Abstract
Numb functions in progenitor cell fate determination and early development but it is also expressed in post-developmental tissues and cancers where its role is unclear. In this study, we report that a targeted knockdown of Numb expression causes a G2/M arrest and reduced cell growth in human melanoma cells. Co-immunoprecipitation and co-localization studies demonstrated that Numb interacts with the serine/threonine polo-like kinase Plk1 and Numb cycles in a cell cycle-dependent fashion along with this mitotic regulator. Interestingly, Numb expression was required for Plk1 protein stability and localization to the spindle poles during mitosis. Reduction in Numb expression resulted in mislocalization of Plk1 at both metaphase and anaphase, leading to disorganized gamma-tubulin recruitment in centrosomes. Together, our findings present a novel function for Numb during symmetric cell division. We suggest that dysregulation of Numb expression results in mislocalized Plk1 and poor centrosomal gamma-tubulin recruitment, potentially contributing to mitotic errors, aneuploidy, and cancer development.
Introduction
Proper regulation of the cell cycle, particularly DNA replication, segregation and their respective checkpoints, are crucial for correct cell ploidy and genomic integrity. An important step that ensures a proper cell division leading to two identical daughter cells is bi-polar spindle formation. A mono-polar cell is unable to divide and a multi-polar cell divides resulting in sister cells obtaining an incorrect complement of DNA, if division occurs at all. An improper cell division with immature spindle poles or with incomplete connection of the microtubules to the kinetochores can lead to the formation of aneuploid daughter cells with a possibility of cancer development (1).
The serine/threonine kinase polo-like kinase 1 (Plk1) has been shown to regulate proper spindle pole formation and maturation. Plk1 has been implicated in regulating the localization of a variety of essential centrosomal associated proteins (2). Further, Plk1 also regulates the localization of Aurora A to the centrosomes for proper maturation (3–5). Severe Plk1 inhibition results in a monopolar phenotype whereas a weaker inhibition can result in phenotypes ranging from multipolar cells, misaligned microtubules, and improper DNA condensation (6–11). Therefore, understanding the requirement and regulators of Plk1 during centrosome development will further our understanding of proper cell division and may lead to novel means for the management of cancer.
The role of Numb during asymmetric self-renewal and progenitor cell fate determination and development is well studied; principally Numb localizes asymmetrically thereby directing the two sister cells along their separate developmental paths based on their respective inheritance or absence of Numb (12–15). This is achieved through Numb’s inhibition of Notch in the sister cell containing Numb versus continued Notch activity in the Numb lacking sister cell. However, expression of Numb is also detectable in adult tissues and possible roles for Numb in these tissues are only beginning to be explored. Recent studies have shown that Numb is capable of stabilizing p53 protein (15–17), is involved in endocytosis of different molecules (18–22), and regulates cell migration (23, 24). However, increasing evidence suggests a potential tumor suppressor function for Numb in differentiated cells, including its stabilization of p53 (15–17), Numb’s antagonistic role for dysplastic promoting protein, Notch (18–20), and the loss of Numb in a variety of tumor types (25–27).
Here, for the first time, employing human melanoma cells we found that Numb is required for Plk1 regulation during mitosis. Surprisingly, in the absence of Numb, Plk1 protein half-life is reduced and its localization during metaphase is altered, thereby resulting in disorganized centrosomal γ-tubulin recruitment and aster formation. Our data suggest that Numb is required for Plk1 stability and localization to ensure proper γ-tubulin recruitment to the centrosomes, where Plk1 is of vital importance. Our study unravels a hitherto unknown function for Numb during symmetric cell division outside progenitor cell fate determination, and a potential contributing mechanism in tumors with reduced Numb expression.
Materials and Methods
Cell culture
Human melanoma cell lines A375 and HS294T; human embryonic kidney cell line, HEK 293T (ATCC; Manassas, VA) and normal human fibroblasts, TiG (a kind gift from H. Tahara) were maintained in Eagle’s Minimum Essential Medium (ATCC, VA) or Dulbecco’s Modified Eagle’s Medium (Invitrogen, CA) with 10% FBS and 1% penicillin/streptomycin at standard cell culture conditions (37°C, 5% CO2 in humidified incubator). The melanoma cells obtained were authenticated by ATCC.
Lentiviral production and transduction
For viral creation, HEK 293T cells were transfected using the CaPO4 method. Five targeting shRNAs were purchased and tested for knockdown efficiency with the clone demonstrating the most knockdown being chosen for further study. Briefly, 8 μg shRNA plasmid DNA (nonsense-NS (SHC002, Sequence: CCGGCAACAAGATGAAGAGCACC AACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTT), Plk1 (Sequence: CCGGCCCGAGGTGCTGAGCAAGAAACTCGAGTTTCTTGCTCAGCACCTCGGG TTTTTG, TRC number: TRCN0000121325, Clone ID: NM 005030.3-711s1c1) or Numb (Sequence: CCGGGCAGCTTTCAATGGTGTAGATCTCGAGATCTACACCATTGAAAGCTGCTTTTT, TRC number: TRCN0000007225, Clone ID: NM_003744.3–1996s1c1) targeting shRNA) (Sigma Aldrich, MO), 5 μg VSV-G and 6 μg Δ8.2 plasmids were mixed with sterile ddH2O to a final volume of 450 μl and mixed with 50 μl of 2.5 M CaCl2. The DNA mix was bubbled and 500 μl 2X HBS (pH 7.05) added drop wise. The transfection mixture was added to its respective plates and incubated overnight. After 24 hours, transfection media was removed and fresh media added. Cell media containing shRNA lentivirus was collected 48 and 72 hours after initial transfection and filtered for use.
For target cell transduction, viral media was added to cells with 8 μg/ml polybrene four times over two days. After 72 hours of transduction, viral media was removed and cells were collected for further experiments.
Brightfield imaging
Following lentiviral treatments, A375 cells were imaged for cell morphology under brightfield conditions using a Nikon Eclipse Ti inverted microscope (Nikon Instruments, Inc., NY) with a S Plan Fluor ELWD 20x Ph1 ADM lens at 1.5X zoom. Images were captured with a Nikon Digital Sight DS-Fi1 camera using NIS Elements AR 3.1 software. Images were converted to grayscale using Photoshop (Adobe, CA).
Cell cycle analysis
Following treatments, cells were trypsinized, washed with PBS and fixed in ice cold 100% ethanol. Cells were then washed with PBS and stained with propidium iodide (Invitrogen, CA) and analyzed on a FACScan bench top cytometer at the University of Wisconsin Paul P. Carbone Comprehensive Cancer Center (UWCCC) Flow cytometry facility. Cell cycle distribution was analyzed using CellQuest software (BD Biosciences, CA).
