Abstract
Meroterpenoids are mixed polyketide-terpenoid natural products with a broad range of biological activities. Herein, we present the structures of four new meroterpenoid antibiotics, merochlorins A–D, produced by the marine bacterium Streptomyces sp. strain CNH-189, which possess novel chemical skeletons unrelated to known bacterial agents. Draft genome sequencing, mutagenesis and heterologous biosynthesis in the genome-minimized model actinomycete Streptomyces coelicolor provided a 57.6 kb gene cluster that contains two genes encoding rare bacterial vanadium-dependent haloperoxidase (VHPO) genes. Pathway expression of two different fosmid clones that differ largely by the presence or absence of the VHPO gene mcl40, resulted in the differential biosynthesis of merochlorin C, suggesting that Mcl40 catalyzes an unprecedented 15-membered chloronium-induced macrocyclization reaction converting merochlorin D to merochlorin C.
The molecular logic of biosynthetic halogenation has illuminated four oxidative enzyme families that deliver halogen ions to a diversity of organic scaffolds.1–3 Vanadium-dependent haloperoxidases (VHPOs) are rare bacterial halogenating enzymes that were first characterized from fungi and macroalgae where they catalyze the oxidative halogenation of electron-rich organic substrates.4,5 We recently showed that the napyradiomycin antibiotics6–8 are distinctive streptomycete meroterpenoids, whose linear isoprenoid side chains undergo intramolecular VHPO-mediated cyclization reactions.9,10 Herein, we report the discovery of a new natural product group of chlorinated meroterpenoid antibiotics, merochlorins A–D (1–4, Figure 1), that we demonstrate through in vivo biosynthetic experiments are unique examples of VHPO-derived products.
Figure 1.
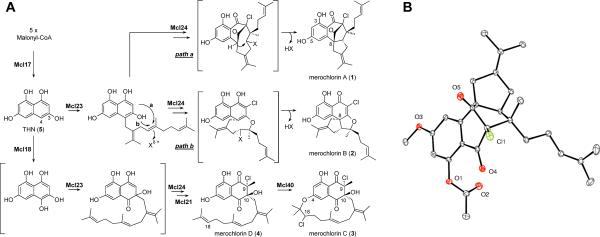
(A) Proposed biosynthesis of merochlorins A–D (1–4). X = oxygen or halogen. (B) Oak Ridge Thermal Ellipsoid Plot (ORTEP) representation of the X-ray structure of 3-acetyl-5-methyl-merochlorin A. The absolute configuration of 1 was assigned based on the diffraction anisotropy of the chlorine atom of the derivatized metabolite.
During our screening campaign to find new anti-cancer and anti-infective agents from marine microorganisms, we isolated Streptomyces sp. strain CNH-189 from a marine sediment sample collected near Oceanside, California.11 We previously reported the discovery of the ansamycin polyketide ansalactam A from this strain,11 and more recently the preliminary characterization of the meroterpenoid antibiotic merochlorin A (1),12 which consists of a highly unusual bicyclo[2.2.1]-heptanone ring system and a propan-2-ylidenecyclopentane moiety. To unambiguously establish its absolute configuration, we solved the crystal structure of a methylated and acetylated derivative of 1 (Figure 1B).
Bioassay-guided fractionation of the crude organic extract of strain CNH-189 facilitated the isolation of two new merochlorin analogues, 2 and 3 (Figure 2). Notably, 1 and 2 are highly active against clinically relevant methicillin-resistant Staphylococcus aureus strains with MICs in the range of 2–4 μg/mL, whereas 3 showed only weak antibiotic properties (Table S4). High-resolution MS (HR-MS) and NMR spectral analysis allowed the complete structural assignment of merochlorins B (2) and C (3), which possess distinctive carbon skeletons unrelated to known natural products [Figure 1; Supporting Information (SI)]. Although 1 and 2 share the same molecular formula, C25H29ClO4, 2 has a unique 6-5-5-fused ring system, composed of dihydroxynaphthalenone and propan-2-ylidenecyclopentane moieties. We established the planar structure of 2 and the fusion of the unusually configured terpenoid residue with the naphthyl unit by extensive two-dimensional NMR spectroscopy. The relative configuration was further determined by interpretation of ROESY correlations as 8S*, 9R*, and 10S*.
Figure 2.
