Abstract
Mitochondrial diseases are frequently caused by heteroplasmic mtDNA mutations. Because these mutations express themselves only at high relative ratios, any approach able to manipulate mtDNA heteroplasmy can potentially be curative. In this study we developed a system to manipulate mtDNA heteroplasmy in all skeletal muscles from neonate mice. We selected muscle because it is one of the most clinically affected tissues in mitochondrial disorders. A mitochondria targeted restriction endonuclease (mito-ApaLI) expressed from AAV9 particles was delivered either by intraperitoneal or intravenous injection in neonate mice harboring two mtDNA haplotypes, only one of which was susceptible to ApaLI digestion. A single injection was able to elicit a predictable and marked change in mtDNA heteroplasmy in all striated muscles analyzed, including heart. No health problems or reduction in mtDNA levels were observed in treated mice, suggesting that this approach could have clinical applications for mitochondrial myopathies.
Introduction
Optimal expression of genes encoded by mitochondrial DNA (mtDNA) is required for the biogenesis of the oxidative phosphorylation system (OXPHOS), impairment of which is central to the etiology of most mitochondrial disorders. Heteroplasmic mtDNA mutations cause a variety of mitochondrial disorders, which can become clinically apparent during infancy 1,2 and generally include cardiovascular, skeletal muscle or neurological manifestations. Among the genetic therapies being considered as strategies for the treatment of such disorders, manipulating mtDNA heteroplasmy to reduce mutation loads has been a goal of our group and others for a number of years.
Cells contain hundred of copies of mtDNA for every copy of the nuclear genome. High heteroplasmic mtDNA mutation loads, generally above 80 %, are required to trigger OXPHOS defects in specific tissues 3,4. Reducing mutation loads below these threshold levels, can restore OXPHOS function, and a complete clearance of mutant mtDNA is unnecessary. For this reason, endonuclease-mediated heteroplasmy shift is an attractive strategy for genetic-therapy. In situations where a heteroplasmic mtDNA mutation results in a unique restriction site, targeting the appropriate restriction enzyme (RE) to mitochondria causes a heteroplasmy shift by reducing the proportion of mutated mtDNA with the restriction site. We have demonstrated this mechanism in a number of model systems 5,6. SmaI, targeted to mitochondria of cells harboring the T8399G NARP/LHON mutation drastically reduced the mutated mtDNA, followed by repopulation by the wild-type mtDNA and restoration of normal intracellular ATP levels and mitochondrial membrane potential 7,8. In vivo work has shown the efficacy of the system in mouse models when viral vectors carrying the recombinant RE were administered focally to muscle and brain 6 or to target specific organs such as liver (with adenovirus) or heart (with AAV6) 9,10. However, manipulation of mtDNA heteroplasmy in all skeletal muscles in the body remained elusive.
The prevalence of mitochondrial disease in the pediatric population has been difficult to determine but is believed to be similar to that seen in the adult population 11. We decided therefore to test whether we could accomplish endonuclease-mediated heteroplasmy shift in all skeletal muscles at early ages using a well-characterized heteroplasmic mouse model containing two polymorphic mtDNA sequence variants, NZB and BALB/c 12. In these mice, both mtDNA haplotypes behave as neutral variants except that with age there is an increase in the percentage of NZB mtDNA in liver and slight reduction in spleen 10,13–15. NZB mtDNA levels in cardiac and skeletal muscle do not change with age. The BALB/c mtDNA variant harbors a unique ApaLI site that is not present in the NZB variant. Our goal was to deliver the recombinant RE to all muscles in the body after a single injection. To accomplish this we produced recombinant AAV9 carrying the mito-RE, ApaLI, and delivered the virus via intraperoneal (IP) or temporal vein (TV) injection in neonates. Our studies were based on previous reports that robust transduction in the skeletal muscle could be achieved in mice after delivery of AAV9 vectors by systemic injection 16,17.
Results
Mito-ApaLI-HA expressed from AAV9 vectors is targeted to mitochondria
Cultured mouse hepatocytes were infected with 1,000 vg/cell of rAAV9[mito-ApaLI-HA] and analyzed 10 days post-transduction by immunocytochemistry. Cells transduced with rAAV9[mito-ApaLI-HA] showed HA expression that co-localized with the mitochondrial dye Mito Tracker Red CMXRos (Supplementary Figure S1).
AAV9[mito-ApaLI-HA] efficiently transduces neonatal mouse muscle
Initially, we administered 5×1011 viral genomes (vg)/mouse intraperitoneal (IP) at P2-P3. The expression of rAAV9 [mito-ApaLI-HA] and a control vector expressing alkaline phosphatase, AAV9[AP], were analyzed at 6, 12 and 24 weeks post-injection. We also tested the efficacy of temporal vein (TV) injection with 5×1011 vg/mouse at P2-P3. Animals were also euthanized at the same time points.
