Abstract
Structural modeling of macromolecular complexes greatly benefits from interactive visualization capabilities. Here we present the integration of several modeling tools into UCSF Chimera. These include comparative modeling by MODELLER, IMP simultaneous fitting of multiple components into electron microscopy density maps by IMP MultiFit, computing of small-angle X-ray scattering profiles and fitting of the corresponding experimental profile by IMP FoXS, and assessment of amino acid sidechain conformations based on rotamer probabilities and local interactions by Chimera.
Keywords: Integrative structural modeling, restraint-based modeling, electron microscopy, small-angle X-ray scattering, interactive molecular visualization
Introduction
Proteins carry out their functions through interactions with other molecules. Of particular interest here are assemblies of multiple proteins, which are often large, dynamic, flexible, and fragile, contributing to the difficulty of determining their structures. Even when single structure determination methods fail, however, atomic models of assemblies can be determined by combining multiple types of experimental data, including those from X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy, electron microscopy (EM), small-angle X-ray scattering (SAXS), cross-linking, mass spectrometry (MS), and affinity purification (Alber et al., 2008; Lasker et al., 2010a; Robinson et al., 2007). Computational integration of diverse experimental data into an ensemble of models that best satisfy the data is not yet an entirely automated process. Therefore, visualization software, used to setup calculations, assess the results, and troubleshoot problems, is essential for the quality and efficiency of iterative integrative structure modeling.
A common structure determination approach is the fitting of crystal structures and comparative models into an EM map of the full molecular assembly. The structures can be fit as rigid bodies by sampling globally (Fabiola and Chapman, 2005) or locally (Goddard et al., 2007; Pintilie et al., 2010). Methods of flexible fitting include molecular dynamics (Trabuco et al., 2008), Monte Carlo (Topf et al., 2008), normal mode analysis (Tama et al., 2004), and morphing (Wriggers, 2010; Wriggers and Chacón, 2001). Restraints such as symmetry (Navaza et al., 2002) and intermolecular distances (Rossmann et al., 2001) can be incorporated into the fitting process. Which available method is best depends on many factors, including the resolution and symmetry of the density map, the availability of additional restraints, and the accuracy of component models.
SAXS profiles have been used widely for low-resolution structural characterization of molecules in solution (Petoukhov and Svergun, 2007; Putnam et al., 2007; Schneidman-Duhovny et al., 2010). While a SAXS profile can be converted into an assembly envelope that can in turn be used directly for fitting component molecules (Svergun, 1999), the SAXS measurement has a relatively low information content - the rotationally averaged scattering intensity versus the scattering angle approximately determines only the system’s radial distribution function. Thus, a good use of an experimental SAXS profile is to compare it to a profile computed from a 3D structural model that was derived from other data (Pons et al., 2010; Schneidman-Duhovny et al., 2010; Svergun et al., 1995). Also, changes in assembly conformation or composition under variations of pH, salt, temperature, cofactors, and drugs can be recognized, and candidate models ranked by comparison of experimental and model-derived SAXS profiles.
Atomic assembly models often generate invaluable testable hypotheses. For example, models predict which residues are in contact at intermolecular interfaces and thus may be essential for assembly formation and function. In models built from individual X-ray crystal structures, the sidechain conformations may not reflect those in the complete assembly, either because of the induced fit or modeling errors. Thus, analysis of sidechain rotamers is useful for assessing residue interactions in the complex (Guharoy et al., 2010).
We have recently integrated comparative (homology) protein structure modeling by MODELLER (Fiser et al., 2000; Marti-Renom et al., 2000; Sali and Blundell, 1993), multiple simultaneous fitting into EM maps by IMP MultiFit (Lasker et al., 2010a; Lasker et al., 2009), SAXS profile fitting by IMP FoXS (Forster et al., 2008; Schneidman-Duhovny et al., 2010), and evaluation of sidechain conformations from backbone-dependent and backbone-independent rotamer libraries (Dunbrack, 2002; Lovell et al., 2000) into the UCSF Chimera molecular visualization package. These capabilities augment over 100 tools already provided by Chimera for the interactive analysis of atomic models, density maps, and protein sequences (Couch et al., 2006; Goddard et al., 2005; Goddard et al., 2007; Meng et al., 2006; Morris et al., 2007; Pettersen et al., 2004; Pintilie et al., 2010). Chimera provides graphical user interfaces to simplify setting up input data and parameters for the fitting process, evaluating results, and performing cycles of refinement for building models of macromolecular assemblies. The homology modeling, EM fitting, and SAXS calculations are launched from Chimera and executed remotely via MODELLER- and IMP-based web services (Russel et al., 2011), with results displayed in the molecular visualization environment as they become available. The web service approach allows incorporation of improvements without the user installing new software, and can provide transparent access to more powerful computing resources. Optionally, calculations can also be performed using locally installed copies of MODELLER and IMP. MultiFit and FoXS are part of the Integrated Modeling Platform (IMP) package (Russel et al., 2011) that performs simultaneous optimization of multiple restraint types to generate ensembles of assembly structures consistent with diverse experimental data (Alber et al., 2008; Lasker et al., 2010b; Robinson et al., 2007). The Chimera user interfaces described here are a first step towards a more comprehensive graphical user interface (GUI) to control and visualize results from this suite of tools.