Trypan blue exclusion assay
Seventy-two hours post lentiviral shRNA transduction, cells were trypsinized and collected in a 1.5 ml tube, pelleted by centrifugation and re-suspended in PBS. A 10 μl aliquot of cell suspension was removed and an equivalent 10 μl of Trypan Blue was added to the cells and they were counted for determining total cell growth.
Immunoblot analysis
Following treatments, cells were trypsinized, washed with ice-cold PBS and lysed with RIPA buffer with PMSF and protease inhibitor cocktail (Pierce, IL). Protein concentration was measured with BCA Protein Assay (Pierce, IL). For immunoblot analysis, 30 μg of protein was subjected to SDS-PAGE using 10% Tris-HCl gels and transferred onto nitrocellulose membrane. Blots were blocked with 5% nonfat dry milk followed by probing with the desired primary antibodies (Plk1, Upstate, MA; Numb and Cyclin B1, Cell Signaling, MA), and then appropriate HRP-conjugated secondary antibodies followed by enhanced chemiluminescent detection. Blots were subsequently stripped and re-probed with goat anti-β-actin (Santa Cruz, CA) primary antibody followed by appropriate secondary and chemiluminescent detection as a loading control. The quantification of protein was performed by a digital analyses of protein bands (TIFF images) using UN-SCAN-IT software (Silk Scientific, Inc., UT).
Protein half-life and proteasome inhibition
The cells were treated with shNS or shNumb as above and then exposed to 75 μg/mL cycloheximide (Sigma, MO) to inhibit new protein translation. Lysates were collected at time points zero, two, four, and six hours and changes to Plk1 and Numb protein levels were evaluated using immunoblot analysis. For proteasome inhibition, cells were treated with shNS or shNumb then exposed to 10 μM MG132 (Sigma, MO) for eight hours. Lysates were collected as previously described and analyzed by immunoblot analysis.
Luciferase activity
A375 cells were co-transfected with shNS or shNumb targeting DNA in the presence of one of three luciferase reporter plasmids and β-galactosidase for normalization using Lipofectamine 2000 (Invitrogen; CA) following vendor’s protocol. The luciferase reporter plasmids consisted of Hes-Luciferase measuring Notch activity, p21-Luciferase for p53 activity, and Gli-Luciferase to measure Hedgehog signaling. Luciferase activity was normalized to β-galactosidase activity.
Co-Immunoprecipitation
The cells were trypsinized, washed with ice-cold PBS, lysed with a non-denaturing 1.0% NP-40 lysis buffer plus PMSF and protease inhibitor cocktail (Pierce, IL) and cleared by centrifugation. To immunoprecipitate endogenous proteins lysate (500 μg) was mixed with 5 μg Plk1 (Upstate, MA), Numb (Cell Signaling Technology, MA) or respective IgG (Cell Signaling Technology, MA) antibodies overnight at 4°C. The next day, UltraLink Immobilized Protein A beads (Pierce, IL) were added to the antigen-antibody complex and incubated for two hours at room temperature with rotation. Beads were recovered by low-speed centrifugation and washed with immunoprecipitation buffer (20 mM sodium phosphate (pH=7.5), 500 mM NaCl, 0.1% SDS, 1% NP-40, 0.02% sodium azide). Loading dye was added to each sample and incubated at 95°C for 5 minutes, centrifuged and subjected to SDS-PAGE. After appropriate separation, gels were transferred and analyzed for co-immunoprecipitation using standard immunoblotting techniques as detailed above.
In vitro transcription and translation
Wild type human Numb (Open Biosystems, AL) was PCR amplified, adding an N-terminal Flag-tag and then cloning the construct into pCDNA3.1 (Invitrogen, CA). Using the TNT-T7 coupled rabbit reticulocyte system (Promega, WI) Flag-Numb or empty pCDNA were transcribed in the presence of L-[35S]-Methionine EasyTag (PerkinElmer, MA) according to manufacturer’s protocol. After in vitro translation, 5% was removed to confirm translation and the remainder was mixed with 0.5 μg recombinant His-Plk1 (Invitrogen, CA) for 30 minutes. The mixture was diluted with 100 mM Tris and mixed with MagneHis Ni-particles (Promega, WI), washed and eluted as per the manufacturer’s protocol. Eluted samples were run on SDS-PAGE, dried and analyzed by autoradiography.
Double thymidine arrest and release
For double thymidine arrest and release, asynchronous cells were arrested for 16 hours with 2 mM thymidine in DMEM. Cells were thoroughly washed with PBS and released into DMEM without thymidine for 6 hours. Final synchronization was achieved with 2 mM thymidine for an additional 16 hours. For final release, cells were thoroughly washed and released into DMEM without thymidine. Cells were collected for immunoblot and cell cycle analysis as detailed above every two hours after release.
Immunofluorescence studies
For immunofluorescence staining, cells were plated and grown on BD Falcon Culture Slides (BD Biosciences, CA) and treated with shNS, shPlk1, or shNumb as previously described. The cells were fixed with 100% methanol and permeabalized with 1% Triton-X 100 in PBS and then blocked for 1 hour at room temperature in 2% bovine serum albumin in PBS plus 0.1% Triton-X 100 (blocking buffer). Following blocking, primary Plk1 (Santa Cruz, CA), Numb (Cell Signaling Technology, MA), α-tubulin or γ-tubulin (Santa Cruz, CA) antibodies (1:500 in blocking buffer) were added and allowed to incubate for 1 hour at room temperature. Primary antibody was removed, wells washed and secondary Alexa Fluor 488 or 568 (Molecular Probes, OR) (1:500 in blocking buffer) were added and incubated for 1 hour at room temperature. Cells were then washed and incubated with the second target antibody (Plk1, Numb or γ-tubulin) as above, then their respective secondary to evaluate all possible combinations. 4′, 6-diamidino-2-phenylindole, dihydrochloride (DAPI) (Pierce, IL) (1 μg/ml) counterstain was used for nuclear staining. Cells were mounted with ProLong anti-fade kit as per vendor’s protocol (Molecular Probes, OR) and examined under Nikon Eclipse Ti inverted microscope (Nikon Instruments, Inc., NY) with a Nikon Plan Fluor 60X DIC H/N2 Oil lens and an additional magnification of 1.5X. Images were acquired using a Photometrics CoolSNAP HQ2 monochrome camera and NIS Elements AR 3.1 software. To visualize Plk1 mislocalization in shNumb treated samples the Plk1 channel (either Red or Green) was increased by 50%. For mitotic error quantitation, at least 100 metaphase cells were scored for proper Plk1 localization to the spindle poles and metaphase plate. Cells were only scored as “mislocalized” when a Plk1 positive signal at least as strong as that seen at the spindle poles or mid-body was seen away from these structures.