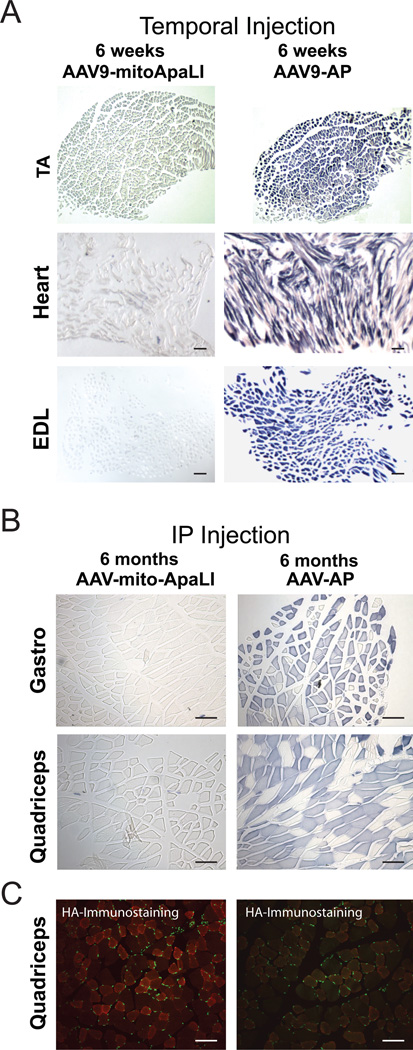
TV injection of AAV9[AP], resulted in increased AP activity in skeletal and cardiac muscle, as illustrated in tibialis anterior (TA), heart and extensor digitorum longus (EDL) after 6 weeks (Figure 1A) and after 12 and 24 weeks (not shown). Only a few cells were stained in liver and no activity was observed in spleen, lung, brain or kidney (not shown). The same pattern of expression was observed after IP injection with AAV9[AP] at 6, 12 and 24 weeks post injection (Figure 1B). Likewise, after IP-injection, high expression of AAV9[mito-ApaLI-HA] was observed by western blotting in all skeletal and cardiac muscles analyzed, including: gastrocnemius, quadriceps, soleus, heart, tibialis anterior (Figure 2) and extensor digitorum longus (not shown), with only a weak signal in liver and no detectable expression in spleen, brain, lung or kidney, at 6 weeks (Figure 2), 12 and 24 weeks post-injection (Supplementary Figure S2A). Transgene expression of AAV9[mito-ApaLI-HA] in muscle was also confirmed after IP injection by immunocytochemistry staining (Figure 1C). TV injection also proved to be an efficient way to deliver the transgene to all muscles as HA-expression was also observed by western blotting in skeletal and cardiac muscle after 6 weeks (Supplementary Figure S2B).
Figure 1. Expression of AAV9[AP] in targeted tissues.
Alkaline phosphatase activity staining reveals high expression of AP in skeletal muscle and heart after delivery of AAV9[AP]. 1A, temporal vein (TV) injections: Tibialis anterior (TA), heart and extensor digitorum longus (EDL) at 6 weeks post injection. 1B, intraperitoneal (IP) injections: gastrocnemius (gastro) and quadriceps at 24 weeks. A similar pattern of expression was observed for Mito-ApaLI-HA in quadriceps at 24 weeks post-delivery of AAV9[ApaLI-HA] using anti-HA immunostaining of in 20 µm cryo-sections counterstained using DAPI (1C). Bar = 100 µm.
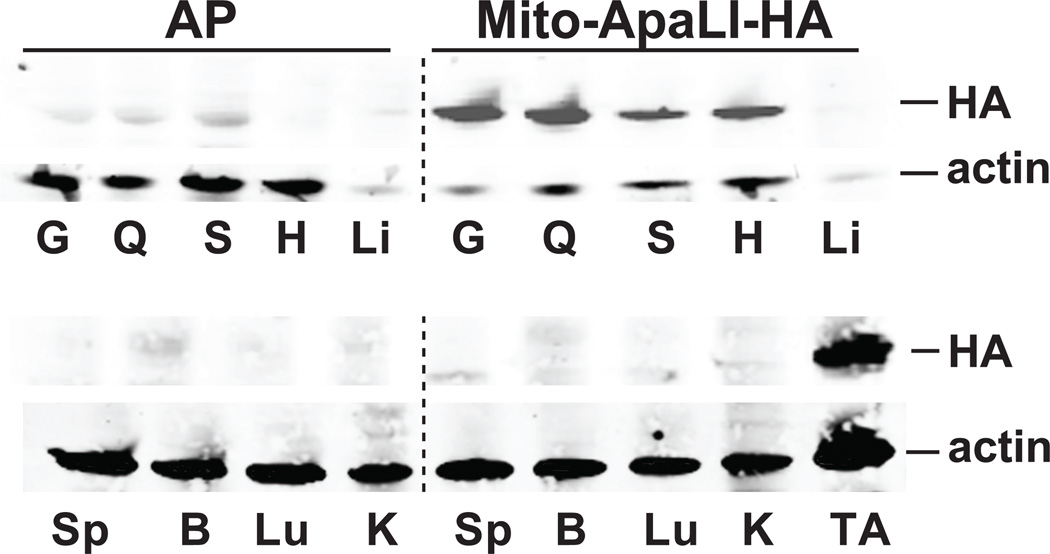
Figure 2. Expression of Mito-ApaLI-HA in targeted tissues.
The expression of ApaLI-HA was analyzed by anti-HA western blotting 6 weeks after IP injection in P2-P3 mice. Homogenates from skeletal muscles, gastrocnemius (G), quadriceps (Q), soleus (S), heart (H) and tibialis anterior (TA) showed high expression. No expression was observed in spleen (Sp), brain (B), lung (Lu) or kidney (K), with weak expression in liver (Li). Actin was used as a loading control.
