remotely Operated Vehicle Partnership using Existing Industrial Technology).
Funding Statement
This research was funded by the Australian Research Council (ARC) linkage grant ARC LP0775183 and constitutes a portion of the Ph.D. thesis of DOC. SJS was funded by an ARC Laureate Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Layman CA, Arrington DA, Montana CG, Post DM (2007) Can stable isotope ratios provide for community-wide measures of trophic structure? Ecology 88: 42–48. [DOI] [PubMed] [Google Scholar]
- 2.Vander Zanden MJ, Hulshof M, Ridgway MS, Rasmussen JB (1998) Application of stable isotope techniques to trophic studies of age-0 smallmouth bass. Transactions of the American Fisheries Society 127: 729–739. [Google Scholar]
- 3.Post DM (2002) Using stable isotopes to estimate trophic position: Models, methods, and assumptions. Ecology 83: 703–718. [Google Scholar]
- 4.Feranec RS, MacFadden KBJ (2000) Evolution of the grazing niche in Pleistocene mammals from Florida: Evidence from stable isotopes. Palaeogeography Palaeoclimatology Palaeoecology 162: 155–169. [Google Scholar]
- 5.Cherel Y, Le Corre M, Jaquemet S, Menard F, Richard P, et al. (2008) Resource partitioning within a tropical seabird community: New information from stable isotopes. Marine Ecology Progress Series 366: 281–291. [Google Scholar]
- 6.Burton RK, Koch PL (1999) Isotopic tracking of foraging and long-distance migration in northeastern Pacific pinnipeds. Oecologia 119: 578–585. [DOI] [PubMed] [Google Scholar]
- 7.Mooney KA, Tillberg CV (2005) Temporal and spatial variation to ant omnivory in pine forests. Ecology 86: 1225–1235. [Google Scholar]
- 8.Darimont CT, Paquet PC, Reimchen TE (2007) Stable isotopic niche predicts fitness of prey in a wolf-deer system. Biological Journal of the Linnean Society 90: 125–137. [Google Scholar]
- 9.Aurioles-Gamboa D, Newsome SD, Salazar-Pico S, Koch PL (2009) Stable isotope differences between sea lions (zalophus) from the Gulf of California and Galapagos Islands. Journal of Mammalogy 90: 1410–1420. [Google Scholar]
- 10.Gross A, Kiszka J, Van Canneyt O, Richard P, Ridoux V (2009) A preliminary study of habitat and resource partitioning among co-occurring tropical dolphins around Mayotte, southwest Indian Ocean. Estuarine Coastal and Shelf Science 84: 367–374. [Google Scholar]
- 11.Van Valen L (1965) Morphological Variation and Width of Ecological Niche. American Naturalist 99: 377–390. [Google Scholar]
- 12.Bearhop S, Adams CE, Waldron S, Fuller RA, Macleod H (2004) Determining trophic niche width: a novel approach using stable isotope analysis. Journal of Animal Ecology 73: 1007–1012. [Google Scholar]
- 13.Raubenheimer D, Simpson SJ (2003) Nutrient balancing in grasshoppers: behavioural and physiological correlates of dietary breadth. Journal of Experimental Biology 206: 1669–1681. [DOI] [PubMed] [Google Scholar]
- 14.Bolnick DI, Svanback R, Araujo MS, Persson L (2007) Comparative support for the niche variation hypothesis that more generalized populations also are more heterogeneous. Proceedings of the National Academy of Sciences of the United States of America 104: 10075–10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flaherty EA, Ben-David M (2010) Overlap and partitioning of the ecological and isotopic niches. Oikos 119: 1409–1416. [Google Scholar]
- 16.Matthews B, Mazumder A (2004) A critical evaluation of intrapopulation variation of δ13C and isotopic evidence of individual specialization. Oecologia 140: 361–371. [DOI] [PubMed] [Google Scholar]