Statistics
All results are expressed as the mean plus or minus standard deviation. Statistical analyses were performed by two-tailed Student’s T-Test. For all analyses, a P-value <0.01 was considered statistically significant.
Results and Discussion
Numb inhibition causes mitotic arrest and reduction in cell growth in melanoma cells
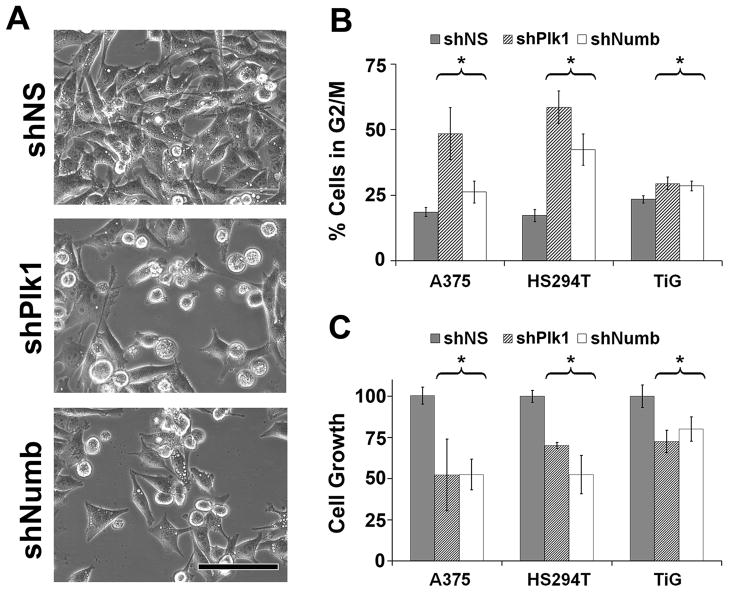
The role for Numb in asymmetric cell division is well studied (12–14, 23, 24, 28). However, Numb has displayed a gamut of tumor suppressor roles in differentiated tissues and its role in symmetric mitosis has not been explored. We first evaluated the consequence of short hairpin (sh) RNA mediated knockdown of Numb in A375 human melanoma cells and found an accumulation of rounded up, mitotic cells similar to that seen upon Plk1 inhibition (Figure 1A). Therefore, we next determined the consequence of Numb inhibition on cell cycle profile and cell growth in multiple cell types viz. A375 and HS294T melanoma cells and diploid lung fibroblasts TiG. As shown in Figure 1, similar to Plk1 inhibition, Numb inhibition resulted in an accumulation of cells in G2/M phase (Figure 1B) as well as a reduction of cell growth in all the cell lines tested (Figure 1C). This reduction in cell growth was also confirmed using colony formation assay as shown in Supplementary Figure 1.
Figure 1. Numb inhibition causes mitotic phenotype similar to Plk1 inhibition.
A) Numb knockdown cell morphology resembles that of Plk1 inhibition. A375 cells were transfected using nonsense (NS), Plk1 or Numb targeting shRNA and imaged under brightfield conditions. Scale bar = 100 μm. B) Inhibition of Numb results in a G2/M cell cycle arrest. A375 and HS294T melanoma cells and the diploid fibroblast cells, TiG, were treated with nonsense (NS), Plk1 or Numb targeting shRNA. Cells were collected and analyzed for cell cycle profile using FACS analysis. Data represents mean ± standard deviation of three separate experiments with similar results (*p<0.01). * indicates significant change in G2/M population in shPlk and shNumb treated cells versus shNS. C) Inhibition of Numb results in a reduced cell growth. A375 and HS294T melanoma cells and the diploid fibroblast cells, TiG, were treated with nonsense (NS), Plk1 or Numb targeting shRNA. Cells were collected and analyzed via trypan blue exclusion analysis. Data represents mean ± standard deviation of three separate experiments with similar results (*p<0.01). * represents significant decrease in growth in shPlk1 and shNumb treated cells versus shNS.
Numb is mitotically regulated
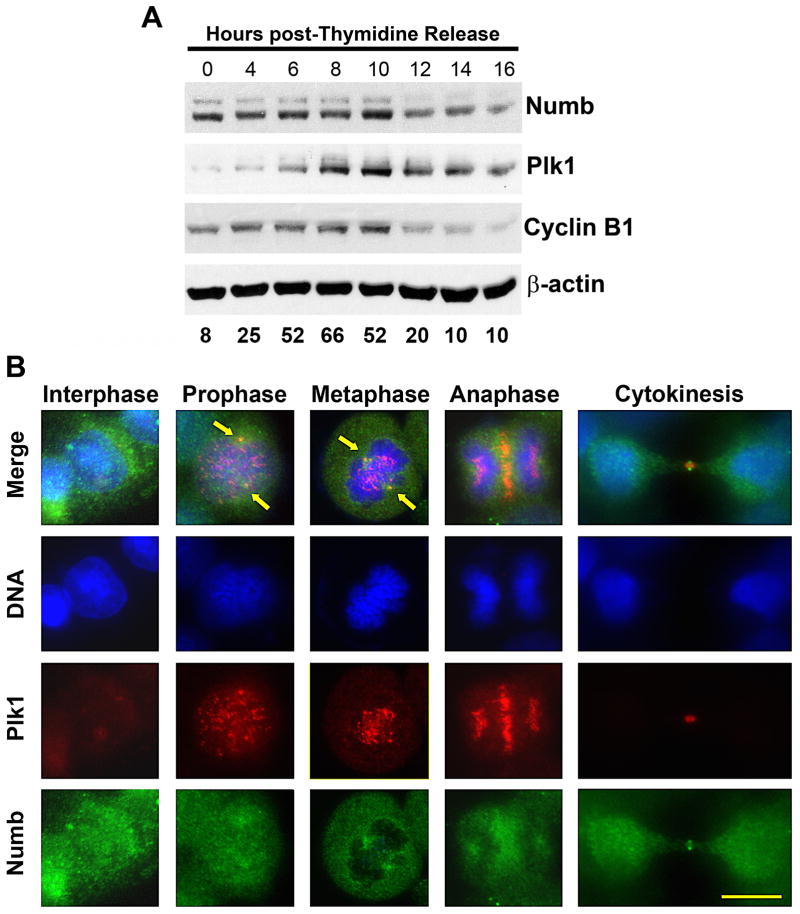
The similar cell cycle distributions seen after Plk1 and Numb knockdown would suggest a possible cell cycle dependent regulation of Numb as well. To explore this possibility, we evaluated the expression of Numb during mitosis employing thymidine arrest and release. We found that Numb protein expression rises and falls almost parallel to Plk1 expression pattern during mitotic entry and exit in A375 cells (Figure 2A) as well as in HS294T cells and TiG fibroblasts (Supplementary Figure 2). This data further supports a potential role for Numb in symmetric mitosis. However, given other established roles of Numb (21, 29), it is not expected to be solely a mitotic protein. This is supported by the expression profile of Numb protein during thymidine block and release showing that Numb is indeed expressed throughout the cell cycle, albeit at fluctuating rates and peaking with Plk1 expression and decreasing at mitotic exit.