Systemic delivery of AAV9[mito-ApaLI-HA] induces shifts in mtDNA heteroplasmy
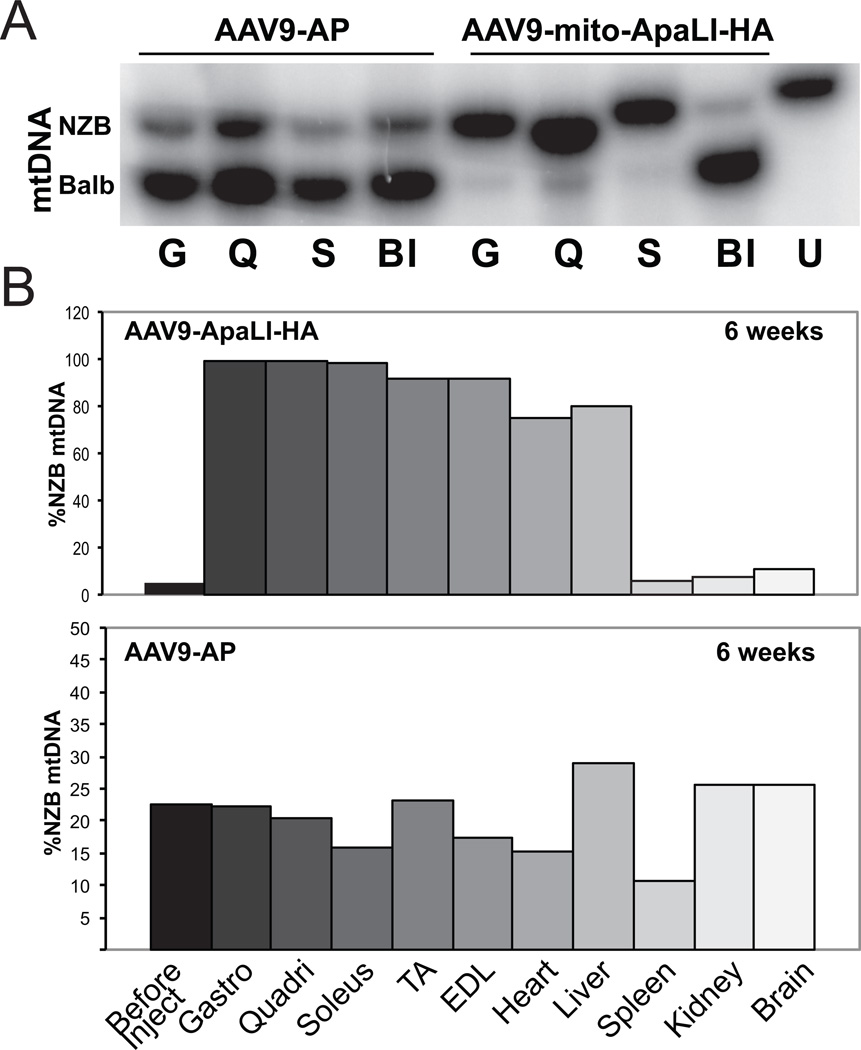
Changes in the relative levels of NZB and BALB/c mtDNA were evaluated using ApaLI restriction fragment polymorphism in tissue samples taken at 6, 12 and 24 weeks post-injection (Figures 3 and 4). As shown graphically in Figure 3 (gel images in Supplementary Figure S3), a significant increase was observed in the percentage of NZB mtDNA in all skeletal and cardiac tissues analyzed in mice injected with AAV9[mito-ApaLI-HA], when compared to the percentage of NZB mtDNA present in tail samples from the same mice before injection (BI). A significant increase in the levels of NZB mtDNA was also observed in liver. No change in the percentage of NZB mtDNA was seen in other organs. Likewise, no changes in heteroplasmy levels were observed in mice injected with AAV9[AP] at any time point (Figure 3 and 4). Long term transduction at 24 weeks post-delivery of AAV9[mito-ApaLI-HA] showed lower changes in NZB levels compared to those recorded at 6 weeks or 12 weeks post-delivery of the transgene (Figures 3 and 4).
Figure 3. AAV9[mito-ApaLI-HA] induces a shift in mtDNA heteroplasmy in muscle.
NZB/BALB/c mtDNA heteroplasmy quantified using last-cycle hot PCR/RFLP with DNA samples from mice injected with AAV9[mito-ApaLI-HA] at 6 weeks post IP injection. Increases in the percentage of NZB mtDNA were observed in all skeletal muscle and heart tissues when compared to the samples obtained from tail before injection. 3A, representative phospho-image of radiolabeled RFLP gel, Gastrocnemius (G), quadriceps (Q), soleus (S), before injection (BI), uncut DNA (U); 3B, quantification of phosphor-imager data, following injection of AAV9[mito-ApaLI-HA] (upper panel) and AAV9[AP] (lower panel); tibialis anterior (TA), extensor digitorum longus (EDL). No change in the percentage of NZB mtDNA was observed in spleen, brain, and kidney; a shift was observed in liver when compared to the AAV9[AP] injected sample that represents the age related shift in heteroplasmy in this tissue. Each bar corresponds to a different tissue from a single mouse.
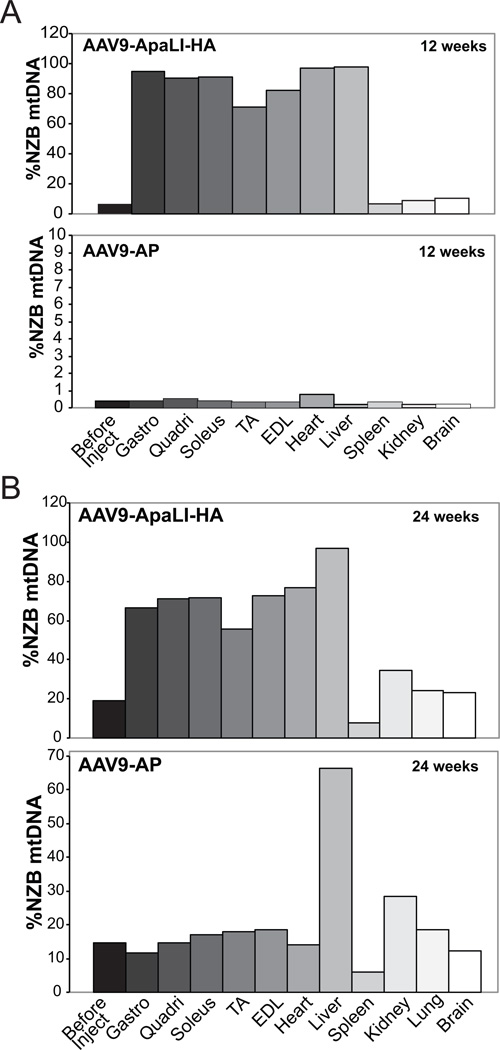
Figure 4. AAV9[mito-ApaLI-HA] induces a long-term shift in mtDNA heteroplasmy.
