Abstract
Brown adipose tissue dissipates energy through heat and functions as a defense against cold and obesity. PPAR ligands have been shown to induce the browning of white adipocytes; however, the underlying mechanisms remain unclear. Here we show that PPAR ligands require full agonism to induce a brown fat gene program preferentially in subcutaneous white adipose. These effects require expression of PRDM16, a factor that controls the development of classical brown fat. Depletion of PRDM16 blunts the effects of the PPAR agonist rosiglitazone on the induced brown fat gene program. Conversely, PRDM16 and rosiglitazone synergistically activate the brown fat gene program in vivo. This synergy is tightly associated with an increased accumulation of PRDM16 protein, due in large measure to an increase in the half-life of the protein in agonist treated cells. Identifying compounds that stabilize PRDM16 protein may represent a plausible therapeutic pathway for the treatment of obesity and diabetes.
Introduction
White adipose tissue (WAT) functions to store excess energy as triglycerides while brown adipose tissue (BAT) is specialized to dissipate chemical energy as heat. The developmental and transcriptional control of BAT has received much attention over the last several years, mainly because of its potential role in the defense against obesity and obesity-associated disorders (reviewed in (Enerback, 2010). Recent findings using 18fluoro-labeled 2-deoxy-glucose positron emission tomography (18FDG-PET) scanning have clearly shown that most if not all normal adult humans have distinct brown fat deposits. The thermogenic activity of this tissue correlates inversely with overall adiposity, raising the possibility that variation in the amount or activity of BAT may contribute to the propensity for weight gain in humans (Cypess et al., 2009; Nedergaard et al., 2007; Saito et al., 2009; van Marken Lichtenbelt et al., 2009; Virtanen et al., 2009; Yoneshiro et al., 2011). Recent research has also identified several dominant transcriptional regulators of brown adipocyte development and function including PGC1α (peroxisome proliferator activated receptor gamma coactivator 1α), FoxC2 (Forkhead box C2) and PRDM16 (PRD1-BF-1-RIZ1 homologous domain containing protein-16) (reviewed in (Kajimura et al., 2010). Genetic loss of PGC1α and PRDM16 in mice clearly interferes with the function or/and development of BAT. Detailed knowledge of these pathways may offer promising opportunities to manipulate BAT in vivo for therapeutic regimens to counteract obesity and type 2 diabetes.
It is now clear that two different types of brown adipocytes exist and these have distinct developmental origins. Classical brown adipocytes that reside in the interscapular and perirenal regions develop from myoblastic-like Myf5 positive precursors (Seale et al., 2008). These Myf5 positive myoblast-like cells differentiate into brown adipocytes through the action of the transcriptional regulators PRDM16 and C/EBPβ (Kajimura et al., 2009; Seale et al., 2008). Furthermore, global gene expression analyses indicate that classical interscapular brown fat precursors have a gene profile overlapping that of skeletal muscle cells (Timmons et al., 2007). On the other hand, pockets of a second, distinct type of UCP1-positive adipocytes are found sporadically in the WAT of adult animals that have been exposed to chronic cold or β-adrenergic agonists. These inducible brown-like adipocytes (beige or brite cells) possess many of the biochemical and morphological characteristics of classical brown adipocytes, including the presence of multilocular lipid droplets (Frontini and Cinti, 2010). However, they arise from a non-Myf5 cell lineage and hence, have distinct origins from the classical brown adipocytes. Indeed, it has been previously shown that epidydimal WAT-derived “brite” cells that are induced by rosiglitazone do not express myocyte-enriched genes (Petrovic et al., 2009). In addition, adipogenic Sca-1+/CD45−/Mac1−progenitors from different adipose depots showed unique molecular signatures (Schulz et al., 2011).
Because the emergence of inducible-brown adipocytes in WAT is associated with a protection against obesity and metabolic diseases in rodent models (Cederberg et al., 2001; Leonardsson et al., 2004; Seale et al., 2011), an important challenge is to understand the molecular mechanisms by which environmental cues stimulate the development of these beige/brite cells. In this regard, it has been shown that activation of PPARγ (peroxisome proliferator-activated receptor-γ) by synthetic ligands induces a brown fat-like gene program in WAT (Fukui et al., 2000; Petrovic et al., 2009; Rong et al., 2007; Sell et al., 2004; Tai et al., 1996; Vernochet et al., 2009; Wilson-Fritch et al., 2004). Mechanistically, these drugs function by directly binding to and activating PPARγ and PPAR-response elements (PPREs) on the promoter and/or enhancer of brown fat-selective genes (Sears et al., 1996; Tai et al., 1996; Viswakarma et al., 2007). However, this explanation concerning the white to brown adipocyte transition cannot be considered adequate or complete for the following reasons. First, PPARγ functions as a master regulator of both white and brown adipocytes (reviewed in (Farmer, 2006). PPARγ is expressed abundantly and equally in white fat and brown fat, and is required for the development of both cell types (He et al., 2003; Imai et al., 2004). Notably, overexpression of PPARγ in white adipocytes does not induce a white-to-brown fat conversion (Sugii et al., 2009). Hence, it is not clear why activation of PPARγ by synthetic compounds should give such browning effects. Second, treatment with PPARγ ligands for multiple days appears to be required for this effect (Petrovic et al., 2009). Such slow kinetics is certainly not consistent with a direct action through PPAR response elements, which would be expected to occur within hours. Hence, it is essential to understand the structural and molecular bases of the white-to-brown fat conversion achieved through PPARγ ligands in closer detail.
In this study, we find that PPARγ ligands require full agonism to induce a brown (beige/brite) fat gene program in subcutaneous WAT and they do so through the activation of the PRDM16 pathway. Surprisingly, such effects are due in large measure to stabilization and accumulation of the PRDM16 protein, independent of PRDM16 mRNA levels. Thus, this may provide a potential insight into designing and developing synthetic compounds to increase energy expenditure by activating new brown adipocyte (beige/brite cell) development.