Figure 2. Numb expression is mitotically regulated.
A) Numb expression levels peak at mitosis, following a pattern similar to Plk1 and Cyclin B1. Using a double thymidine arrest and release protocol, A375 cells were collected for immunoblot and FACS analysis every two hours. Lysates were evaluated for Numb, Plk1 and Cyclin B1. Equal loading was confirmed by re-probing the blots for β-actin. The data represent three independent experiments where average G2/M percentage at each time point is shown. B) Plk1 and Numb co-localize during mitotic progression. To determine what role Numb may have during its peak at mitosis, the asynchronous A375 cells at various stages of mitosis were evaluated using immunofluorescence analysis. Cells were fixed and stained for Plk1 (red), Numb (green) and DNA (blue). Arrows indicate the co-localization at the spindle poles during prophase and metaphase. Scale bar = 10 μm.
Numb and Plk1 co-localize during mitosis in human melanoma cells
One of the most striking aspects of Plk1 is its dynamic localization during mitosis. To evaluate if there is a specific location and role for the interaction between Numb and Plk1 during mitosis, we conducted immunofluorescence studies to assess the localization of two proteins at various stages of mitosis in A375 cells. Interestingly, we found a defined co-localization of Numb and Plk1 at the spindle poles during mitosis (Figure 2B). During interphase, Numb was diffusely localized throughout the cytoplasm and associated with the cell membrane, which is consistent with its known role in endocytosis. Plk1, as expected, was barely detectable during interphase. As cells enter prophase, Plk1 began to localize to the kinetochores and spindle poles and Numb showed an increasing localization overlapping Plk1 at the spindle poles. During metaphase, Plk1 localization was maintained at the spindle poles and a pronounced increase in Numb localization was seen. Interestingly, whereas Plk1 localizes along the metaphase plate to fulfill its role in the spindle assembly checkpoint and chromosome segregation, Numb localization did not overlap DNA to any detectable level, suggesting that the regulation of Plk1 by Numb is restricted to Plk1’s role at the spindle poles. Further, as chromosome segregation occurred in anaphase, Plk1 expression was maintained at the mid-body but the punctate signal at the spindle poles was lost with a diffuse expression maintained in each sister cell. Numb localization was similar, with localization extending into the cytoplasm surrounding the mid-body and a diffuse expression remaining in each sister cell. Finally, during cytokinesis both Plk1 and Numb signals were detected at the mid-body. The localization of Numb to the spindle poles appears to be conserved in mouse oocyte meiosis (30), and based on our data we believe this localization is conserved across both mitotic and meiotic cell division.
Numb regulates Plk1 protein
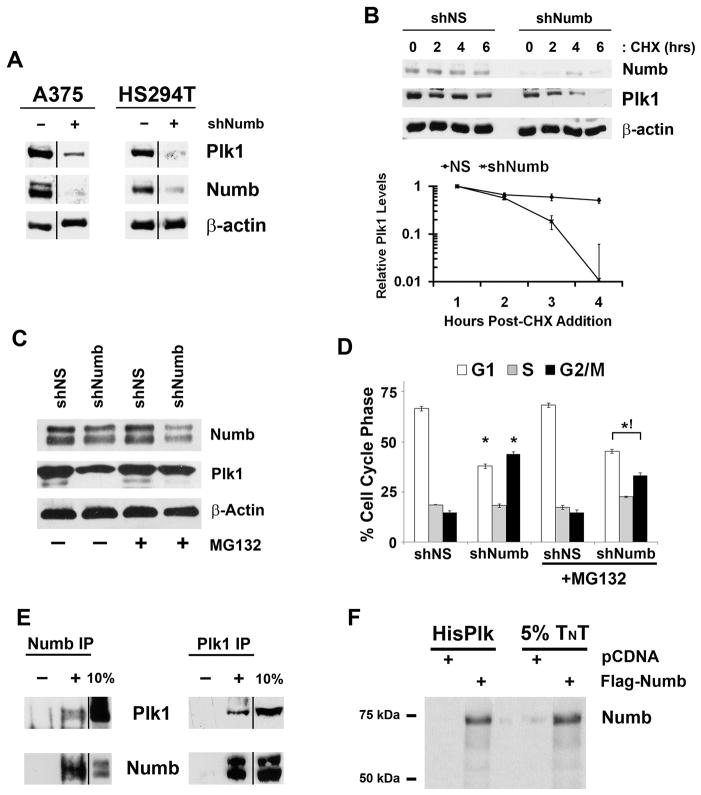
The cell cycle distribution after Numb inhibition closely resembled to that seen upon Plk1 inhibition (Figure 1B). Therefore, we evaluated the effects of Numb inhibition on Plk1 protein expression (Figure 3A). Because Numb inhibition caused a G2/M phase cell cycle arrest we expected an increase in Plk1 protein levels similar to that seen with a G2/M enriched cell population (Figure 2A, ten hours after thymidine release). Surprisingly, we found that Numb knockdown resulted in a significant reduction in Plk1 protein expression in A375 and HS294T cells (Figure 3A). To determine if this effect was due to changes in Plk1 protein stability, we treated cells with cycloheximide (CHX) after Numb knockdown and found that loss of Numb significantly reduced Plk1 protein half-life in A375 cells (Figure 3B) as well as HS294T cells (Supplementary Figure 3). The observed down-regulation of Plk1 is possibly the factor contributing to the cell cycle arrest because Plk1 inhibition has been shown to cause a G2/M phase arrest and apoptosis in multiple cell lines (8–11, 31). To confirm that the reduction in Plk1 protein half-life is not an indirect consequence of Numb’s other non-asymmetric roles (such as Notch and Hedgehog endocytosis and p53 stabilization (15–20)), we assesed the transcriptional activity of these three pathways following Numb knockdown and did not find any appreciable change in their respective promoter activities (Supplementary Figure 4). Plk1 protein is regulated by ubiquitin dependent degradation. To evaluate if Numb is stabilizing Plk1 levels through blocking Plk1 degradation we knocked down Numb in the presence and absence of the proteasomal inhibitor MG132. Figure 3C demonstrates that the addition of MG132 results in Plk1 protein levels similar to that seen in shNS controls. Additionally, the normalization of Plk1 levels with MG132 addition slightly, though significantly, reverses the effects of the cell cycle profile to that seen in control (Figure 3D). Next, employing immunoprecipitation studies, we assessed if Numb and Plk1 were interacting with each other. Our data demonstrated that endogenous Numb and Plk1 reciprocally co-immunoprecipitate, whereas matched IgG control immunoprecipitations are negative (Figure 3E). Additionally, in vitro translated Numb was found to be co-immunoprecipitated with His-tagged recombinant Plk1 (Figure 3F). These data suggested that Numb directly binds to and is required for Plk1 stabilization.