NZB/BALB/c heteroplasmy quantified using last-cycle hot PCR/RFLP using muscle DNA from mice injected with AAV9[mito-ApaLI-HA] at 12 weeks (4A) and 24 weeks (4B) post IP injection. Tissue abbreviations as in Figure 3. Increased percentage of NZB mtDNA was observed in all the skeletal muscles and heart when compared to before injection samples from tails. Again no change in the percentage of NZB mtDNA was observed in spleen, kidney, lung or brain, with slight shift in liver. Each bar corresponds to a different tissue from a single mouse.
AAV9[mito-ApaLI-HA] expression in muscle does not induce mtDNA depletion
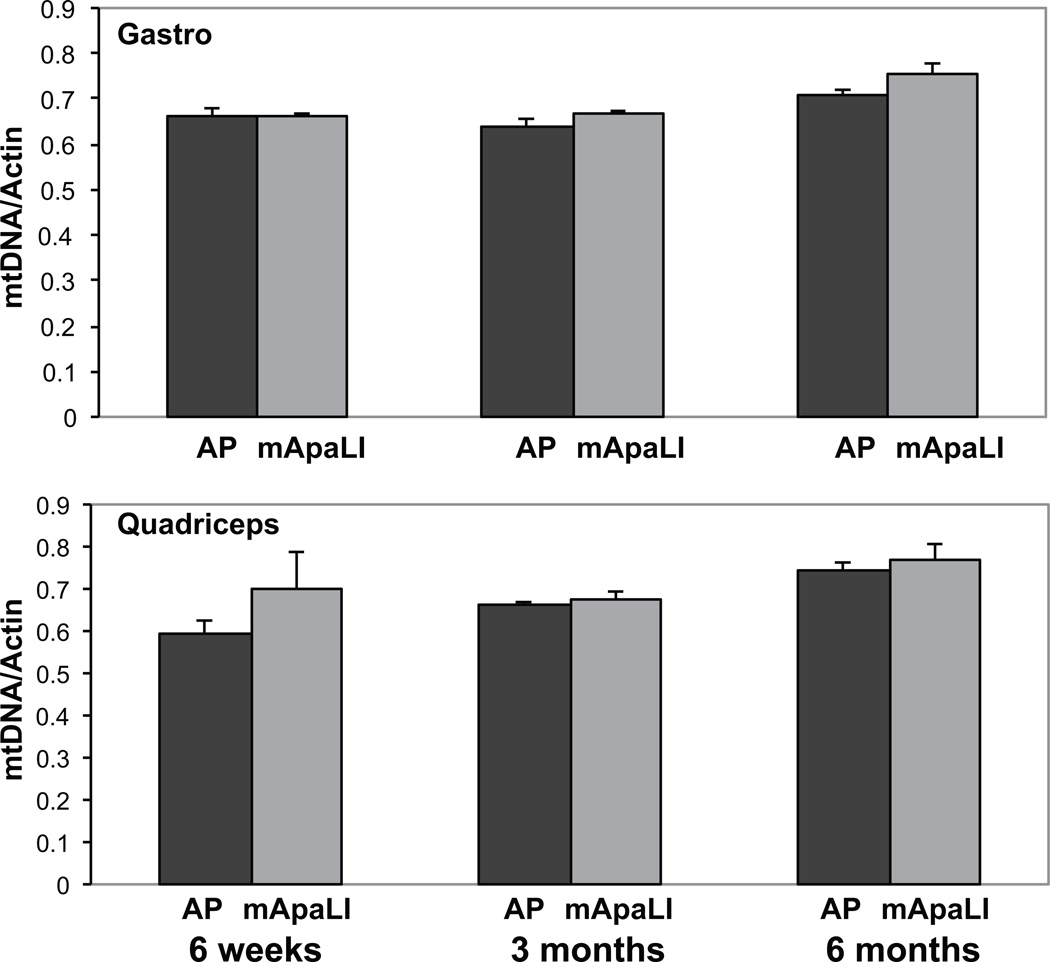
qPCR of DNA from gastrocnemius, quadriceps (Figure 5), heart and liver (not shown) was used to determine whether transgene expression induced undesirable mtDNA depletion. The levels of mtDNA were quantified using amplicons of the mtDNA ND1 gene and the nuclear coded actin (Figure 5). The results showed no change in the levels of mtDNA at any time-point analyzed. Similar results were observed when GAPDH was used as a normalization control (data not shown).
Figure 5. AAV9[mito-ApaLI-HA] expression in muscle does not induce mtDNA depletion.
Quantitative PCR of gastrocnemius and quadriceps DNA showed no sign of mtDNA depletion at 6, 12 and 24 weeks post injection (three independent samples from the each tissue *P<0.05).
Discussion
Restriction endonuclease as a tool for gene therapy
Individuals affected with mitochondrial diseases commonly harbor heteroplasmic mtDNA mutations. Heteroplasmic mtDNA mutations can both be inherited from asymptomatic maternal carriers or result from spontaneous mutation and asymmetries during meiosis. This can result in oocytes with different mtDNA mutation loads, which can lead to children with different degrees of mitochondrial dysfunction 18. Clinical disease is higly influenced by mtDNA heteroplasmy, which is a dynamic phenomenon. The level of heteroplasmy may vary considerably from tissue to tissue or even from cell to cell 19. Threshold effects also influence the penetrance of heteroplasmic mutations 20. Mitochondrial disease commonly presents with a combination of muscle and brain involvement. Both tissues are post-mitotic and have high metabolic requirements, which may explain why they are frequently affected. Other organs may be involved depending on the proportion of mutated mtDNA present and their individual threshold for the mutation. Consequently management and prevention of mtDNA diseases has lagged far behind the understanding of their cause 21. Substitution of the mutated mtDNA by wild-type mtDNA would be a powerful therapeutic option to change mtDNA heteroplasmy. To achieve this goal, we have pursued the removal of mutated mtDNA by mitochondria targeted endonucleases.