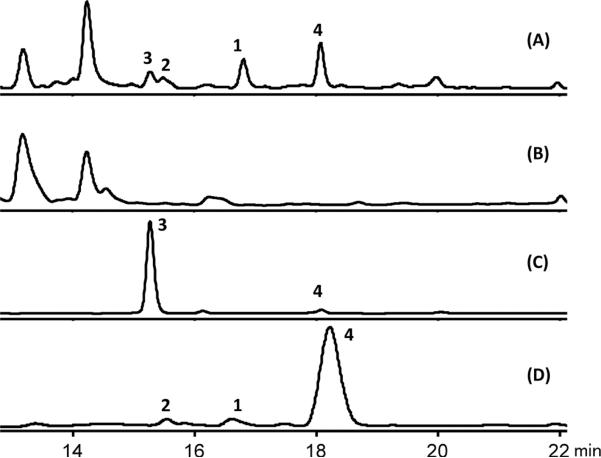
Metabolite profiles of (A) CNH-189 cultured in A1 medium, (B) CNH-189Δmcl17 in A1 medium, (C) CNH-189 in M1 medium, (D) S. coelicolor M1154/merLK01 in R5 medium. The production level of merochlorins was highly variable in seawater-based A1 medium. Culture conditions were optimized, and M1 medium (C) was shown to reproducibly yield consistent quantities (10 mg/L) of merochlorin C (3). The traces represent reversed-phase HPLC chromatograms acquired by detection at 254 nm.
The structure of 3, on the other hand, was clearly distinct from 1 and 2 based on its molecular formula of C26H32Cl2O5, containing additional carbon and chlorine atoms. HMBC correlation data not only revealed a different attachment of the terpenoid residue to the naphthyl unit at C-10, but also exposed a second attachment site through an ether linkage between C-19 and C-4 via four-bond correlation from the singlet methyl proton H-21 to C-14. The macrocyclized sesquiterpenoid residue is thus attached on both ends of the tetrahydronaphthoquinone core molecule (Figure 1). Although we established the relative configuration of C-9 and C-10 as 9R* and 10S*, we have not yet assigned the configuration of the isolated C-18 chloro group.
The unique structures of the merochlorins suggested that they are constructed via unusual biotransformations, possibly involving VHPO-dependent catalysis of a chloroetherification reaction similar to napyradiomycin formation.10 To gain access to the biosynthetic gene cluster, we subjected CNH-189 genomic DNA to short-read, paired-end Illumina sequencing. Targeted assembly of portions of the genome with homology to napyradiomycin biosynthesis (nap) genes yielded three contigs with significant sequence similarity to known meroterpenoid gene clusters (Figure S17).9,13–15 A 15.8-kb contig that contained a putative 1,3,6,8-tetrahydroxynaphthalene (THN, 5) synthase16,17 gene (mcl17) was found on fosmid 3D9 in a pCC1FOS-based genomic DNA library of strain CHN-189. We next replaced mcl17 with an apramycin resistance cassette in CNH-189 to link the isolated genes to merochlorin production.18 A comparison of culture extracts from CNH-189Δmcl17 and the wild-type strain by HPLC showed that the mutant was unable to produce merochlorins 1–3 (Figure 2). This confirmed that the THN synthase, encoded by mcl17, is essential for merochlorin production and that the genes identified on fosmid 3D9 likely represent part of the merochlorin biosynthetic gene cluster.
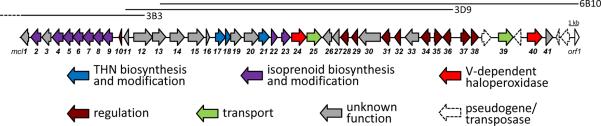
We obtained the complete gene cluster (57.6 kb, Figure 3) from total sequencing of 3D9, isolation of overlapping fosmids, and primer walking. Forty-one open reading frames (ORFs), mcl1–41, were assigned to the cluster and attributed to biosynthetic, resistance, transport, and regulatory functions (Table S6). Several genes have sequence homology to genes from other meroterpenoid clusters, such as mcl2–mcl9 that putatively encode the mevalonate pathway for isoprene biosynthesis;19mcl18, a putative THN monooxygenase gene;20mcl21, a putative methyltransferase gene; and mcl23, a putative aromatic prenyltransferase gene.21 In addition, the mcl cluster includes numerous structural genes that are unprecedented in meroterpenoid clusters, e.g., a putative polyprenyl synthase, mcl22, and a putative iron–sulfur cluster-containing protein, mcl30. Intriguingly, two genes, mcl24 and mcl40, were found to be highly homologous to the nap VHPO genes (Table S6).9
Figure 3.
Merochlorin biosynthetic gene cluster color-coded according to the biosynthetic function of different genes. Overlapping fosmids are indicated by solid lines. A table listing the size and putative function of each mcl gene product appears in the SI (Table S6).