Figure 3. Numb regulates Plk1 protein levels through direct interaction.
A) Numb knockdown decreases Plk1 expression. A375 and HS294T cells were treated with nonsense (NS), Plk1 or Numb targeting shRNA, lysates were collected and evaluated via immunoblot analysis for Plk1 and Numb. Equal loading was confirmed by re-probing the blots for β-actin. Results shown are from the same membrane, line denotes removal of a single lane between the two samples. B) Numb is required for Plk1 protein stability. To evaluate if Numb positively regulates Plk1 protein half-life, A375 cells were treated with nonsense (NS) or Numb targeting shRNA then with cycloheximide (CHX) to inhibit new protein synthesis. Following treatment with CHX (0, 2, 4 and 6 hours), lysates were collected and analyzed by immunoblot analysis for Plk1 and Numb. Equal loading was confirmed by re-probing the blots for β-actin. The protein levels were quantitated by a densitometric analysis of protein bands. The data (relative density normalized to β-actin) is expressed as mean ± standard deviation of three experiments on a log10 scale. C) Numb stabilizes Plk1 protein by blocking proteasomal degradation. A375 cells were treated with nonsense (NS) or Numb targeting shRNA in the presence or absence of 10 μM MG132 to block proteasome mediated degradation for 8 hours. Following treatment, lysates were collected and analyzed by immunoblot analysis for Plk1, Numb and β-actin as a loading control. D) Proteasome inhibition normalizes the cell cycle abnormalities of Numb knockdown. After treatments described in Figure 2C, cells were collected and analyzed for cell cycle profile using FACS analysis. Data represents mean ± standard deviation of three separate experiments with similar results (*p<0.01 relative to shNS within respective vehicle or MG132 treatment, !p<0.01 relative to shNumb of vehicle treated cells). E) Endogenous Plk1 and Numb co-immunoprecipitate in vivo. Cell lysates were prepared from actively dividing A375 cells and immunoprecipitations were performed with Plk1, Numb or their respective IgG control antibodies. The immunoprecipitates were separated by SDS-PAGE with a 10% aliquot of untreated lysate and probed for the converse target using immunoblot analysis. “−” denotes addition of animal specific IgG inclusion as control, “+” denotes antibody included in immunoprecipitation. Results shown here are from same membrane, the line denotes the removal of extraneous lanes. F) Plk1 and Numb directly interact in vitro. To confirm if the interaction between Plk1 and Numb is a direct interaction or requires a third priming kinase or mediator we in vitro translated and transcribed Flag-Numb plus [35S]-Methionine using Promega’s TNT system. The TNT product was mixed with His-Tagged recombinant Plk1 and purified using Ni-Magne-His purification system. Purified lysates were separated using SDS- PAGE along with 5% of the TNT reactions, dried and analyzed by autoradiography.
Numb regulates Plk1 localization during metaphase
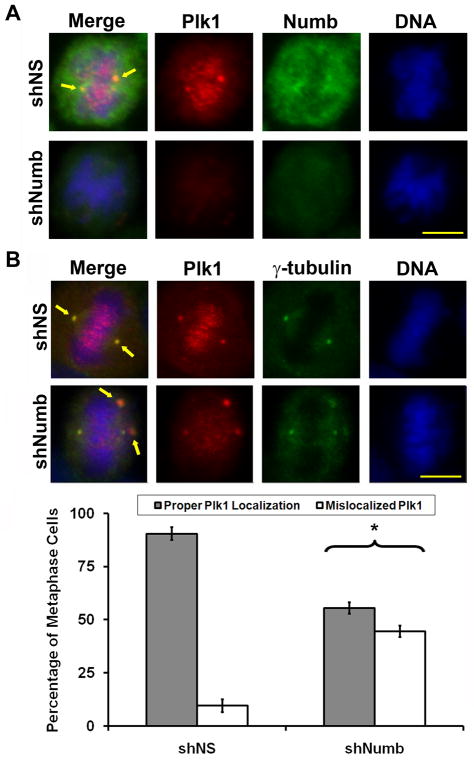
With co-localization of Numb and Plk1 to two vital Plk1 locales during mitosis, we wanted to evaluate the mitotic changes that occur following the loss of Numb. We determined the effect of Numb knockdown on Plk1 localization at various stages of mitosis. Immunofluorescent staining of Plk1 after Numb knockdown showed reduced levels of Plk1 intensity, as expected (Figure 4A). There was no change in the number of mono-polar prophase cells suggesting that Numb is not required for centrosome duplication and separation (Data not shown). However, evaluation of the remaining detectable Plk1 demonstrates a striking increase in mislocalized Plk1 during metaphase upon Numb knockdown (see Figure 4B, Plk1 intensity enhanced to visualize mislocalization). The staining resembles a phenotype consistent with a multi-polar cell. To test if this mislocalized Plk1 was a consequence of staining a multi-polar phenotype or is truly mislocalized Plk1, we performed immunofluorescence staining of A375 and TiG cells for both Plk1 and γ-tubulin following Numb inhibition (Figure 4B and Supplementary Figure 5, Plk1 intensity enhanced to visualize mislocalization). We did not find a change in the number of mono-, bi-, or multi-polar cells. However, a significant increase in the number of mitotic cells showing diffuse and disorganized centrosomal γ-tubulin recruitment was observed (Figure 4B and Supplementary Figure 5). We also evaluated changes in microtubule structure by staining for α-tubulin after Numb knockdown (Figure 5). Interestingly we found that microtubule formation and orientation does not seem to be affected by the mislocalized Plk1, as bipolar and kinetochore orientation and attachment appear to be present. However, treatment with shNumb lentivirus results in reduced aster microtubule formation. These results support the idea that γ-tubulin recruitment to the centrosomes is reduced after Numb inhibition (Figure 4B) as γ-tubulin is required for proper mitotic aster formation (32).