Results
Full agonism of PPARγ is required to activate a thermogenic brown fat gene program in subcutaneous white fat
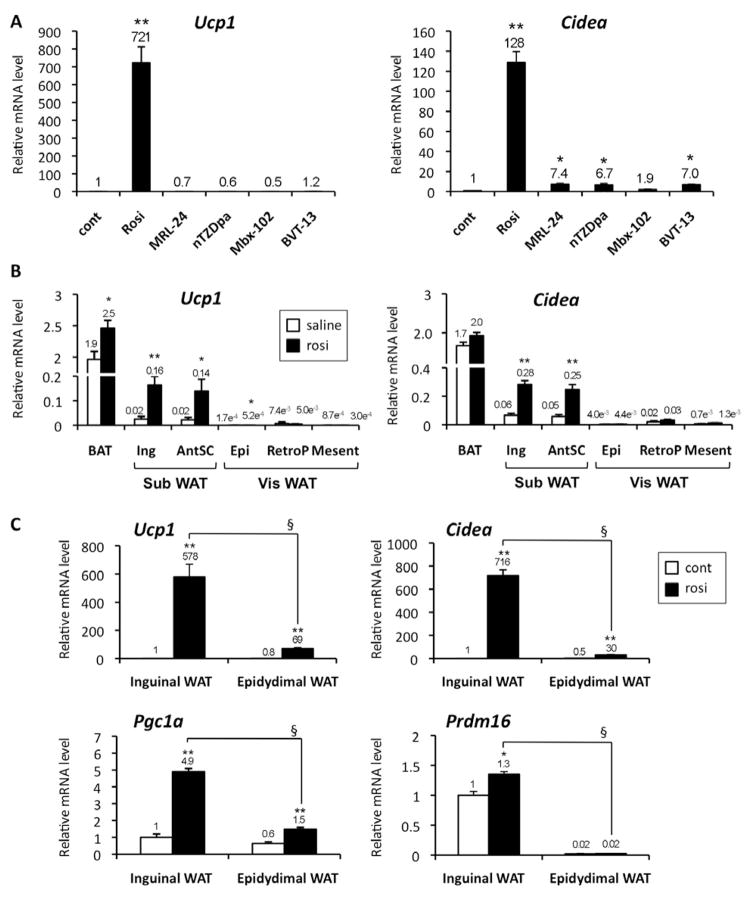
Anti-diabetic PPARγ ligand drugs, such as rosiglitazone, have been shown to have the ability to turn on a thermogenic gene program in brown fat and activate a “browning” of white adipose tissues (Fukui et al., 2000; Petrovic et al., 2009; Rong et al., 2007; Sell et al., 2004; Tai et al., 1996; Vernochet et al., 2009; Wilson-Fritch et al., 2004). Although these drugs are full agonists for PPARγ, they have recently been shown to have a second activity, in addition to classical agonism: blocking the phosphorylation of PPARγ by cdk 5 at serine 273 (Choi et al., 2010). We therefore asked if the actions of PPARγ ligands on brown fat-selective gene expression were correlated with classical receptor transcriptional agonism per se. To this end, we compared the “browning” effect by a well-known agonist thiazolidinedione (TZD) compound, rosiglitazone, with the effects of several anti-diabetic PPARγ ligands with weak or partial agonism. These include non-TZD compounds such as MRL24, nTZDpa, Mbx-102, and BVT.13, that have been shown to have relatively weak agonist properties (Berger et al., 2003; Gregoire et al., 2009; Ostberg et al., 2004). As shown in Fig. 1A, chronic treatment of primary adipocytes derived from inguinal WAT with 1μM rosiglitazone robustly induced mRNA expression for several brown fat-selective genes, including ucp1 (721-fold) and cidea (128-fold). This effect was not due to an enhancement of adipogenesis per se, since the induction was still significant when the mRNA levels for these genes were normalized to those of fabp4, an adipogenic marker gene (Supplementary Fig. 1A). In contrast, the synthetic PPARγ ligands with weak agonism, at doses well above their respective KDs, had very modest or no effects on the expression of these brown fat-selective genes. These effects were highly correlated with the transcriptional activity of PPARγ on PPREs, as assessed by a luciferase assay (Bruning et al., 2007; Choi et al., 2010). These results strongly suggest that the effects of these compounds on ucp1 and other brown-fat selective genes require strong classical agonism.
Fig. 1. Preferential browning effects in subcutaneous WAT requires full agonism of PPARγ.
(A) Primary inguinal white preadipocytes were differentiated in the presence of full or partial agonists for PPARγ, such as rosiglitazone (rosi), MRL24, nTZDpa, Mbx-102, and BVT.13. mRNA levels for brown fat-selective genes (ucp1 and cidea) were analyzed by qRT-PCR. * P<0.05, ** P<0.01 relative to control. (B) Wild type C57BL/6J mice were injected IP with saline or rosiglitazone at 10mg/kg for 10days. mRNA levels for ucp1and cidea were measured by qRT-PCR in interscapular BAT, inguinal WAT (Ing), anterior subcutaneous WAT (AntSC), epidydimal WAT (Epi), retroperitoneal WAT (retroP) and mesenteric WAT (Mesent). *P<0.05, **P<0.01 relative to saline control. (C) mRNA levels for ucp1, cidea, pgc1a and prdm16 were measured by qRT-PCR in primary adipocytes differentiated from SVF derived from inguinal WAT and from epidydimal WAT in the presence or absence of rosiglitazone. *P<0.05, **P<0.01 relative to control. § P<0.01 relative to epidydimal cells treated with rosiglitazone. Data are expressed as means ± SEM.
We next asked if rosiglitazone had a preferential effect on the brown-fat gene program in either the visceral or subcutaneous WAT depot in vivo. As shown in Fig. 1B and Supplementary Fig. 1B, treatment with rosiglitazone at 10 mg/kg for 10 days preferentially increased brown fat-selective genes such as ucp1, cidea, and cox8b in two different subcutaneous WAT depots, inguinal WAT and anterior subcutaneous (SC) WAT. On the other hand, this effect was modest or minimal in three distinct visceral WAT depots, including epidydimal, retroperitoneal, and mesenteric WAT. Importantly, the preferential browning effects by rosiglitazone on the subcutaneous WAT compared to visceral WAT appear to be cell autonomous, in that it occurred in cultured adipocytes. As shown in Fig. 1C, treatment with rosiglitazone at 1μM robustly increased the brown fat gene expression of ucp1, cidea, and pgc1a in the primary adipocytes differentiated from the stromal-vascular fraction (SVF) of the inguinal WAT. This induction was much smaller in the primary adipocytes derived from the SVF of the epidydimal WAT depot. Interestingly, mRNA for prdm16, a dominant regulator of classical brown fat development, was highly enriched in the inguinal adipocytes but was only modestly increased by the rosiglitazone treatment. There was no significant difference in the expression of rb, a known negative regulator of brown fat cell fate (Hansen et al., 2004) (Supplementary Fig. 1C).