Based on the bioinformatic analysis of the mcl cluster, we concluded that most of the unique biosynthetic capacity for merochlorin production was encoded on fosmid 3D9, except for the putative VHPO Mcl40 (Figure 3). To investigate whether the genes found on 3D9 are sufficient for the formation of the merochlorins and to explore the role of mcl40, we expressed the fosmid heterologously in the genome minimized strain, Streptomyces coelicolor M1154.22 Therefore, 3D9 was redesigned for stable integration into Streptomyces chromosomes via attB/attP-site-specific recombination.23 Culture extracts of the generated S. coelicolor M1154/merLK01 strain were analyzed by HPLC-MS.
Compounds 1 and 2 were accumulated by the mutant, but 3 was not produced (Figures 2, S18). However, we observed a previously unrecognized metabolite, merochlorin D (4). HPLC chromatographic comparison with the CNH-189 wild-type strain also revealed the production of 4 (Figure 2). To obtain sufficient quantities of 4 for structure elucidation, we optimized the culture conditions of CNH-189 to yield approximately 5 mg/L. HR-MS and NMR spectral analysis revealed that 4 was structurally similar to 3, except for the presence of an olefinic proton at C-18 and only one chlorine atom instead of two (Figure 1A). HMBC correlation data confirmed the dimethyl terminal olefin unit, and ROESY data gave the same relative configuration as in 3. Thus, the heterologous pathway expression showed that a 36-kb fragment of the 57.6-kb gene cluster encodes all of the enzymes required for the biosynthesis of 1, 2, and 4. However, the absence of 3 in the mutant extract suggested that one or more essential genes required for its formation are missing on fosmid 3D9. We speculate that 4 is a direct precursor to 3 and that the VHPO Mcl40 may catalyze the unprecedented macromolecular chlorination–cyclization reaction to give the 15-membered ring. In the biosynthesis of the napyradiomycins, NapH1 supports a chloronium-assisted stereoselective cyclization to yield a 6-membered ring, harboring a chlorinated gem-dimethyl ether.10 Similarly, the putative VHPO Mcl40 might use 4 or a less substituted derivative as a substrate to afford the massive 15-membered cyclic ether ring in 3.
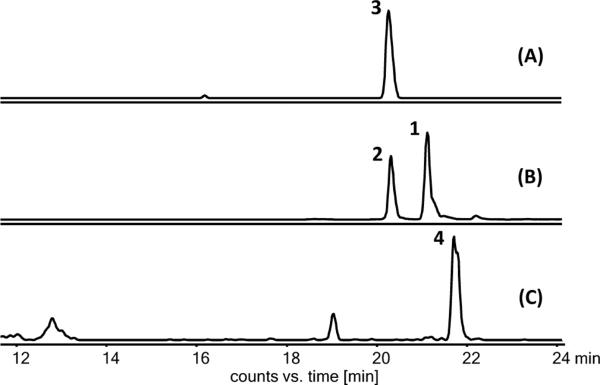
To explore whether Mcl40 can indeed support the biosynthesis of 3, we heterologously expressed fosmid clone 6B10, which contains a larger portion of the mcl locus and includes mcl40 (Figure 3). HPLC-MS analysis of the resulting S. coelicolor M1152/merLK30 strain revealed the production of all natural merochlorins, including 3 (Figure 4), suggesting that Mcl40 is involved in the chloronium-assisted macrocyclization of 4 to 3 (Figure 1A). However, the production yields of 3 and 4 were significantly lower compared with the original fosmid clone 3D9, which produced 4 but not 3. We speculate that the loss of the mcl12/13 genes of unknown function in 6B10 may lead to the observed lower merochlorin production levels. With respect to the second homologous VHPO in the mcl cluster, Mcl24, we hypothesize that it may catalyze a chlorination step of an early pathway intermediate to install the polyketide chlorine atom that can be found on all characterized merochlorin molecules.
Figure 4.
HPLC-HR-MS analysis of culture extracts from S. coelicolor M1152/merLK30. Extracted ion chromatograms for (A) m/z 495.17 [M+H]+ (3); (B) m/z 429.18 [M+H]+ (1 and 2); and (C) m/z 461.21 [M+H]+ (4).