Figure 4. Numb regulates the localization of Plk1 during metaphase.
A) Plk1 localization is dysregulated after Numb knockdown. A375 melanoma cells were treated with nonsense (NS) or Numb targeting shRNA then fixed and stained for Plk1 (red), Numb (green) or DNA (blue). Representative metaphase images from each treatment are shown. Arrows indicate the typical overlap of Plk1 and Numb staining. B) Mislocalization of Plk1 during Numb knockdown results in disorganized centrosomal γ-tubulin recruitment. To evaluate if the mislocalized Plk1 after Numb knockdown is a result of a multipolar phenotype, A375 melanoma cells were treated with nonsense (NS) or Numb targeting shRNA, fixed and stained for Plk1 (red), γ-tubulin (green), or DNA (blue). Because Numb knockdown results in decreased Plk1 staining intensity the Red channel (Plk1) was increased by 50% to visualize mislocalized Plk1. At least 100 metaphase cells in three independent trials (non mono-polar) were counted and scored as possessing proper Plk1 localization (aligned at the centrosomes with γ-tubulin) or mislocalized Plk1 (increased punctate signal off the centrosome or metaphase plate). Data represents the mean ± standard deviation of three independent trials (*p<0.01). Arrows indicate normal Plk1 and γ-tubulin overlap in first row and mislocalized Plk1 away from the centrosome and/or off the metaphase plate in the second row. Scale bar = 10 μm.
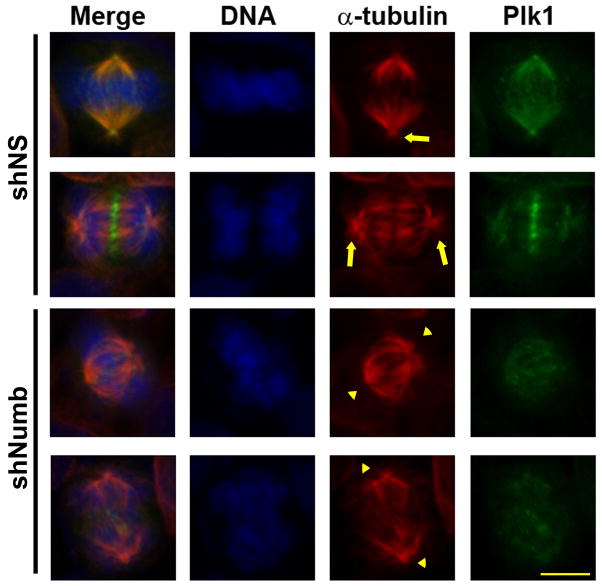
Figure 5. Reduced γ-tubulin recruitment results in loss of mitotic aster formation.
To determine if loss of Numb affects α-tubulin dynamics A375 melanoma cells were treated with nonsense (NS) or Numb targeting shRNA, fixed and stained for Plk1 (green), α-tubulin (red), or DNA (blue). Because Numb knockdown results in decreased Plk1 staining intensity the Green channel (Plk1) was increased by 50% to visualize mislocalized Plk1. Arrows indicate presence of aster microtubules and arrowheads highlight the lack of aster microtubules. Scale bar = 10μM.
Evidence in Drosophila has shown that polo, a Plk1 homolog, regulates the localization of numb during asymmetric division of sensory progenitors (28, 31). In that study, however, polo was shown to regulate the localization of Numb through the mediator partner of numb. Here we demonstrate that Numb expression is required for Plk1 stability and localization during symmetric cell division.
Gamma-tubulin recruitment is essential for proper centrosome maturation, mitotic aster formation, and microtubule nucleation (33) and Plk1 has been shown to play an important role in localization of γ-tubulin to the spindle poles (6, 34). Immature centrosome development can lead to dysregulated cell division and aneuploidy (35). Our data demonstrates a previously unknown function for Numb through proper localization of Plk1 and γ-tubulin during mitosis. Numb controls Plk1 through two mechanisms. First, Numb stabilizes Plk1 protein to ensure its presence during mitosis. Second, Numb binding to Plk1 is needed for proper localization to the centrosomes for sufficient γ-tubulin recruitment needed for centrosome maturation. After Numb knockdown, there is reduced Plk1 expression and mislocalization causing a reduction in γ-tubulin recruitment to the centrosomes and improper mitotic aster formation. A potential consequence of this is the cell cycle arrest observed in our study. However, progression of cells through mitosis with immature centrosomes enhances the risk for an aneuploid phenotype (35). We propose that loss of Numb may contribute to tumorigenesis through dysregulation of Plk1 and γ-tubulin recruitment leading to chromosome abnormalities and aneuploidy.
A tumor suppressor role for Numb has been suggested previously. Numb has been shown to form an intricate complex with the tumor suppressor p53 and the oncoprotein Mdm2 (15–17). Though the complete mechanism has not been elucidated, it appears that Numb stabilizes p53 protein through blocking Mdm2 binding and ubiquitination of p53 (16). Additionally, Mdm2 is capable of targeting Numb for degradation, though the upstream signals for Numb to release p53 allowing Mdm2 targeted ubiquitination, and for Mdm2 ubiquitination of Numb itself, are still unclear (15, 17). Also, Plk1 is a negative regulator of p53 upon DNA damage checkpoint exit, both directly through binding and phosphorylation (36), and indirectly through Plk1’s regulation of Mdm2 and Topors, two regulators of p53 turnover (37–39). Though important for tumorigenesis, and another possible convergence of Plk1 and Numb, the intricate dynamics between the DNA damage checkpoint transitioning into mitosis and the spindle assembly checkpoint are beyond the scope of our study. However, it raises some important questions with regards to the possibility of premature exit from the DNA damage checkpoint into mitosis further reducing cellular integrity. It may be important to determine p53 expression and activity following knockdown of both Numb and Plk1. Thus, if Numb stabilizes p53, and Plk1 counters this via increasing p53 turnover, it will be interesting to determine the consequence of loss of both Numb and Plk1. Does this reduce p53 functional fidelity? And if so, does it result in a premature entry into mitosis further abrogating the potential for aneuploidy? Numb also demonstrates tumor suppressor roles through recruitment of the ubiquitin ligase Itch to target both Notch and Gli1 for degradation, two transcription factors shown to contribute to tumorigenesis (18–20). However, this does not explain the mitotic phenotypes presented above and our data confirms p53, Notch, and Hedgehog transcriptional activity are unchanged in the presence of Numb knockdown (Supplementary Figure 4).