PRDM16 is required for the development of rosiglitazone-inducible brown adipocyte
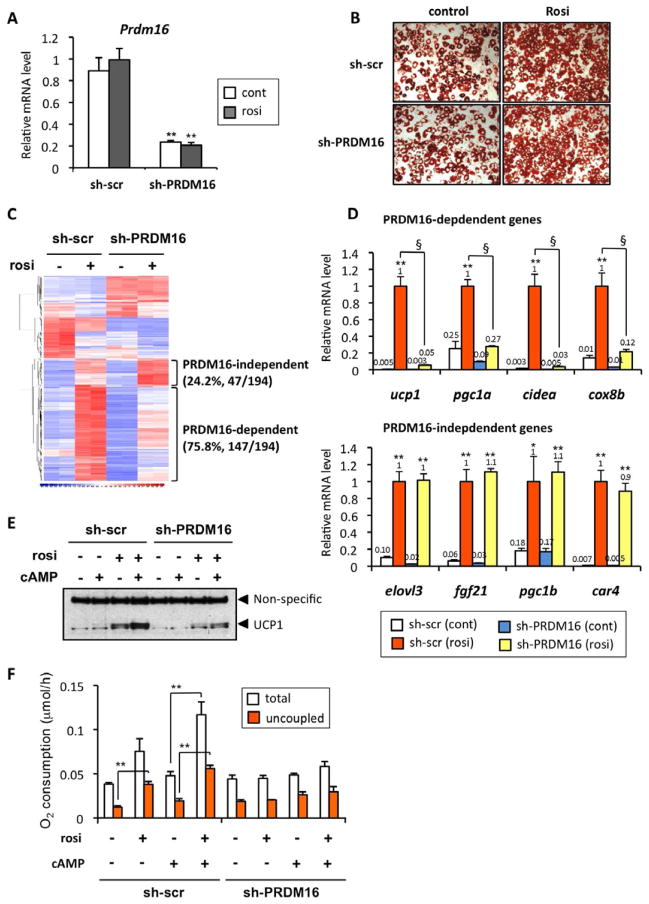
We next asked if PRDM16 was required for the rosiglitazone-induced activation of the white-to-brown fat (beige/brite) cell conversion. To do this, primary preadipocytes from inguinal WAT were infected with adenoviral vectors expressing short-hairpin RNA targeting PRDM16 (sh-PRDM16) or a scramble control (sh-scr), in the presence or absence of rosiglitazone (Fig. 2A). As shown in Fig. 2B, rosiglitazone treatment slightly enhanced adipogenesis; however, knockdown of PRDM16 did not affect adipogenesis per se, as shown by Oil-Red-O staining of accumulated lipid. Next we systematically analyzed how the rosiglitazone-induced gene expression in these cells was affected by depletion of PRDM16 using Affimetrix microarray analyses. As shown in Fig. 2C and D, we identified 194 genes in total that were significantly increased by rosiglitazone by 2 fold or greater (P<0.05). Importantly, expression of 147 of these genes (75.8%) was significantly blunted or completely abolished by the PRDM16 shRNA (PRDM16-dependent gene set, Supplementary Table 1). No significant change was observed in 47 genes (PRDM16-independent gene set, 24.2%, shown in Supplementary Table 2). The induction of key brown fat/thermogenic genes such as ucp1, cidea, pgc1a and cox8b were all significantly reduced by the PRDM16 knockdown. In contrast, expression of elovl3, fgf21, pgc1b and car4 was not affected by depletion of PRDM16 (Fig. 2D). Western blot analysis showed that UCP1 protein levels were severely reduced by loss of PRDM16 both in untreated and cAMP-stimulated adipocytes (Fig. 2E).
Fig. 2. PRDM16 is required for the browning of white adipocytes in response to rosiglitazone.
(A) mRNA levels for Prdm16 were measured by qRT-PCR in primary inguinal adipocytes infected with a control scramble shRNA (sh-scr) or shRNA targeting PRDM16 (sh-PRDM16). **P<0.01 relative to sh-scr. (B) Oil-red O staining of primary inguinal cells at day 7 of differentiation in the presence or absence of rosiglitazone. (C) Microarray analysis of the differentiated inguinal adipocytes expressing sh-scr or sh-PRDM16 in the presence or absence of rosiglitazone. (D) mRNA levels for the indicated genes were analyzed by qRT-PCR. * P<0.05, **P<0.01 relative to control cells. § P<0.01 relative to the cells expressing sh-PRDM16 treated with rosiglitazone. (E) UCP1 protein levels were analyzed by Western blotting. The cells were treated with or without forskolin (cAMP), at 10 μM for 6 hrs. (F) Total and uncoupled cellular respiration were measured in primary inguinal adipocytes expressing sh-scr or sh-PRDM16 in the presence or absence of rosiglitazone and/or. dibutyryl cAMP. ** P<0.01. Data are expressed as means ± SEM.
Next, to examine whether rosiglitazone alters the function of beige/brite cells to dissipate energy in a PRDM16-dependent manner, oxygen consumption in control and PRDM16-depleted cells were measured in the presence or absence of rosiglitazone. As shown in Fig. 2F, rosiglitazone significantly increased uncoupled respiration at the basal state. Furthermore, the rosiglitazone-treated cells had greater total and uncoupled respiration in response to cAMP stimulation. In contrast, no significant change was observed in the PRDM16-depleted cells in response to rosiglitazone and/or cAMP. These results clearly indicate that PRDM16 is required for the rosiglitazone-induced brown fat gene program and thermogenic function of subcutaneous WAT.