Although 1–4 have different molecular architectures, they share common structural features, suggesting that they may also share early biosynthetic intermediates (Figure 1A). For example, the aromatic portion of 1–4 is likely derived from THN, similar to other hybrid polyketide–terpenoid metabolites.16 Thus, we propose that THN is the general precursor for two distinct metabolic pathways. The first pathway may proceed through prenylation, chlorination, and two divergent cyclization reactions to produce 1 and 2. The second pathway would generate 3 and 4 and may involve oxidation, methylation, prenylation, and two chlorination reactions. The order of the proposed biotransformations as outlined in Figure 1 is speculative and serves as a model for interrogating the function of merochlorin biosynthetic enzymes that may catalyze new interesting biochemical reactions. This divergent pathway that we propose represents a rare example where a single gene cluster encodes the formation of several architecturally unique metabolites.24
The merochlorins appear to be constructed with the same, rearranged C15-isoprene unit. To the best of our knowledge, the sesquilavandulyl carbon skeleton is unprecedented in bacteria. However, this type of skeleton has been infrequently encountered in plant natural products such as the xanthane roeperanone from Hypericum roeperanu.25 The prenylation substrate may be a preformed sesquilavandulyl diphosphate generated by Mcl22, that shows sequence similarity to undecaprenyl diphosphate synthase. Alternatively, the isoprenoid moiety may simply be formed by the sequential addition of dimethylallyl and geranyl units to a THN-based intermediate.
Only one putative prenyltransferase, Mcl23, is encoded in the mcl cluster. Merochlorins A/B and C/D are likely prenylated at two different positions, suggesting that Mcl23 catalyzes the prenylation at two adjacent carbon atoms at C-8 to give 1 and 2 and at C-10 to furnish 3 and 4 (the carbon atom numbering scheme adopted for the final natural product, Figure 1A). The oxidation state of THN (Figure 1A, 5) supports prenylation at C-4 to give an intermediate that can lead to 1 and 2. However, the same oxidation state does not allow C-3 alkylation to produce 3 and 4. We therefore propose that 5 is first partially oxidized by Mcl18 to the flaviolin hydroquinone, or directly to flaviolin followed by reduction, to produce an intermediate that is activated for alkylation at the C-3 position, which ultimately provides 3.
The genetic basis for the terpene cyclizations involved in the biosynthesis of 1 and 2 remains highly speculative. Notably, homologues to known terpene cyclases are not encoded in the cluster. We hypothesize that the internal olefin on the sesquiterpene attached to 5 undergoes epoxidation or halogenation, activating the molecule for an intramolecular cyclization. Nucleophilic attack by a carbon to form a C–C bond would lead to 1, whereas nucleophilic attack by oxygen to form a C–O bond would lead to 2 (Figure 1A, Paths a and b). In both scenarios, a second cyclization event would follow with concomitant elimination of water or HCl to yield 1 and 2. Because both VHPOs seem to be involved in chlorination reactions, we suspect that an oxidative transformation is more likely to occur in the final steps of 1 and 2 biosynthesis.
In conclusion, we discovered the merochlorins as new streptomycete antibiotics with diverse molecular architectures that derive from a novel pathway involving polyketide and terpenoid biochemistry. Our in vivo biosynthetic results established the genetic basis for merochlorin construction and support the involvement of just the second example of VHPOs in bacterial natural product assembly.
Supplementary Material
ACKNOWLEDGMENT
We gratefully acknowledge Michael Fischbach and Joseph DeRisi for helpful discussions, Juan-Pablo Gomez-Escribano and Mervyn Bibb for providing S. coelicolor M1152/M1154, Alessandra Eustaquio for preliminary work on the integration cassette, and Yongxuan Su at the UCSD Molecular Mass Spectrometry Facility for HR-MS assistance that was partially funded by the NIH (SRR0256362). This work was generously supported by research grants from the NIH (AI047818 to B.S.M and CA044848 to W.F.) and the California Sea Grant Program (R/NMP-100 to B.S.M. and P.R.J.). We also acknowledge postdoctoral fellowships from the NIH (GM096711) to P.B., the Alexander von Humboldt-Foundation to L.K, and the DFG to S.L. as well as a predoctoral fellowship from the PhRMA Foundation to P.S.-C.