- 17.Jackson AL, Inger R, Parnell AC, Bearhop S (2011) Comparing isotopic niche widths among and within communities: SIBER – Stable Isotope Bayesian Ellipses in R. Journal of Animal Ecology. 80: 595–602. [DOI] [PubMed] [Google Scholar]
- 18.Semmens BX, Ward EJ, Moore JW, Darimont CT (2009) Quantifying Inter- and Intra-Population Niche Variability Using Hierarchical Bayesian Stable Isotope Mixing Models. PLoS ONE 4: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newsome SD, del Rio CM, Bearhop S, Phillips DL (2007) A niche for isotopic ecology. Frontiers in Ecology and the Environment 5: 429–436. [Google Scholar]
- 20.Cummings DO, Booth DJ, Lee RW, Simpson SJ, Pile AJ (2010) Ontogenetic diet shifts in the reef fish Pseudanthias rubrizonatus from isolated populations on the North-West Shelf of Australia. Marine Ecology Progress Series 419: 211–222. [Google Scholar]
- 21.Yohannes E, Hansson B, Lee RW, Waldenstrom J, Westerdahl H, et al. (2008) Isotope signatures in winter moulted feathers predict malaria prevalence in a breeding avian host. Oecologia 158: 299–306. [DOI] [PubMed] [Google Scholar]
- 22.Minagawa M, Wada E (1984) Stepwise enrichment of 15N along food-chains – further evidence and the relation between delta δ15N and animal age. Geochimica Et Cosmochimica Acta 48: 1135–1140. [Google Scholar]
- 23.Barreiros JP, Morato T, Santos RS, de Borba AE (2003) Interannual changes in the diet of the almaco jack, Seriola rivoliana (Perciformes: Carangidae) from the Azores. Cybium 27: 37–40. [Google Scholar]
- 24.Andaloro F, Pipitone C (1997) Food and feeding habits of the amberjack, Seriola dumerili in the Central Mediterranean Sea during the spawning season. Cahiers de Biologie Marine 38: 91–96. [Google Scholar]
- 25.Matallanas J, Casadevall M, Carrasson M, Boix J, Fernandez V (1995) The food of Seriola dumerili (Pisces, Carangidae) in the Catalan Sea (Western Mediterranean). Journal of the Marine Biological Association of the United Kingdom 75: 257–260. [Google Scholar]
- 26.Gillanders BM, Ferrell DJ, Andrew NL (2001) Estimates of movement and life-history parameters of yellowtail kingfish (Seriola lalandi): how useful are data from a cooperative tagging programme? Marine and Freshwater Research 52: 179–192. [Google Scholar]
- 27.Tanaka S (1972) Migration of yellowtails along pacific coast of Japan observed by tagging experiments. 2. considerations from catch statistics and length composition. Bulletin of the Japanese Society of Scientific Fisheries 38: 93–&. [Google Scholar]
- 28.Cummings DO, Lee RW, Simpson SJ, Booth DJ, Pile AJ, et al. (2011) Resource partitioning amongst co-occurring decapods on wellheads from Australia's North-West shelf. An analysis of carbon and nitrogen stable isotopes. Journal of Experimental Marine Biology and Ecology 409: 186–193. [Google Scholar]
- 29.O'Brien RG (1981) A simple test for variance effects in experimental-designs. Psychological Bulletin 89: 570–574. [Google Scholar]
- 30.Quinn GP, Keough MJ (2002) Experimental design and data analysis for biologists. Cambridge: Cambridge University Press. [Google Scholar]
- 31.Erhardt EB (2010) SISUS: Stable isotope sourcing using sampling [dt]. [Google Scholar]
- 32.Phillips DL, Gregg JW (2003) Source partitioning using stable isotopes: coping with too many sources. Oecologia 136: 261–269. [DOI] [PubMed] [Google Scholar]