PRDM16 and rosiglitazone synergistically promote the development of inducible-brown adipocyte
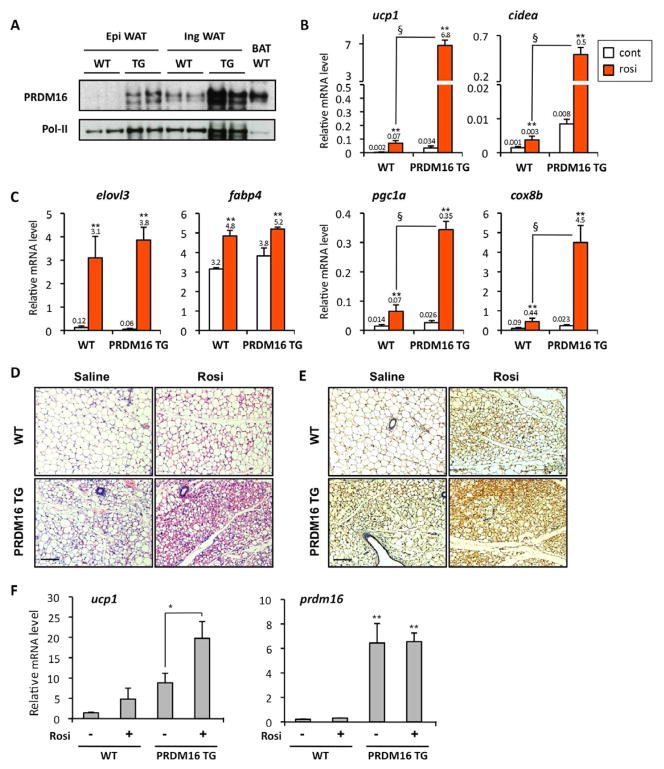
Since visceral fat expresses less PRDM16 mRNA than the subcutaneous fat, we next asked if enhanced PRDM16 expression would be sufficient to sensitize the visceral white adipocytes to turn on the brown fat gene program with rosiglitazone treatment. To test this idea, we used fabp4-PRDM16 transgenic mice in which transgenic expression of prdm16 was driven by the −5kb fabp4 (aP2) promoter/enhancer (Seale et al., 2007). Importantly, PRDM16 protein levels in the epididymal WAT of PRDM16 transgenic mice were nearly equivalent to the endogenous PRDM16 levels observed in the inguinal WAT of wild type mice (Fig. 3A). Primary visceral preadipocytes were isolated from epidydimal WAT of wild type and PRDM16 transgenic mice, and were differentiated into mature adipocytes in the presence or absence of rosiglitazone at 1μM. As shown in Fig. 3B, rosiglitazone-mediated induction of brown fat-selective genes including ucp1, cidea, pgc1a and cox8b were all robustly enhanced in visceral adipocytes derived from SVF cultures of the PRDM16 transgenic mice, compared to the controls. On the other hand, mRNA expression of elovl3 and an adipogenic marker fabp4 were not affected by transgenic expression of PRDM16 (Fig. 3C).
Fig. 3. PRDM16 and rosiglitazone synergistically promote the browning of white adipocytes.
(A) PRDM16 protein levels were analyzed by Western blotting in epidydimal WAT, inguinal WAT and interscapular BAT from wild type and PRDM16 transgenic mice. Pol-II was blotted as a loading control. (B) mRNA levels for ucp1, cidea, pgc1a, and cox8b were measured by qRT-PCR in primary epidydimal adipocytes from wild type and PRDM16 transgenic mice. **P<0.01 relative to control. § P<0.01 relative to wild type cells treated with rosiglitazone. (C) mRNA levels for elovl3 and fabp4 were measured by qRT-PCR. (D) Wild type mice and PRDM16 transgenic mice were injected IP with saline or rosiglitazone at 10mg/kg for 10days. The inguinal WAT depots were fixed and stained by H&E. Scale bar, 100 μm. (E) Immunohistochemistry for UCP1 expression from samples in (D). Scale bar, 100 μm. (F) mRNA levels for ucp1 and prdm16 were quantified by qRT-PCR from samples in (D). *P<0.05, **P<0.01. Data are expressed as means ± SEM.
To further test if PRDM16 synergistically enhanced the rosiglitazone-induced browning action in vivo, wild type and PRDM16 transgenic mice were treated with saline or rosiglitazone at 10mg/kg for 10 days. As shown in Fig. 3D, multilocular adipocytes, a morphological characteristic of brown adipocytes, were sporadically observed in the wild type mice treated with rosiglitazone or in the PRDM16 transgenic mice treated with saline. When PRDM16 transgenic mice were treated with rosiglitazone, the emergence of multilocular adipocytes was dramatically induced (Fig. 3D, bottom right panel). These multilocular cells were indeed UCP1-positive adipocytes, as shown by immunohistochemistry (Fig. 3E). Gene expression analyses also illustrated that transgenic expression of PRDM16, together with rosiglitazone, synergistically induced ucp1 gene expression in vivo (Fig. 3F). These data clearly indicate that PRDM16 synergistically activates the inducible-brown fat (beige/brite) gene program together with the PPARγ agonist.
Rosiglitazone stimulates a powerful stabilization of the PRDM16 protein
A key issue is the molecular mechanism by which the PPARγ agonist activates a brown fat phenotype in a PRDM16-dependent manner. We have previously shown that PRDM16 directly interacted with PPARγ and coactivated the transcriptional activity of PPARγ (Seale et al., 2008). However, this PRDM16-PPARγ interaction was not enhanced by rosiglitazone (Supplementary Fig. 2A), suggesting that alternative mechanisms were operative.
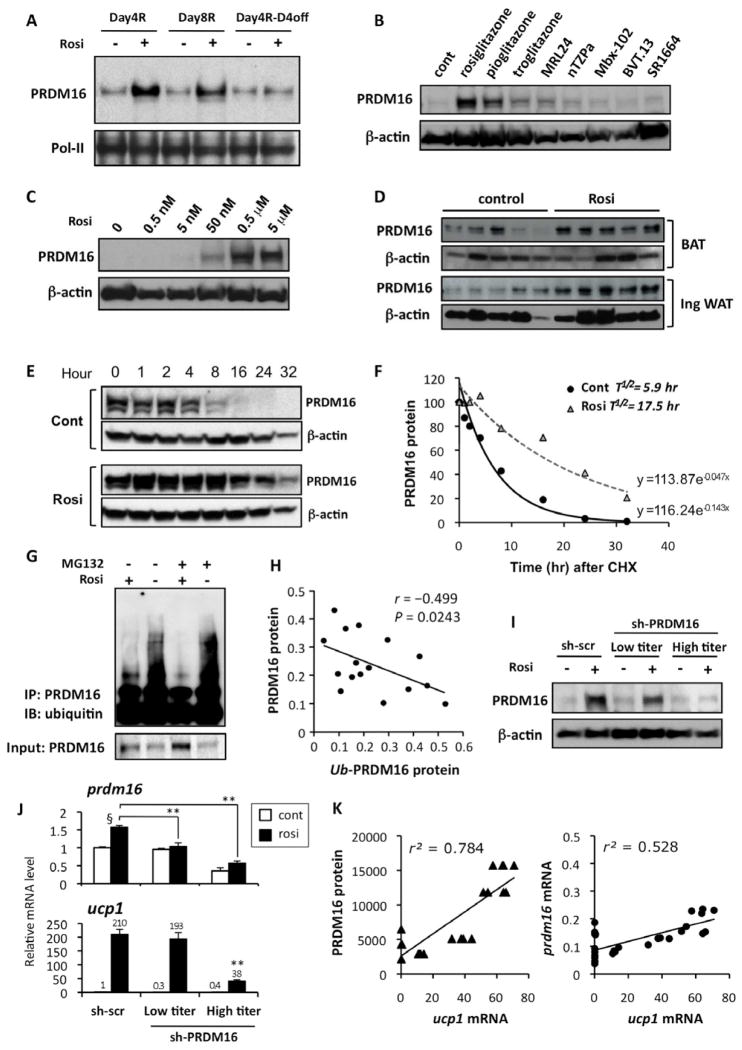
We examined PRDM16 protein levels during the rosiglitazone-induced white-to-brown fat conversion in inguinal adipocytes. As shown in Supplementary Fig. 2B, no significant increase was found in endogenous PRDM16 mRNA expression in cells with a 4 day treatment. Minor but statistically significant increases were observed in PRDM16 mRNA with a 8 day treatment, and with a 4 day treatment followed by a 4 day removal. However, a very striking increase in PRDM16 protein level was observed with rosiglitazone treatment at days 4 and 8; this correlated very well with the induction of the brown/beige fat genes such as ucp1 and pgc1α (Fig. 4A, and Supplementary Fig. 2C). Similarly, a time course experiment showed that induction of this beige/brite fat gene program required at least a 3 day treatment with rosiglitazone. This correlated well with the PRDM16 protein accumulation but not with the induction of a well-known PPARγ target gene, fabp4 (Supplementary Fig. 2D and E).