Footnotes
Supporting Information. Experimental section, supporting tables, detailed compound characterization, and crystallographic data are provided. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Vaillancourt FH, Yeh E, Vosburg DA, Garneau-Tsodikova S, Walsh CT. Chem. Rev. 2006;106:3364–3378. doi: 10.1021/cr050313i. [DOI] [PubMed] [Google Scholar]
- (2).Wagner C, El Omari M, Konig GM. J. Nat. Prod. 2009;72:540–553. doi: 10.1021/np800651m. [DOI] [PubMed] [Google Scholar]
- (3).Butler A, Sandy M. Nature. 2009;460:848–854. doi: 10.1038/nature08303. [DOI] [PubMed] [Google Scholar]
- (4).Butler A, Carter-Franklin JN. Nat. Prod. Rep. 2004;21:180–188. doi: 10.1039/b302337k. [DOI] [PubMed] [Google Scholar]
- (5).Winter JM, Moore BS. J. Biol. Chem. 2009;284:18577–18581. doi: 10.1074/jbc.R109.001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Soria-Mercado IE, Prieto-Davo A, Jensen PR, Fenical W. J. Nat. Prod. 2005;68:904–910. doi: 10.1021/np058011z. [DOI] [PubMed] [Google Scholar]
- (7).Shiomi K, Nakamura H, Iinuma H, Naganawa H, Isshiki K, Takeuchi T, Umezawa H, Iitaka Y. J. Antibiot. 1986;39:494–501. doi: 10.7164/antibiotics.39.494. [DOI] [PubMed] [Google Scholar]
- (8).Gomi S, Ohuchi S, Sasaki T, Itoh J, Sezaki M. J. Antibiot. 1987;40:740–749. doi: 10.7164/antibiotics.40.740. [DOI] [PubMed] [Google Scholar]
- (9).Winter JM, Moffitt MC, Zazopoulos E, McAlpine JB, Dorrestein PC, Moore BS. J. Biol. Chem. 2007;282:16362–16368. doi: 10.1074/jbc.M611046200. [DOI] [PubMed] [Google Scholar]
- (10).Bernhardt P, Okino T, Winter JM, Miyanaga A, Moore BS. J. Am. Chem. Soc. 2011;133:4268–4270. doi: 10.1021/ja201088k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Wilson MC, Nam SJ, Gulder TA, Kauffman CA, Jensen PR, Fenical W, Moore BS. J. Am. Chem. Soc. 2011;133:1971–1977. doi: 10.1021/ja109226s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Sakoulas G, Nam SJ, Loesgen S, Fenical W, Jensen PR, Nizet V, Hensler M. PLoS ONE. 2012;7:e29439. doi: 10.1371/journal.pone.0029439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Gallagher KA, Fenical W, Jensen PR. Curr. Opin. Biotechnol. 2010;21:794–800. doi: 10.1016/j.copbio.2010.09.010. [DOI] [PubMed] [Google Scholar]
- (14).Haagen Y, Gluck K, Fay K, Kammerer B, Gust B, Heide L. Chembiochem. 2006;7:2016–2027. doi: 10.1002/cbic.200600338. [DOI] [PubMed] [Google Scholar]
- (15).Kawasaki T, Hayashi Y, Kuzuyama T, Furihata K, Itoh N, Seto H, Dairi T. J. Bacteriol. 2006;188:1236–1244. doi: 10.1128/JB.188.4.1236-1244.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Funa N, Ohnishi Y, Fujii I, Shibuya M, Ebizuka Y, Horinouchi S. Nature. 1999;400:897–899. doi: 10.1038/23748. [DOI] [PubMed] [Google Scholar]
- (17).Austin MB, Izumikawa M, Bowman ME, Udwary DW, Ferrer JL, Moore BS, Noel JP. J. Biol. Chem. 2004;279:45162–45174. doi: 10.1074/jbc.M406567200. [DOI] [PubMed] [Google Scholar]
- (18).Gust B, Challis GL, Fowler K, Kieser T, Chater KF. Proc. Natl. Acad. Sci. U.S.A. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Takagi M, Kuzuyama T, Takahashi S, Seto H. J. Bacteriol. 2000;182:4153–4157. doi: 10.1128/jb.182.15.4153-4157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Funa N, Funabashi M, Yoshimura E, Horinouchi S. J. Biol. Chem. 2005;280:14514–14523. doi: 10.1074/jbc.M500190200. [DOI] [PubMed] [Google Scholar]
- (21).Kuzuyama T, Noel JP, Richard SB. Nature. 2005;435:983–987. doi: 10.1038/nature03668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Gomez-Escribano JP, Bibb MJ. Microb. Biotechnol. 2011;4:207–215. doi: 10.1111/j.1751-7915.2010.00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Bierman M, Logan R, O'Brien K, Seno ET, Rao RN, Schoner BE. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- (24).Bernhardt P, O'Connor SE. Curr. Opin. Chem. Biol. 2009;13:35–42. doi: 10.1016/j.cbpa.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Rath G, Potterat O, Mavi S, Hostettmann K. Phytochemistry. 1996;43:513–520. doi: 10.1016/0031-9422(96)00284-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.