A group of diseases that is amenable to this approach is the one caused by the m.8993T>G MT-ATP6 mutation. This mtDNA mutation has been associated with Leigh or NARP (neuropathy, ataxia, and retinitis pigmentosa) syndromes, which are part of a continuum of progressive neurodegenerative disorders. The mutation creates a unique XmaI site in the mtDNA of patients that could be a suitable target for the restriction endonuclease treatment8. Muscle weakness is commonly observed in patients with NARP22 but to address the CNS involvement would likely require combination treatment with other vectors. However, as describe below the development of endonucleases with new specificities will be crucial to expand the approach to other pathogenic mtDNA mutations.
rAAV9 for systemic skeletal muscle transduction in mitochondrial myopathies
Gene therapy for mitochondrial myopathies requires transduction of all skeletal muscles in the body. To achieve the goal of systemic skeletal muscle transduction, we delivered AAV serotype 9 vectors via IP or TV injection. Robust transduction of skeletal muscle and heart has been described in mice after delivery of AAV9 vectors 16,17,23. We achieved high levels of expression of AAV9 transgenes in skeletal muscle as rapidly as 6 weeks, with long-term expression until 24 weeks post-delivery. In concordance with the above reports, we also obtained efficient transduction of the heart at 6 weeks post-delivery. Our results are consistent with those published by Ghosh et al 17, in that AAV9 targeted striated muscle, however, we did not detect any signs of expression in lung. Our pattern of expression in neonates was very similar to the preferential tropism of AAV9 described by Zincarelli et al 24, where high levels of luciferase expression were observed mainly in skeletal muscles and remained strong nine month after IV (tail vein) injection. We observed very few cells transduced in liver compared to their reports of high expression. However, our results are consistent with the findings of Bostick et al where liver, aorta and kidney were preferentially transduced in adult mice, but not neonates, demonstrating that systemic transduction of AAV9 in mice is influenced by age but not the route of administration 25.
Despite the apparent low, but clearly detectable expression of mito-ApaLI in liver, IP and TV injection of AAV9[mito-ApaLI-HA] induced a shift in mtDNA heteroplasmy, showing that these low levels of expression may be enough to promote a partial shift in mtDNA heteroplasmy in liver. This robust change may be due to the fact that liver has a natural tendency to increase NZB mtDNA levels, and may not apply to any mtDNA polymorphism. We could not detect expression in brain, although Foust et al described an apparent tropism of AAV9 for motor neurons, especially those within the spinal cord, in neonates 26. This discrepancy may be due to the use of the β-actin promoter in that study, while we used a CMV promoter. We did not find any difference in the pattern of expression between IP and TV injection. Intraperitoneal administration was previously shown to be an efficient route used for gene transfer of rAAV in neonates27.
Although the high titers required for in vivo transduction and the current viral production techniques limited the number of mice analyzed, all mice used in this study showed essentially identical dramatic shifts in mtDNA heteroplasmy towards the NZB mtDNA, providing unambiguous proof that it is possible to affect mtDNA heteroplasmy in neonate’s muscle after a single systemic injection of rAAV9 expressing a mitochondria targeted restriction endonuclease.”
Treating mitochondrial myopathies in neonates
We decided to test our approach in newborn mice to investigate the feasibility of targeting muscle mtDNA at a young age. Our results showed high expression of the transgene as early as 6 weeks and no evidence of associated pathology was observed in the mice at any studied time-point. It has been shown that AAV2 preferentially transduces slow-twitch muscle fibers while AAV6 transduces both slow- and fast-twitch fiber types efficiently 28–30. It is not clear whether AAV9 displays a fiber type preference 25. Although we observed weaker AP staining in muscles with predominant low-twitch fibers (e.g. soleus), as previously described 25, we did not observe differences in the increases in NZB mtDNA levels in any of the different skeletal muscles analyzed.
Prenatal diagnosis of mtDNA mutations has been accomplished 3,31. Therefore, mito-restriction endonuclease approach could be used in patients affected by mtDNA mutations with disease onset in childhood or young adulthood that can be diagnosed before birth.
AAV9 mito-ApaLI promoted efficient shift in mtDNA heteroplasmy in all skeletal muscles
A small shift in mtDNA heteroplasmy would typically be enough to restore OXPHOS function in most tissues, as the threshold for a biochemical manifestation of a mtDNA mutation is typically > 80% mutant, < 20% wild-type 32. In our experiments, we observed increases in the percentage of NZB mtDNA to > 60 % as early as 6 weeks after injection that persisted for the duration of our experiment (24 weeks). Thus the heteroplasmy shift we induced would correspond to a reduction in “undesired” mtDNA loads to < 40%, well below the threshold levels for mitochondrial myopathy observed in patients. Because of the limited numbers of mice used we could not do an extensive analysis of the effect of the initial heteroplasmy on the change, but it was clear that even very low levels of resistant mtDNA would be amplified by this procedure. Sequencing through the mitochondrial ApaLI restriction site in similar studies has previously confirmed that observed heteroplasmy shift is not due to cleavage-induced mutations 10. The highest percentage heteroplasmy change was observed in mice with low levels of NZB mtDNA before AAV9[mito-ApaLI-HA] administration. However, because we analyzed few mice, we cannot be certain that this pattern of heteroplasmy change will be consistent.