Fig. 4. Browning of the white adipocytes is tightly linked to the rosiglitazone-induced PRDM16 protein stabilization.
(A) Primary inguinal cells from SVF were differentiated in the presence or absence of rosiglitazone at 1μM for 4 days (Day4R), for 8 days (Day8R), and for 4 days with rosiglitazone and additional 4 days without rosiglitazone (Day4R-D4off). PRDM16 protein levels were analyzed by Western blotting. Pol-II was blotted as a loading control. (B) Primary inguinal WAT-derived adipocytes were treated with full or partial agonists for PPARγ. PRDM16 protein levels were analyzed by Western blotting. β-actin was blotted as a loading control. (C) PRDM16 protein levels were analyzed by Western blotting in primary adipocytes treated with rosiglitazone at different doses. (D) PRDM16 protein levels were analyzed by Western blotting in BAT and inguinal WAT isolated from wild-type mice. Mice were injected IP with saline or rosiglitazone at 10mg/kg for 10 days. (E) PRDM16 protein levels in a cycloheximide chase experiment were analyzed by Western blotting. Top panel: control cells, Bottom panel: rosiglitazone treated cells (F) Regression analysis of PRDM16 protein stability in (E). (G) F442A cells expressing flag-tagged PRDM16 were differentiated in the presence or absence of rosiglitazone. Cells were treated with a proteasome inhibitor MG132 (10 μM) for 16 hours prior to harvesting the cells. PRDM16 was immunoprecipitated using flag antibody, and ubiquitin was detected by Western blotting. Total PRDM16 protein is shown in the bottom panel. (H) Correlation analysis between PRDM16 protein levels and ubiquitinated PRDM16 levels. (I) PRDM16 protein levels were analyzed by Western blotting in primary inguinal adipocytes infected with control scramble shRNA or shRNA targeting PRDM16 at low- or high-tires adenovirus. (J) mRNA levels for prdm16 and ucp1 were measured by qRT-PCR in (I). **P<0.01 relative to sh-scr control cells treated with rosiglitazone. § P<0.01 relative to cells without rosiglitazone treatment. (K) Correlation analyses between ucp1 mRNA levels and PRDM16 protein levels (left panel), and between ucp1 mRNA levels and prdm16 mRNA levels (right panel). Data are expressed as means ± SEM.
To further investigate how PRDM16 protein level is regulated by PPARγ ligands, we analyzed PRDM16 protein levels in the primary white adipocytes treated with a variety of full and partial PPARγ ligands including SR1664, which has previously been shown to block the Cdk5-mediated phosphorylation of PPARγ at S273 with no agonism (Choi et al., 2011). As shown in Fig. 4B, PRDM16 protein levels were robustly induced by two strong PPARγ agonists, rosiglitazone and pioglitazone. On the other hand, other weaker agonists had modest or no effects on the PRDM16 protein. No induction was observed by the treatment with SR1664. The induction of the PRDM16 protein was highly correlated with the browning effects, as judged by the induction of ucp1 mRNA expression (Supplementary Fig. 3A). In addition, a dose response experiment showed that rosiglitazone at doses of 50 nM and higher significantly increased PRDM16 protein levels (Fig. 4C), in consistent with the previous report that rosiglitazone binds to the ligand-binding domain of PPARγ with a KD of 43nM (Lehmann et al., 1995). These effects were also correlated well with the induction of ucp1 mRNA levels (Supplementary Fig. 3B). Furthermore, the rosiglitazone-mediated induction of the PRDM16 protein was largely blocked by PPARγ antagonists (Supplementary Fig. 3C). Importantly, PRDM16 protein was highly increased by rosiglitazone in vivo, both in BAT and inguinal WAT from wild-type mice (Fig. 4D) and from PRDM16 transgenic mice (Supplementary Fig. 3D), while there was modest or no effect on PRDM16 mRNA levels (Supplementary Fig. 3E).
We next asked if this increased protein accumulation for PRDM16 was due to changes in the rate of protein degradation or altered mRNA translation. To this end, the stability of PRDM16 protein was investigated with a cycloheximide chase experiment. The data in Figure 4E and F clearly showed that rosiglitazone treatment of cells treated with cycloheximide dramatically extended PRDM16 half-life, from 5.9 hr to 17.5 hr.
To further investigate the extent to which PRDM16 protein stability is regulated by PPARγ ligands, we examined whether PRDM16 was ubiquitinated in response to rosiglitazone. PRDM16 ubiquitination was detected by immunoprecipitation of PRDM16 followed by immunoblotting with an antibody against ubiquitin. As shown in Fig. 4G, ubiquitination of PRDM16 was strikingly reduced by rosiglitazone, in the presence or absence of a proteasome inhibitor MG132. Importantly, there was a statically significant inverse correlation between the changes in the PRDM16 protein levels and the changes in the ubiquitinated PRDM16 protein levels in response to rosiglitazone (r=−0.499, P=0.0243, t-test, Fig. 4H). Furthermore, addition of MG132 largely blocked the protein degradation of PRDM16 after the removal of rosiglitazone (Supplementary Fig. 3F). These results strongly indicate that prolongation of the half-life of PRDM16 protein by PPARγ ligands is regulated, in part, through the ubiquitin-proteasome pathway.