In this study the shRNA reduces expression of all four isoforms of Numb. This is important because there is evidence in Drosophila that overexpression of the PRRS isoforms induces differentiation, conversely overexpression of the PRRL isoforms promotes proliferation (40). These findings are confirmed by the same group overexpressing the different isoforms in mammalian cortical cultures (41). However, conflicting evidence shows no difference in differentiation and proliferation when the isoforms are expressed during mammalian embryogenesis (13). Therefore, more research needs to be done to determine if the mitotic phenotype demonstrated here is due to the loss of one or all Numb isoforms.
A study by Bric et al screened for and found ten candidate tumor suppressor genes that contributed to increased lymphomagenesis using a high throughput in vivo shRNA approach (42). In this study, Numb was found to be one of the potential tumor suppressors contributing to increased tumorigenesis. Indeed, the mechanism by which loss of Numb contributes to increased tumor development was not evaluated in this study and its regulation of p53 cannot be excluded. However, based on our findings we believe that the loss of Numb’s regulation on Plk1 stabilization and localization during cell division may be one mechanism, in addition to its role in p53 stability, contributing to tumor development by retarding the maturation process of the spindle poles necessary for proper cell division.
Supplementary Material
Acknowledgments
This work was partially supported by funding from the NIH (NIEHS T32 ES007015 to TLS; and NIAMS R01AR059130 to NA) and the Department of Veterans Affairs (Merit Review funding to NA).
Footnotes
Conflict of Interest: No Conflict of Interest
References
- 1.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–87. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol. 2009;10:265–75. doi: 10.1038/nrm2653. [DOI] [PubMed] [Google Scholar]
- 3.Chan EH, Santamaria A, Sillje HH, Nigg EA. Plk1 regulates mitotic Aurora A function through betaTrCP-dependent degradation of hBora. Chromosoma. 2008;117:457–69. doi: 10.1007/s00412-008-0165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Luca M, Lavia P, Guarguaglini G. A functional interplay between Aurora-A, Plk1 and TPX2 at spindle poles: Plk1 controls centrosomal localization of Aurora-A and TPX2 spindle association. Cell Cycle. 2006;5:296–303. doi: 10.4161/cc.5.3.2392. [DOI] [PubMed] [Google Scholar]
- 5.van Leuken R, Clijsters L, van ZW, Lim D, Yao X, Wolthuis RM, et al. Polo-like kinase-1 controls Aurora A destruction by activating APC/C-Cdh1. PLoSOne. 2009;4:e5282. doi: 10.1371/journal.pone.0005282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lane HA, Nigg EA. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J Cell Biol. 1996;135:1701–13. doi: 10.1083/jcb.135.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Llamazares S, Moreira A, Tavares A, Girdham C, Spruce BA, Gonzalez C, et al. polo encodes a protein kinase homolog required for mitosis in Drosophila. Genes Dev. 1991;5:2153–65. doi: 10.1101/gad.5.12a.2153. [DOI] [PubMed] [Google Scholar]
- 8.McInnes C, Mazumdar A, Mezna M, Meades C, Midgley C, Scaerou F, et al. Inhibitors of Polo-like kinase reveal roles in spindle-pole maintenance. Nat Chem Biol. 2006;2:608–17. doi: 10.1038/nchembio825. [DOI] [PubMed] [Google Scholar]
- 9.Schmit TL, Zhong W, Setaluri V, Spiegelman VS, Ahmad N. Targeted depletion of Polo-like kinase (Plk) 1 through lentiviral shRNA or a small-molecule inhibitor causes mitotic catastrophe and induction of apoptosis in human melanoma cells. J Invest Dermatol. 2009;129:2843–53. doi: 10.1038/jid.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spankuch-Schmitt B, Bereiter-Hahn J, Kaufmann M, Strebhardt K. Effect of RNA silencing of polo-like kinase-1 (PLK1) on apoptosis and spindle formation in human cancer cells. J Natl Cancer Inst. 2002;94:1863–77. doi: 10.1093/jnci/94.24.1863. [DOI] [PubMed] [Google Scholar]
- 11.Spankuch-Schmitt B, Wolf G, Solbach C, Loibl S, Knecht R, Stegmuller M, et al. Downregulation of human polo-like kinase activity by antisense oligonucleotides induces growth inhibition in cancer cells. Oncogene. 2002;21:3162–71. doi: 10.1038/sj.onc.1205412. [DOI] [PubMed] [Google Scholar]
- 12.Gulino A, Di ML, Screpanti I. The multiple functions of Numb. Exp Cell Res. 2010;316:900–6. doi: 10.1016/j.yexcr.2009.11.017. [DOI] [PubMed] [Google Scholar]
- 13.Petersen PH, Tang H, Zou K, Zhong W. The enigma of the numb-Notch relationship during mammalian embryogenesis. Dev Neurosci. 2006;28:156–68. doi: 10.1159/000090761. [DOI] [PubMed] [Google Scholar]
- 14.Petersen PH, Zou K, Krauss S, Zhong W. Continuing role for mouse Numb and Numbl in maintaining progenitor cells during cortical neurogenesis. Nat Neurosci. 2004;7:803–11. doi: 10.1038/nn1289. [DOI] [PubMed] [Google Scholar]
- 15.Yogosawa S, Miyauchi Y, Honda R, Tanaka H, Yasuda H. Mammalian Numb is a target protein of Mdm2, ubiquitin ligase. Biochem Biophys Res Commun. 2003;302:869–72. doi: 10.1016/s0006-291x(03)00282-1. [DOI] [PubMed] [Google Scholar]
- 16.Colaluca IN, Tosoni D, Nuciforo P, Senic-Matuglia F, Galimberti V, Viale G, et al. NUMB controls p53 tumour suppressor activity. Nature. 2008;451:76–80. doi: 10.1038/nature06412. [DOI] [PubMed] [Google Scholar]
- 17.Juven-Gershon T, Shifman O, Unger T, Elkeles A, Haupt Y, Oren M. The Mdm2 oncoprotein interacts with the cell fate regulator Numb. Mol Cell Biol. 1998;18:3974–82. doi: 10.1128/mcb.18.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di ML, Ferretti E, Greco A, De SE, Po A, Sico MA, et al. Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nat Cell Biol. 2006;8:1415–23. doi: 10.1038/ncb1510. [DOI] [PubMed] [Google Scholar]