No mtDNA depletion was observed after AAV-9 mito-ApaLI transduction
The degree and efficiency of mtDNA heteroplasmy shift depends on the presence of a residual population of undigested mtDNA. One concern related to this approach is the risk of inducing mtDNA depletion 9. In the cases where the overall mutant load is very high, the wild-type levels would have to increase rapidly to compensate for the loss of mutant mtDNA and OXPHOS restore function. This was the case in our experiments as the initial level of BALB/c the target of ApaLI was > 80% in most of the mice analyzed. We could not detect mtDNA depletion at any time point. Moreover, we did not detect any change in behavior of treated mice.
Prospects
Among potential therapies to treat mitochondrial diseases caused by mtDNA mutations, reducing mutant load through endonuclease-mediated heteroplasmy shift is arguably one of the most promising approaches. It has proven to be efficient and safe in our mouse models. There are few rodent models heteroplasmic for pathogenic mtDNA mutations, making it difficult to model mitochondrial gene therapies in vivo 2. However, it is well established that the biochemical phenotype is directly related to the mutant mtDNA load in cells 33 and we have been able to induce considerable changes in heteroplasmy in muscle following systemic injection of AAV9[mito-ApaLI-HA]. The use of bacterial restriction nucleases is limited by the requirement that a unique site must be created by the mutation, however zinc-finger nucleases have been designed to target mtDNA mutations 34 and we and others are trying to take advantage of these new approaches to expand the tools for clinical applications. In theory, any DNA sequence can be specifically targeted by modular zinc finger nucleases 35. The lack of deleterious side-effects in our experiments with rAAV9 together with the efficient systemic muscle delivery is encouraging and provides a basis for future work on therapies for mitochondrial myopathies caused by heteroplasmic mtDNA mutations.
Material and Methods
Mito-ApaLI-HA constructs
A synthetic gene coding for the ApaLI RE with a C-terminal HA (Hemagglutinin antigen) tag was purchased from Integrated DNA Technologies (Coralville, IA, USA) with codon usage was optimized for mammalian translation. An N-terminal mitochondrial import signal derived from COX8A was added, and the resulting construct was cloned into a cis plasmid for the AAV9 backbone vector under the control of the cytomegalovirus promoter (CMV). The poly adenylation sequence was from the human growth hormone gene. The cis plasmid for AAV9-AP (Alkaline Phosphatase) has been reported before 17. Recombinant AAV9 stock was produced using a triple plasmid transfection protocol followed by two rounds of isopycnic ultracentrifugation in calcium chloride gradient as described previously 17,25. Viral titer was determined by quantitative PCR and confirmed by slot blot.
NZB/BALB/c mice
Female NZB/BALB/c heteroplasmic founders were a kind gift from Eric Shoubridge (McGill University). These mice carry variable levels of the NZB and BALB/c murine mtDNA haplotypes in a BALB/c genetic background and do not display any form of mitochondrial dysfunction 12,36
Virus administration and sample preparation
To achieve systemic delivery, mice at P2-P3 were subject to intraperitoneal (IP) or temporal vein (TV) injection. Each mouse was injected with 5×1011 viral genomes (vg) in a 50ul final volume in PBS of either AAV9[mito-ApaLI-HA] or control AAV[AP] (Alkaline Phospathase). Injection was carried out using a short insulin syringe with a 31G needle (Becton Dickenson). Tail tissue was obtained before performing the injections to determine day 0 heteroplasmy levels. At 6,12 and 24 weeks post-injection, mice were anesthetized and perfused with chilled PBS and skeletal and skeletal and cardiac tissues, liver, kidney, spleen, brain and lung samples were taken. The University of Miami IACUC approved studies reported here.
Heteroplasmy analysis
Total DNA from tissue samples was obtained after phenol-chloroform extraction. mtDNA heteroplasmy was determined by ‘Last-cycle hot’ PCR 37 using mouse mtDNA primers (5228–5250; 5665–5690). The PCR product was digested with ApaLI, which digests BALB/c mtDNA at position 5461 and subjected to electrophoreses in an 8% polyacrylamide gel 6. The radioactive signal was quantified using a Cyclone phosphorimaging system (Perkin-Elmer, Waltham, MA, USA) as described 9.
Western blotting
Fifty µg of total protein from tissue homogenates was electrophoresed using 4–20% Tris-HCl polyacrylamide gels (Bio-Rad) and transferred to nitrocellulose membranes (Bio-Rad). Rat anti-HA antibody obtained from Roche Biochemicals and a donkey anti-rat IgG IRDye 800-conjugated secondary antibody from Rockland (Gilbertsville, PA, USA) were used. Odyssey Infrared Imaging System (LI-COR) was used to scan the blots. A polyclonal anti-actin antibody (Sigma Co.) was used as loading control throughout.
Colorimetric and Immunohistochemical studies
NZB/BALB/c mouse hepatocytes were plated onto coverslips and infected with 1×103 viral genomes per cell of AAV9[mito-ApaLI-HA] for 10 days. Cells were then incubated for 30 min at 37°C with 200nM Mito Tracker Red CMXRos (Invitrogen, Carlsbad, CA, USA) and fixed with 2% paraformaldehyde (PFA) in PBS for 20 min. After a brief treatment with methanol (5 min), a primary anti-HA antibody (Roche) in 2% bovine serum albumin (BSA)/PBS was left overnight at 4°C followed by 2 h incubation at room temperature with an Alexa Fluor 488-conjugated secondary antibody (Molecular Probes, Invitrogen, Carlsbad, CA, USA). Images were recorded using a confocal microscope LSM510 from Carl Zeiss.