- 33.Parnell AC, Inger R, Bearhop S, Jackson AL (2010) Source Partitioning Using Stable Isotopes: Coping with Too Much Variation. PLoS ONE 5: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore JW, Semmens BX (2008) Incorporating uncertainty and prior information into stable isotope mixing models. Ecology Letters 11: 470–480. [DOI] [PubMed] [Google Scholar]
- 35.Bolnick DI, Yang LH, Fordyce JA, Davis JM, Svanback R (2002) Measuring individual-level resource specialization. Ecology 83: 2936–2941. [Google Scholar]
- 36.Araujo MS, Bolnick DI, Machado G, Giaretta AA, dos Reis SF (2007) Using delta C-13 stable isotopes to quantify individual-level diet variation. Oecologia 152: 643–654. [DOI] [PubMed] [Google Scholar]
- 37.Rau GH, Sweeney RE, Kaplan IR (1982) Plankton 13C:12C ratio changes with latitude: differences between northern and southern oceans. Deep Sea Research (Part I, Oceanographic Research Papers) 29: 1035–1039. [Google Scholar]
- 38.Post DM (2003) Individual variation in the timing of ontogenetic niche shifts in largemouth bass. Ecology 84: 1298–1310. [Google Scholar]
- 39.Barnes C, Jennings S, Polunin NVC, Lancaster JE (2008) The importance of quantifying inherent variability when interpreting stable isotope field data. Oecologia 155: 227–235. [DOI] [PubMed] [Google Scholar]
- 40.Willson JD, Winne CT, Pilgrim MA, Romanek CS, Gibbons JW (2010) Seasonal variation in terrestrial resource subsidies influences trophic niche width and overlap in two aquatic snake species: a stable isotope approach. Oikos 119: 1161–1171. [Google Scholar]
- 41.Olsson K, Stenroth P, Nystrom P, Graneli W (2009) Invasions and niche width: does niche width of an introduced crayfish differ from a native crayfish? Freshwater Biology 54: 1731–1740. [Google Scholar]
- 42.Frederich B, Fabri G, Lepoint G, Vandewalle P, Parmentier E (2009) Trophic niches of thirteen damselfishes (Pomacentridae) at the Grand R, cif of Toliara, Madagascar. Ichthyological Research 56: 10–17. [Google Scholar]
- 43.Woo KJ, Elliott KH, Davidson M, Gaston AJ, Davoren GK (2008) Individual specialization in diet by a generalist marine predator reflects specialization in foraging behaviour. Journal of Animal Ecology 77: 1082–1091. [DOI] [PubMed] [Google Scholar]
- 44.Chen G, Wu ZH, Gu BH, Liu DY, Li X, et al. (2011) Isotopic niche overlap of two planktivorous fish in southern China. Limnology 12: 151–155. [Google Scholar]
- 45.Hammerschlag-Peyer CM, Yeager LA, Araujo MS, Layman CA (2011) A Hypothesis-Testing Framework for Studies Investigating Ontogenetic Niche Shifts Using Stable Isotope Ratios. PLoS ONE 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson RM, Chanton J, Lewis G, Nowacek D (2009) Combining Organic Matter Source and Relative Trophic Position Determinations to Explore Trophic Structure. Estuaries and Coasts 32: 999–1010. [Google Scholar]
- 47.Layman CA, Quattrochi JP, Peyer CM, Allgeier JE (2007) Niche width collapse in a resilient top predator following ecosystem fragmentation. Ecology Letters 10: 937–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soule M, Stewart BR (1970) The “Niche-Variation” Hypothesis: A test and alternatives. American Naturalist 104: 85–97. [Google Scholar]
- 49.Bolnick DI, Svanback R, Fordyce JA, Yang LH, Davis JM, et al. (2003) The ecology of individuals: Incidence and implications of individual specialization. American Naturalist 161: 1–28. [DOI] [PubMed] [Google Scholar]
- 50.Charnov EL (1976) Optimal foraging, marginal value theorem. Theoretical Population Biology 9: 129–136. [DOI] [PubMed] [Google Scholar]