Finally, we investigated whether the browning effect of rosiglitazone could be more closely associated with these effects on levels of the PRDM16 mRNA or protein. As shown in Fig. 4I and J, PRDM16 mRNA levels were modestly but significantly reduced by low titers of the adenoviral vectors expressing the PRDM16 shRNA but its protein level reached a level almost equivalent to that in control cells treated with rosiglitazone. On the other hand, high-titers of the adenovirus of PRDM16 shRNA reduced PRDM16 mRNA and also completely abolished the PRDM16 protein accumulation. Induction of brown fat gene program by rosiglitazone, as judged by ucp1 mRNA expressions, correlated much more tightly with the PRDM16 protein levels than with PRDM16 mRNA levels. Indeed, there was a powerful positive correlation (r2=0.784, P<0.0001) between the changes of ucp1 mRNA levels and the changes of PRDM16 protein levels caused by rosiglitazone treatment (Fig. 4K, left). Despite a significant correlation between the changes of ucp1 mRNA levels and the changes of prdm16 mRNA levels (r2=0.528, P<0.001), the correlation between ucp1 mRNA levels and PRDM16 protein levels was significantly tighter than that of ucp1 mRNA levels and prdm16 mRNA levels (Fig. 4K, right). Population correlation coefficient of these two was significantly different (P<0.001, Z-test). Taken together, these results strongly suggest that rosiglitazone-induced white-to-brown fat conversion is regulated in large measure through an enhanced stability of PRDM16 protein.
Discussion
PRDM16 is a 140kDa zinc-finger protein that was originally identified at a chromosomal breakpoint of t(1;3)(p36;q21)-positive human acute myeloid leukemia cells (Mochizuki et al., 2000). We have previously shown that PRDM16 acts as a molecular switch to induce brown fat development in a subset of Myf5-positive, myoblast-like precursor cells during embryological development. It does this through interactions with several transcription factors such as C/EBPβ, PPARγ, PGC-1α β and C-terminal binding proteins (Kajimura et al., 2009; Kajimura et al., 2008; Seale et al., 2008; Seale et al., 2007). The present studies show that PRDM16 plays a pivotal role in the development of beige/brite cells in subcutaneous WAT induced by PPARγ agonists (Supplementary Fig. 4). A key and unexpected mechanism here is that this drug causes a greatly enhanced accumulation of PRDM16 due to prolongation of the half-life of this protein.
The browning effect on adipose tissues by PPARγ agonists was first suggested to be mediated by the direct activation of the thermogenic genes via PPREs in their promoter/enhancer (Sears et al., 1996; Tai et al., 1996; Viswakarma et al., 2007). While this model still may have some validity, the slow induction of the expression of ucp1 and other thermogenic genes by rosiglitazone (at least 3 days or longer) suggests a less direct mechanism. Most direct gene regulatory effects by nuclear receptors and their agonists occur within several hours after ligand treatment (e.g., induction of fabp4 mRNA expression by rosiglitazone in Supplementary Fig. 2E). On the other hand, the stabilization of the PRDM16 protein by rosiglitazone shown here can explain several aspects of this phenomenon quite well. First, substantially greater expression of PRDM16 mRNA in the subcutaneous fat, compared to the visceral depots, allows a protein stabilization mechanism to produce biologically active amounts of this factor only in the subcutaneous fat. Second, rosiglitazone treatment robustly extends PRDM16 protein half-life from 5.9 hr to 17.5 hr. Accumulation of the PRDM16 protein to biologically active levels, as shown in Fig. 4E&F, would thus be expected to take several half-lives, consistent with the slow pace of cellular browning by rosiglitazone. Furthermore, an inverse correlation between the PRDM16 protein levels and the ubiquitinated PRDM16 levels clearly indicates that PPARγ ligand-mediated RDM16 protein stabilization is regulated, at least in part, through the ubiquitin-proteasome pathway. Future studies will aim at identifying factors that directly control PRDM16 ubiquitination and its protein stability.
The structural basis of PPARγ activation by agonists has been extensively studied. Full PPARγ agonists, such as rosiglitazone, bind to and stabilize helix12 of its ligand binding domain (LBD). This allows interaction with known coactivators such as SRC1 (Nolte et al., 1998). On the other hand, partial agonists such as MRL24 and nTZDpa preferentially affect helix3 and the β-sheet region, with minimum influence on helix12 (Bruning et al., 2007). The results shown here, indicating that these partial agonists had little or no browning effects, implies that structural changes in helix3 and the β-sheet region of the LBD may not be sufficient to induce a white-to-brown fat conversion. Of note, Farmer and colleagues have recently shown that mutations within helix 7 of the PPARγ LBD (E365 and F372) blunted the troglitazone-mediated browning of Swiss 3T3-adipocytes (Vernochet et al., 2009). These mutations also caused impaired regulation of a subset of PPARγ target genes (Vernochet et al., 2010; Vernochet et al., 2009; Wang et al., 2008). In addition, a dominant-negative mutation (P465L) near the activating function-2 (AF-2) domain of helix12, originally discovered in human lipodystrophy patients with extreme insulin resistance, causes an impairment in cAMP-induced recruitment of brown adipocytes in WAT in vivo (Gray et al., 2006). The emerging question is how structural changes in the helix7 or the helix 12 of PPARγ affect browning activity and whether these changes also affect PRDM16 protein stability. If it is possible to develop PPARγ ligands that affect PRDM16 protein accumulation without full agonist activity, such compounds could have plausible therapeutic activity toward obesity and diabetes.
Materials and Methods
Cell Culture
Adipocyte differentiation was induced by treating confluent cells in DMEM/F12 medium (D-glucose 17.51 mM) containing 10% FBS, 0.5 mM isobutylmethylxanthine, 125 nM indomethacin, 1 mM dexamethasone, 850 nM insulin, 1 nM T3. Two days after induction, cells were switched to the maintenance medium containing 10% FBS, 850 nM insulin and 1 nM T3.
Animals
Animal experiments were performed according to procedures approved by Institutional Animal Care and Use Committee at Beth Israel Deaconess Medical Center and at UCSF. Male C57BL/6J mice between 6–8 week-old were intraperitoneally injected daily with 10 mg/kg rosiglitazone for 10 days. For histological analyses, paraffin-embedded sections were incubated with anti-UCP1 antibody (Chemicon), as described previously (Kajimura et al., 2009).
Gene expression analysis
Quantitative real-time PCR (qRT-PCR) was performed with SYBR green fluorescent dye using an ABI9300 PCR machine or ABI ViiA™7. TATA-binding protein (TBP) served as an internal control. Primer sequences are provided in Supplementary Table 3. For a microarray analysis, Affymetrix GeneChip Mouse Genome 430 2.0 array was used according to established methods (Lockhart et al., 1996). The array data were analyzed using the DNA-Chip Analyzer (dChip) software (Li and Wong, 2001). The statistical significance of differences in gene expression was assessed by unpaired t-test (P< 0.05). Microarray data has been deposited in Gene Expression Omnibus (GEO): GSE35011.