Tissue samples were fixed in 4% PFA and cryopreserved using 30% sucrose in PBS before freezing in liquid nitrogen-cooled isopentane and stored at −80C. For Alkaline Phosphatase staining, samples were sectioned using a cryostat (20 µm) and mounted on Superfrost Plus microscope slides (Thermo Fisher Scientific, Walthan, MA, USA). Slices were fixed in 4% PFA, rinsed in cold PBS and pre-heated for 90 min in PBS at 67°C to eliminate the endogenous heat sensitive alkaline phosphatase. Chromogenic staining was achieved by incubation with an NBT/BCIP solution (Sigma-Aldrich, St. Louis, MO, USA). Samples for immunofluorescence were treated with 0.5% Triton X-100, followed by PBS with 2% BSA for 30 min and then stained using anti-HA and DAPI for nuclei staining 10
Quantitative PCR
Total DNA was extracted from snap-frozen tissues as described above and treated with RNAase A at 37°C-30min (1ul/10ug DNA) followed by sodium acetate and ethanol precipitation. Quantitative PCR reactions were performed on the DNA samples in the presence of fluorescent dye (SYBR Green, Fermentas) using mouse ND1 and β-actin or GAPDH primers and ΔΔCT values were calculated.
ND1 3281F: 5’-CAGCCTGACCCATAGCCATA-3’,
ND1 3364B: 5’-ATTCTCCTTCTGTCAGGTCGAA-3’.
β-actin Exon 6F: 5’-GCGCAAGTACTCTGTGTGGA-3’,
β-actin Exon 6B: 5’-CATCGTACTCCTGCTTGCTG-3’
GAPDH Exon 5F: 5’-GCAGTGGCAAAGTGGAGATT-3’
GAPDH Exon 5B: 5’-GAATTTGCCGTGAGTGGAGT-3’
Supplementary Material
Acknowledgements
This work was supported by NIH grant EY010804.
References
- 1.Kyriakouli DS, Boesch P, Taylor RW, Lightowlers RN. Progress and prospects: gene therapy for mitochondrial DNA disease. Gene Ther. 2008;15:1017–1023. doi: 10.1038/gt.2008.91. [DOI] [PubMed] [Google Scholar]
- 2.Cwerman-Thibault H, Sahel JA, Corral-Debrinski M. Mitochondrial medicine: to a new era of gene therapy for mitochondrial DNA mutations. J Inherit Metab Dis. 2011;34:327–344. doi: 10.1007/s10545-010-9131-5. [DOI] [PubMed] [Google Scholar]
- 3.Thorburn DR, Dahl HH. Mitochondrial disorders: genetics, counseling, prenatal diagnosis and reproductive options. Am J Med Genet. 2001;106:102–114. doi: 10.1002/ajmg.1380. [DOI] [PubMed] [Google Scholar]
- 4.DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N Engl J Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 5.Srivastava S, Moraes CT. Manipulating mitochondrial DNA heteroplasmy by a mitochondrially targeted restriction endonuclease. Hum Mol Genet. 2001;10:3093–3099. doi: 10.1093/hmg/10.26.3093. [DOI] [PubMed] [Google Scholar]
- 6.Bayona-Bafaluy MP, Blits B, Battersby BJ, Shoubridge EA, Moraes CT. Rapid directional shift of mitochondrial DNA heteroplasmy in animal tissues by a mitochondrially targeted restriction endonuclease. Proc Natl Acad Sci U S A. 2005;102:14392–14397. doi: 10.1073/pnas.0502896102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka M, Borgeld HJ, Zhang J, Muramatsu S, Gong JS, Yoneda M, et al. Gene therapy for mitochondrial disease by delivering restriction endonuclease SmaI into mitochondria. J Biomed Sci. 2002;9:534–541. doi: 10.1159/000064726. [DOI] [PubMed] [Google Scholar]
- 8.Alexeyev MF, Venediktova N, Pastukh V, Shokolenko I, Bonilla G, Wilson GL. Selective elimination of mutant mitochondrial genomes as therapeutic strategy for the treatment of NARP and MILS syndromes. Gene Ther. 2008;15:516–523. doi: 10.1038/gt.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bacman SR, Williams SL, Hernandez D, Moraes CT. Modulating mtDNA heteroplasmy by mitochondria-targeted restriction endonucleases in a 'differential multiple cleavage-site' model. Gene Ther. 2007;14:1309–1318. doi: 10.1038/sj.gt.3302981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bacman SR, Williams SL, Garcia S, Moraes CT. Organ-specific shifts in mtDNA heteroplasmy following systemic delivery of a mitochondria-targeted restriction endonuclease. Gene Ther. 2010;17:713–720. doi: 10.1038/gt.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uusimaa J, Remes AM, Rantala H, Vainionpaa L, Herva R, Vuopala K, et al. Childhood encephalopathies and myopathies: a prospective study in a defined population to assess the frequency of mitochondrial disorders. Pediatrics. 2000;105:598–603. doi: 10.1542/peds.105.3.598. [DOI] [PubMed] [Google Scholar]
- 12.Jenuth JP, Peterson AC, Shoubridge EA. Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat Genet. 1997;16:93–95. doi: 10.1038/ng0597-93. [DOI] [PubMed] [Google Scholar]
- 13.Battersby BJ, Shoubridge EA. Selection of a mtDNA sequence variant in hepatocytes of heteroplasmic mice is not due to differences in respiratory chain function or efficiency of replication. Hum Mol Genet. 2001;10:2469–2479. doi: 10.1093/hmg/10.22.2469. [DOI] [PubMed] [Google Scholar]
- 14.Battersby BJ, Loredo-Osti JC, Shoubridge EA. Nuclear genetic control of mitochondrial DNA segregation. Nat Genet. 2003;33:183–186. doi: 10.1038/ng1073. [DOI] [PubMed] [Google Scholar]
- 15.Moreno-Loshuertos R, Acin-Perez R, Fernandez-Silva P, Movilla N, Perez-Martos A, Rodriguez de Cordoba S, et al. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat Genet. 2006;38:1261–1268. doi: 10.1038/ng1897. [DOI] [PubMed] [Google Scholar]