- 19.McGill MA, Dho SE, Weinmaster G, McGlade CJ. Numb regulates post-endocytic trafficking and degradation of Notch1. J Biol Chem. 2009;284:26427–38. doi: 10.1074/jbc.M109.014845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J Biol Chem. 2003;278:23196–203. doi: 10.1074/jbc.M302827200. [DOI] [PubMed] [Google Scholar]
- 21.Santolini E, Puri C, Salcini AE, Gagliani MC, Pelicci PG, Tacchetti C, et al. Numb is an endocytic protein. J Cell Biol. 2000;151:1345–52. doi: 10.1083/jcb.151.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sorensen EB, Conner SD. AAK1 regulates Numb function at an early step in clathrin-mediated endocytosis. Traffic. 2008;9:1791–800. doi: 10.1111/j.1600-0854.2008.00790.x. [DOI] [PubMed] [Google Scholar]
- 23.Nishimura T, Kaibuchi K. Numb controls integrin endocytosis for directional cell migration with aPKC and PAR-3. Dev Cell. 2007;13:15–28. doi: 10.1016/j.devcel.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Rasin MR, Gazula VR, Breunig JJ, Kwan KY, Johnson MB, Liu-Chen S, et al. Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat Neurosci. 2007;10:819–27. doi: 10.1038/nn1924. [DOI] [PubMed] [Google Scholar]
- 25.Yan B, Omar FM, Das K, Ng WH, Lim C, Shiuan K, et al. Characterization of Numb expression in astrocytomas. Neuropathology. 2008;28:479–84. doi: 10.1111/j.1440-1789.2008.00907.x. [DOI] [PubMed] [Google Scholar]
- 26.Maiorano E, Favia G, Pece S, Resta L, Maisonneuve P, Di Fiore PP, et al. Prognostic implications of NUMB immunoreactivity in salivary gland carcinomas. Int J Immunopathol Pharmacol. 2007;20:779–89. doi: 10.1177/039463200702000414. [DOI] [PubMed] [Google Scholar]
- 27.Rennstam K, McMichael N, Berglund P, Honeth G, Hegardt C, Ryden L, et al. Numb protein expression correlates with a basal-like phenotype and cancer stem cell markers in primary breast cancer. Breast Cancer Res Treat. 2010;122:315–24. doi: 10.1007/s10549-009-0568-x. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Ouyang Y, Somers WG, Chia W, Lu B. Polo inhibits progenitor self-renewal and regulates Numb asymmetry by phosphorylating Pon. Nature. 2007;449:96–100. doi: 10.1038/nature06056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pece S, Confalonieri S, PRR, Di Fiore PP. NUMB-ing down cancer by more than just a NOTCH. Biochim Biophys Acta. 2011;1815:26–43. doi: 10.1016/j.bbcan.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 30.Lv H, Wang JC, Wu KL, Gao X, Wang LC, You L, et al. Numb regulates meiotic spindle organisation in mouse oocytes. Reprod Fertil Dev. 2010;22:664–72. doi: 10.1071/RD09236. [DOI] [PubMed] [Google Scholar]
- 31.Lenart P, Petronczki M, Steegmaier M, Di FB, Lipp JJ, Hoffmann M, et al. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr Biol. 2007;17:304–15. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 32.Kotani T, Yamashita M. Overexpression of truncated gamma-tubulins disrupts mitotic aster formation in Xenopus oocyte extracts. Biochem J. 2005;389:611–7. doi: 10.1042/BJ20050243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gueth-Hallonet C, Antony C, Aghion J, Santa-Maria A, Lajoie-Mazenc I, Wright M, et al. gamma-Tubulin is present in acentriolar MTOCs during early mouse development. J Cell Sci. 1993;105 ( Pt 1):157–66. doi: 10.1242/jcs.105.1.157. [DOI] [PubMed] [Google Scholar]
- 34.Glover DM. Polo kinase and progression through M phase in Drosophila: a perspective from the spindle poles. Oncogene. 2005;24:230–7. doi: 10.1038/sj.onc.1208279. [DOI] [PubMed] [Google Scholar]
- 35.Lingle WL, Lukasiewicz K, Salisbury JL. Deregulation of the centrosome cycle and the origin of chromosomal instability in cancer. Adv Exp Med Biol. 2005;570:393–421. doi: 10.1007/1-4020-3764-3_14. [DOI] [PubMed] [Google Scholar]
- 36.Ando K, Ozaki T, Yamamoto H, Furuya K, Hosoda M, Hayashi S, et al. Polo-like kinase 1 (Plk1) inhibits p53 function by physical interaction and phosphorylation. J Biol Chem. 2004;279:25549–61. doi: 10.1074/jbc.M314182200. [DOI] [PubMed] [Google Scholar]
- 37.Dias SS, Hogan C, Ochocka AM, Meek DW. Polo-like kinase-1 phosphorylates MDM2 at Ser260 and stimulates MDM2-mediated p53 turnover. FEBS Lett. 2009;583:3543–8. doi: 10.1016/j.febslet.2009.09.057. [DOI] [PubMed] [Google Scholar]
- 38.Yang X, Li H, Zhou Z, Wang WH, Deng A, Andrisani O, et al. Plk1-mediated phosphorylation of Topors regulates p53 stability. J Biol Chem. 2009;284:18588–92. doi: 10.1074/jbc.C109.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie S, Wu H, Wang Q, Cogswell JP, Husain I, Conn C, et al. Plk3 functionally links DNA damage to cell cycle arrest and apoptosis at least in part via the p53 pathway. J Biol Chem. 2001;276:43305–12. doi: 10.1074/jbc.M106050200. [DOI] [PubMed] [Google Scholar]
- 40.Verdi JM, Bashirullah A, Goldhawk DE, Kubu CJ, Jamali M, Meakin SO, et al. Distinct human NUMB isoforms regulate differentiation vs. proliferation in the neuronal lineage. Proc Natl Acad Sci U S A. 1999;96:10472–6. doi: 10.1073/pnas.96.18.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bani-Yaghoub M, Kubu CJ, Cowling R, Rochira J, Nikopoulos GN, Bellum S, et al. A switch in numb isoforms is a critical step in cortical development. Dev Dyn. 2007;236:696–705. doi: 10.1002/dvdy.21072. [DOI] [PubMed] [Google Scholar]
- 42.Bric A, Miething C, Bialucha CU, Scuoppo C, Zender L, Krasnitz A, et al. Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model. Cancer Cell. 2009;16:324–35. doi: 10.1016/j.ccr.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.