Protein Stability Assay and Western Blotting
Primary cells were incubated in medium containing 20 μg/ml cycloheximide in the presence or absence of rosiglitazone. Total cell lysates or nuclear extracts were isolated and separated by SDS-page. Antibodies for PRDM16 (Seale et al., 2011), ubiquitin (Santa Cruz), UCP1 (Abcam), β-actin, M2 flag (Sigma) or Pol-II (Cell Signaling) were used for Western blotting.
Oxygen Consumption Assay
Cellular oxygen consumption was measured as described previously (Kajimura et al., 2009). For cAMP-induced respiration assays, fully differentiated adipocytes were incubated with 0.5 mM dibutyryl cyclic AMP for 12 hours prior to measuring oxygen consumption.
Statistical Analyses
Significant differences between two groups were assessed by two-tailed Student’s t test with unequal variance. Data are expressed as means ± SEM.
Supplementary Material
Acknowledgments
We thank Dr. Patrick Griffin at the Scripps Institute for providing partial PPARγ agonists. We are grateful to Dr. Jang-Hyun Choi, Ms. Emi Tomoda, and Ms. Lauren Ruiz for discussion, technical help and editorial assistance on the manuscript. This work was supported by grants from the NIH to BMS (DK31405 and DK090861) and to S.K (DK087853). We also acknowledge the DERC center grant (NIH P30 DK063720).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Berger JP, Petro AE, Macnaul KL, Kelly LJ, Zhang BB, Richards K, Elbrecht A, Johnson BA, Zhou G, Doebber TW, Biswas C, Parikh M, Sharma N, Tanen MR, Thompson GM, Ventre J, Adams AD, Mosley R, Surwit RS, Moller DE. Distinct properties and advantages of a novel peroxisome proliferator-activated protein [gamma] selective modulator. Molecular endocrinology (Baltimore, Md) 2003;17:662–676. doi: 10.1210/me.2002-0217. [DOI] [PubMed] [Google Scholar]
- Bruning JB, Chalmers MJ, Prasad S, Busby SA, Kamenecka TM, He Y, Nettles KW, Griffin PR. Partial agonists activate PPARgamma using a helix 12 independent mechanism. Structure. 2007;15:1258–1271. doi: 10.1016/j.str.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Cederberg A, Gronning LM, Ahren B, Tasken K, Carlsson P, Enerback S. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell. 2001;106:563–573. doi: 10.1016/s0092-8674(01)00474-3. [DOI] [PubMed] [Google Scholar]
- Choi JH, Banks AS, Estall JL, Kajimura S, Bostrom P, Laznik D, Ruas JL, Chalmers MJ, Kamenecka TM, Bluher M, Griffin PR, Spiegelman BM. Anti-diabetic drugs inhibit obesity-linked phosphorylation of PPARgamma by Cdk5. Nature. 2010;466:451–456. doi: 10.1038/nature09291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Banks AS, Kamenecka TM, Busby SA, Chalmers MJ, Kumar N, Kuruvilla DS, Shin Y, He Y, Bruning JB, Marciano DP, Cameron MD, Laznik D, Jurczak MJ, Schurer SC, Vidovic D, Shulman GI, Spiegelman BM, Griffin PR. Antidiabetic actions of a non-agonist PPARgamma ligand blocking Cdk5-mediated phosphorylation. Nature. 2011 doi: 10.1038/nature10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, Kolodny GM, Kahn CR. Identification and importance of brown adipose tissue in adult humans. The New England journal of medicine. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enerback S. Human brown adipose tissue. Cell metabolism. 2010;11:248–252. doi: 10.1016/j.cmet.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Farmer SR. Transcriptional control of adipocyte formation. Cell metabolism. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frontini A, Cinti S. Distribution and development of brown adipocytes in the murine and human adipose organ. Cell metabolism. 2010;11:253–256. doi: 10.1016/j.cmet.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Fukui Y, Masui S, Osada S, Umesono K, Motojima K. A new thiazolidinedione, NC-2100, which is a weak PPAR-gamma activator, exhibits potent antidiabetic effects and induces uncoupling protein 1 in white adipose tissue of KKAy obese mice. Diabetes. 2000;49:759–767. doi: 10.2337/diabetes.49.5.759. [DOI] [PubMed] [Google Scholar]
- Gray SL, Dalla Nora E, Backlund EC, Manieri M, Virtue S, Noland RC, O’Rahilly S, Cortright RN, Cinti S, Cannon B, Vidal-Puig A. Decreased brown adipocyte recruitment and thermogenic capacity in mice with impaired peroxisome proliferator-activated receptor (P465L PPARgamma) function. Endocrinology. 2006;147:5708–5714. doi: 10.1210/en.2006-0684. [DOI] [PubMed] [Google Scholar]
- Gregoire FM, Zhang F, Clarke HJ, Gustafson TA, Sears DD, Favelyukis S, Lenhard J, Rentzeperis D, Clemens LE, Mu Y, Lavan BE. MBX-102/JNJ39659100, a novel peroxisome proliferator-activated receptor-ligand with weak transactivation activity retains antidiabetic properties in the absence of weight gain and edema. Molecular endocrinology (Baltimore, Md) 2009;23:975–988. doi: 10.1210/me.2008-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JB, Jorgensen C, Petersen RK, Hallenborg P, De Matteis R, Boye HA, Petrovic N, Enerback S, Nedergaard J, Cinti S, te Riele H, Kristiansen K. Retinoblastoma protein functions as a molecular switch determining white versus brown adipocyte differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4112–4117. doi: 10.1073/pnas.0301964101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Barak Y, Hevener A, Olson P, Liao D, Le J, Nelson M, Ong E, Olefsky JM, Evans RM. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Takakuwa R, Marchand S, Dentz E, Bornert JM, Messaddeq N, Wendling O, Mark M, Desvergne B, Wahli W, Chambon P, Metzger D. Peroxisome proliferator-activated receptor gamma is required in mature white and brown adipocytes for their survival in the mouse. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4543–4547. doi: 10.1073/pnas.0400356101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, Spiegelman BM. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Spiegelman BM. Transcriptional Control of Brown Fat Development. Cell metabolism. 2010 doi: 10.1016/j.cmet.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Tomaru T, Erdjument-Bromage H, Cooper MP, Ruas JL, Chin S, Tempst P, Lazar MA, Spiegelman BM. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes & development. 2008;22:1397–1409. doi: 10.1101/gad.1666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann JM, Moore LB, Smith-Oliver TA, Wilkison WO, Willson TM, Kliewer SA. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma) The Journal of biological chemistry. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- Leonardsson G, Steel JH, Christian M, Pocock V, Milligan S, Bell J, So PW, Medina-Gomez G, Vidal-Puig A, White R, Parker MG. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8437–8442. doi: 10.1073/pnas.0401013101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, Brown EL. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nature biotechnology. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Shimizu S, Nagasawa T, Tanaka H, Taniwaki M, Yokota J, Morishita K. A novel gene, MEL1, mapped to 1p36.3 is highly homologous to the MDS1/EVI1 gene and is transcriptionally activated in t(1;3)(p36;q21)-positive leukemia cells. Blood. 2000;96:3209–3214. [PubMed] [Google Scholar]
- Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. American journal of physiology. 2007;293:E444–452. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, Rosenfeld MG, Willson TM, Glass CK, Milburn MV. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- Ostberg T, Svensson S, Selen G, Uppenberg J, Thor M, Sundbom M, Sydow-Backman M, Gustavsson AL, Jendeberg L. A new class of peroxisome proliferator-activated receptor agonists with a novel binding epitope shows antidiabetic effects. The Journal of biological chemistry. 2004;279:41124–41130. doi: 10.1074/jbc.M401552200. [DOI] [PubMed] [Google Scholar]
- Petrovic N, Walden TB, Shabalina IG, Timmons JA, Cannon B, Nedergaard J. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. The Journal of biological chemistry. 2009;285:7153–7164. doi: 10.1074/jbc.M109.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong JX, Qiu Y, Hansen MK, Zhu L, Zhang V, Xie M, Okamoto Y, Mattie MD, Higashiyama H, Asano S, Strum JC, Ryan TE. Adipose mitochondrial biogenesis is suppressed in db/db and high-fat diet-fed mice and improved by rosiglitazone. Diabetes. 2007;56:1751–1760. doi: 10.2337/db06-1135. [DOI] [PubMed] [Google Scholar]
- Saito M, Okamatsu-Ogura Y, Matsushita M, Watanabe K, Yoneshiro T, Nio-Kobayashi J, Iwanaga T, Miyagawa M, Kameya T, Nakada K, Kawai Y, Tsujisaki M. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes. 2009;58:1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TJ, Huang TL, Tran TT, Zhang H, Townsend KL, Shadrach JL, Cerletti M, McDougall LE, Giorgadze N, Tchkonia T, Schrier D, Falb D, Kirkland JL, Wagers AJ, Tseng YH. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:143–148. doi: 10.1073/pnas.1010929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, Tempst P, Rudnicki MA, Beier DR, Spiegelman BM. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, Cohen P, Cinti S, Spiegelman BM. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. The Journal of clinical investigation. 2011;121:96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. Transcriptional control of brown fat determination by PRDM16. Cell metabolism. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears IB, MacGinnitie MA, Kovacs LG, Graves RA. Differentiation-dependent expression of the brown adipocyte uncoupling protein gene: regulation by peroxisome proliferator-activated receptor gamma. Molecular and cellular biology. 1996;16:3410–3419. doi: 10.1128/mcb.16.7.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sell H, Berger JP, Samson P, Castriota G, Lalonde J, Deshaies Y, Richard D. Peroxisome proliferator-activated receptor gamma agonism increases the capacity for sympathetically mediated thermogenesis in lean and ob/ob mice. Endocrinology. 2004;145:3925–3934. doi: 10.1210/en.2004-0321. [DOI] [PubMed] [Google Scholar]
- Sugii S, Olson P, Sears DD, Saberi M, Atkins AR, Barish GD, Hong SH, Castro GL, Yin YQ, Nelson MC, Hsiao G, Greaves DR, Downes M, Yu RT, Olefsky JM, Evans RM. PPARgamma activation in adipocytes is sufficient for systemic insulin sensitization. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:22504–22509. doi: 10.1073/pnas.0912487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai TA, Jennermann C, Brown KK, Oliver BB, MacGinnitie MA, Wilkison WO, Brown HR, Lehmann JM, Kliewer SA, Morris DC, Graves RA. Activation of the nuclear receptor peroxisome proliferator-activated receptor gamma promotes brown adipocyte differentiation. The Journal of biological chemistry. 1996;271:29909–29914. doi: 10.1074/jbc.271.47.29909. [DOI] [PubMed] [Google Scholar]
- Timmons JA, Wennmalm K, Larsson O, Walden TB, Lassmann T, Petrovic N, Hamilton DL, Gimeno RE, Wahlestedt C, Baar K, Nedergaard J, Cannon B. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4401–4406. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJ. Cold-activated brown adipose tissue in healthy men. The New England journal of medicine. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- Vernochet C, Davis KE, Scherer PE, Farmer SR. Mechanisms regulating repression of haptoglobin production by peroxisome proliferator-activated receptor-gamma ligands in adipocytes. Endocrinology. 2010;151:586–594. doi: 10.1210/en.2009-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernochet C, Peres SB, Davis KE, McDonald ME, Qiang L, Wang H, Scherer PE, Farmer SR. C/EBPalpha and the corepressors CtBP1 and CtBP2 regulate repression of select visceral white adipose genes during induction of the brown phenotype in white adipocytes by peroxisome proliferator-activated receptor gamma agonists. Molecular and cellular biology. 2009;29:4714–4728. doi: 10.1128/MCB.01899-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto NJ, Enerback S, Nuutila P. Functional brown adipose tissue in healthy adults. The New England journal of medicine. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- Viswakarma N, Yu S, Naik S, Kashireddy P, Matsumoto K, Sarkar J, Surapureddi S, Jia Y, Rao MS, Reddy JK. Transcriptional regulation of Cidea, mitochondrial cell death-inducing DNA fragmentation factor alpha-like effector A, in mouse liver by peroxisome proliferator-activated receptor alpha and gamma. The Journal of biological chemistry. 2007;282:18613–18624. doi: 10.1074/jbc.M701983200. [DOI] [PubMed] [Google Scholar]
- Wang H, Qiang L, Farmer SR. Identification of a domain within peroxisome proliferator-activated receptor gamma regulating expression of a group of genes containing fibroblast growth factor 21 that are selectively repressed by SIRT1 in adipocytes. Molecular and cellular biology. 2008;28:188–200. doi: 10.1128/MCB.00992-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Fritch L, Nicoloro S, Chouinard M, Lazar MA, Chui PC, Leszyk J, Straubhaar J, Czech MP, Corvera S. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. The Journal of clinical investigation. 2004;114:1281–1289. doi: 10.1172/JCI21752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneshiro T, Aita S, Matsushita M, Kameya T, Nakada K, Kawai Y, Saito M. Brown adipose tissue, whole-body energy expenditure, and thermogenesis in healthy adult men. Obesity (Silver Spring, Md) 2011;19:13–16. doi: 10.1038/oby.2010.105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.