- 51.Krebs JR, Ryan JC, Charnov EL (1974) Hunting by expectation or optimal foraging – study of patch use by chickadees. Animal Behaviour 22: 953–&. [Google Scholar]
- 52.Behmer ST, Cox E, Raubenheimer D, Simpson SJ (2003) Food distance and its effect on nutrient balancing in a mobile insect herbivore. Animal Behaviour 66: 665–675. [Google Scholar]
- 53.Simpson SJ, Raubenheimer D, Charleston MA, Clissold FJ, Network A-NVF (2010) Modelling nutritional interactions: From individuals to communities. Trends in Ecology & Evolution 25: 53–60. [DOI] [PubMed] [Google Scholar]
- 54.Burke CM, Montevecchi WA (2009) The foraging decisions of a central place foraging seabird in response to fluctuations in local prey conditions. Journal of Zoology 278: 354–361. [Google Scholar]
- 55.Costa GC (2009) Predator size, prey size, and dietary niche breadth relationships in marine predators. Ecology 90: 2014–2019. [DOI] [PubMed] [Google Scholar]
- 56.Hobson KA (1999) Tracing origins and migration of wildlife using stable isotopes: a review. Oecologia 120: 314–326. [DOI] [PubMed] [Google Scholar]
- 57.Vander Zanden MJ, Rasmussen JB (1999) Primary consumer δ13C and δ15N and the trophic position of aquatic consumers. Ecology 80: 1395–1404. [Google Scholar]
- 58.Zohary T, Erez J, Gophen M, Bermanfrank I, Stiller M (1994) Seasonality of stable carbon isotopes within the pelagic food-web of Lake Kinneret. Limnology and Oceanography 39: 1030–1043. [Google Scholar]
- 59.Doi H, Kikuchi E, Shikano S, Takagi S (2010) Differences in nitrogen and carbon stable isotopes between planktonic and benthic microalgae. Limnology 11: 185–192. [Google Scholar]
- 60.Phillips DL, Gregg JW (2001) Uncertainty in source partitioning using stable isotopes. Oecologia 127: 171–179. [DOI] [PubMed] [Google Scholar]
- 61.Lubetkin SC, Simenstad CA (2004) Multi-source mixing models to quantify food web sources and pathways. Journal of Applied Ecology 41: 996–1008. [Google Scholar]
- 62.Semmens BX, Moore JW, Ward EJ (2009) Improving Bayesian isotope mixing models: A response to Jackson et al. (2009). Ecology Letters 12: E6–E8. [DOI] [PubMed] [Google Scholar]
- 63.Jackson AL, Inger R, Bearhop S, Parnell A (2009) Erroneous behaviour of MixSIR, a recently published Bayesian isotope mixing model: A discussion of Moore & Semmens (2008). Ecology Letters 12: E1–E5. [DOI] [PubMed] [Google Scholar]
- 64.Kristensen DK, Kristensen E, Mangion P (2010) Food partitioning of leaf-eating mangrove crabs (Sesarminae): Experimental and stable isotope (C-13 and N-15) evidence. Estuarine Coastal and Shelf Science 87: 583–590. [Google Scholar]
- 65.Flaherty EA, Ben-David M, Smith WP (2010) Diet and food availability: implications for foraging and dispersal of Prince of Wales northern flying squirrels across managed landscapes. Journal of Mammalogy 91: 79–91. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data output from simulations of the isotopic signatures for Part 1 from the modelled Almaco Jack in δ-space that were both dietary and habitat specialists (DsHs) for the common.
(TIF)
Data output from simulations of the isotopic signatures for Part 1 from the modelled Almaco Jack in δ-space that were both dietary and habitat specialists (DsHs) for the common species.
(TIF)
Data output from simulations of the isotopic signatures from the modelled Almaco Jack in δ-space. A) Habitat generalists specialising on the common species (DsHg) accounting for distance between sites – Part 2. B) – Habitat specialists feeding on the entire prey assemblages (DgHs) – Part 3.
(TIF)
Detailed model description.
(DOCX)