- 16.Inagaki K, Fuess S, Storm TA, Gibson GA, McTiernan CF, Kay MA, et al. Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Mol Ther. 2006;14:45–53. doi: 10.1016/j.ymthe.2006.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosh A, Yue Y, Long C, Bostick B, Duan D. Efficient whole-body transduction with trans-splicing adeno-associated viral vectors. Mol Ther. 2007;15:750–755. doi: 10.1038/sj.mt.6300081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wenz T, Williams SL, Bacman SR, Moraes CT. Emerging therapeutic approaches to mitochondrial diseases. Dev Disabil Res Rev. 2010;16:219–229. doi: 10.1002/ddrr.109. [DOI] [PubMed] [Google Scholar]
- 19.Lightowlers RN, Chinnery PF, Turnbull DM, Howell N. Mammalian mitochondrial genetics: heredity, heteroplasmy and disease. Trends Genet. 1997;13:450–455. doi: 10.1016/s0168-9525(97)01266-3. [DOI] [PubMed] [Google Scholar]
- 20.Chiaratti MR, Meirelles FV, Wells D, Poulton J. Therapeutic treatments of mtDNA diseases at the earliest stages of human development. Mitochondrion. 2011 doi: 10.1016/j.mito.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Poulton J, Turnbull DM. 74th ENMC international workshop: mitochondrial diseases 19–20 november 1999, Naarden, the netherlands. Neuromuscul Disord. 2000;10:460–462. doi: 10.1016/s0960-8966(00)00101-2. [DOI] [PubMed] [Google Scholar]
- 22.Tsao CY, Mendell JR, Bartholomew D. High mitochondrial DNA T8993G mutation (<90%) without typical features of Leigh's and NARP syndromes. J Child Neurol. 2001;16:533–535. doi: 10.1177/088307380101600716. [DOI] [PubMed] [Google Scholar]
- 23.Pacak CA, Mah CS, Thattaliyath BD, Conlon TJ, Lewis MA, Cloutier DE, et al. Recombinant adeno-associated virus serotype 9 leads to preferential cardiac transduction in vivo. Circ Res. 2006;99:e3–e9. doi: 10.1161/01.RES.0000237661.18885.f6. [DOI] [PubMed] [Google Scholar]
- 24.Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 25.Bostick B, Ghosh A, Yue Y, Long C, Duan D. Systemic AAV-9 transduction in mice is influenced by animal age but not by the route of administration. Gene Ther. 2007;14:1605–1609. doi: 10.1038/sj.gt.3303029. [DOI] [PubMed] [Google Scholar]
- 26.Foust KD, Nurre E, Montgomery CL, Hernandez A, Chan CM, Kaspar BK. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat Biotechnol. 2009;27:59–65. doi: 10.1038/nbt.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogura T, Mizukami H, Mimuro J, Madoiwa S, Okada T, Matsushita T, et al. Utility of intraperitoneal administration as a route of AAV serotype 5 vector-mediated neonatal gene transfer. J Gene Med. 2006;8:990–997. doi: 10.1002/jgm.916. [DOI] [PubMed] [Google Scholar]
- 28.Pruchnic R, Cao B, Peterson ZQ, Xiao X, Li J, Samulski RJ, et al. The use of adeno-associated virus to circumvent the maturation-dependent viral transduction of muscle fibers. Hum Gene Ther. 2000;11:521–536. doi: 10.1089/10430340050015716. [DOI] [PubMed] [Google Scholar]
- 29.Blankinship MJ, Gregorevic P, Allen JM, Harper SQ, Harper H, Halbert CL, et al. Efficient transduction of skeletal muscle using vectors based on adeno-associated virus serotype 6. Mol Ther. 2004;10:671–678. doi: 10.1016/j.ymthe.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 30.Lai Y, Yue Y, Liu M, Ghosh A, Engelhardt JF, Chamberlain JS, et al. Efficient in vivo gene expression by trans-splicing adeno-associated viral vectors. Nat Biotechnol. 2005;23:1435–1439. doi: 10.1038/nbt1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pettman R, Hurley T, Addis J, Robinson B, Scott H, Kronick JB. Prenatal diagnosis by amniocentesis and chorionic villus biopsy of mtDNA mutation 8993T >G. J Inherit Metab Dis. 2007;30:404. doi: 10.1007/s10545-007-0460-y. [DOI] [PubMed] [Google Scholar]
- 32.Tatuch Y, Christodoulou J, Feigenbaum A, Clarke JT, Wherret J, Smith C, et al. Heteroplasmic mtDNA mutation (T----G) at 8993 can cause Leigh disease when the percentage of abnormal mtDNA is high. Am J Hum Genet. 1992;50:852–858. [PMC free article] [PubMed] [Google Scholar]
- 33.Rossignol R, Faustin B, Rocher C, Malgat M, Mazat JP, Letellier T. Mitochondrial threshold effects. Biochem J. 2003;370:751–762. doi: 10.1042/BJ20021594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minczuk M. Engineered zinc finger proteins for manipulation of the human mitochondrial genome. Methods Mol Biol. 2010;649:257–270. doi: 10.1007/978-1-60761-753-2_16. [DOI] [PubMed] [Google Scholar]
- 35.Handel EM, Cathomen T. Zinc-finger nuclease based genome surgery: it's all about specificity. Curr Gene Ther. 2011;11:28–37. doi: 10.2174/156652311794520120. [DOI] [PubMed] [Google Scholar]
- 36.Bacman SR, Williams SL, Moraes CT. Intra- and inter-molecular recombination of mitochondrial DNA after in vivo induction of multiple double-strand breaks. Nucleic Acids Res. 2009;37:4218–4226. doi: 10.1093/nar/gkp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moraes CT, Ricci E, Bonilla E, DiMauro S, Schon EA. The mitochondrial tRNA(Leu(UUR)) mutation in mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes (MELAS): genetic, biochemical, and morphological correlations in skeletal muscle. Am J Hum Genet. 1992;50:934–949. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.