Next, we describe the current assembly modeling tools and then illustrate their range of capabilities on two example systems, GroEL chaperonin and ARP2/3.
Modeling functionalities in Chimera
MODELLER
MODELLER is used for homology or comparative modeling of protein three-dimensional structures (Eswar et al., 2001; Marti-Renom et al., 2000). The user provides an initial alignment of the sequence to be modeled (“target”) to the sequence(s) of one or more known structures (“templates”). MODELLER then calculates a set of plausible structures containing all non-hydrogen atoms. MODELLER implements comparative modeling by satisfaction of spatial restraints (Sali and Blundell, 1993) and can perform many additional tasks, including fold assignment, sequence- and structure-based alignments, de novo modeling of loops, and model assessment (Fiser and Sali, 2003; Fiser et al., 2000).
MODELLER Interface in Chimera
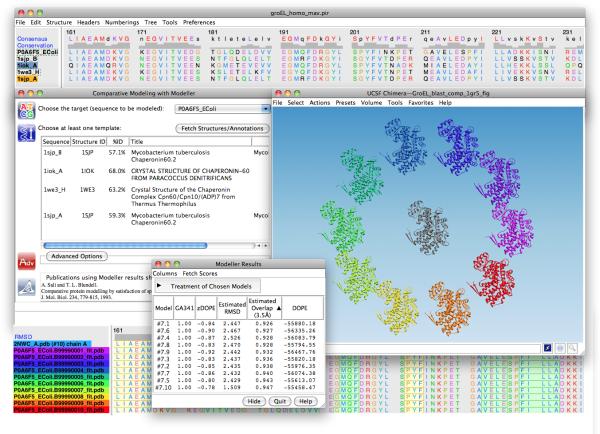
In Chimera, the modeling process can be initiated by input of only the target sequence, or if already available, a sequence alignment including the target and at least one other sequence for which a structure is known. If the input is only the target sequence, BLAST is used via a web service to search the PDB database for potential templates. The user can choose one or more of the hits to be fetched from the PDB and to be included along with the query (target) in a sequence alignment. Sequence alignments in Chimera are displayed in the Multalign Viewer tool (Meng et al., 2006). This tool includes many features, such as automatic sequence-structure communication, calculation of measures of conservation, and simple editing (e.g., adjusting gaps as well as adding and deleting sequences). When the alignment is satisfactory, the user can choose Structure -> Modeller Tools from the Multalign Viewer menu to set up and launch the MODELLER calculation, to be run either locally or via a web service. The process is run in the background and can be monitored with Chimera’s task manager. When the results become available, the models are displayed in Chimera and their associated scores shown in a table (Figure 1). The table lists the GA341 (Melo et al., 2002), zDOPE and DOPE scores (Pieper et al., 2011; Shen and Sali, 2006). Clicking “Fetch Scores” triggers a web service that calculates additional model scores: the estimated RMSD and estimated overlap (Eramian et al., 2008).
Figure 1.
Chimera MODELLER interface and homology models of a monomer of E. coli GroEL. A multiple sequence alignment of the target with hits from a Chimera BLAST query is displayed in Multalign Viewer (top window). In the Comparative Modeling with Modeller dialog (left), the P0A6F5_EColi (GroEL) sequence is selected as the target and sequences 1sjp_A, 1sjp_B, 1iok_A and 1we3_H are designated as templates. The resulting modeled structures are shown in a circle around the experimentally determined structure of E. coli GroEL (PDB: 2NWC) in the UCSF Chimera 3D graphics window. The sequence alignment between the models and 2NWC is also included (bottom window), as are the modeling scores (Modeller Results window).
IMP: MultiFit
The MultiFit module of IMP simultaneously fits atomic structures of components into their assembly EM density map at resolutions as low as 25 Å (Lasker et al., 2009). The component positions and orientations are optimized with respect to a scoring function that includes the quality-of-fit of components in the map, the protrusion of components from the map envelope, and the shape complementarity between pairs of components. The scoring function is optimized by the exact inference optimizer DOMINO that efficiently finds the global minimum in a discrete sampling space. If cyclic symmetry is specified, the symmetry is imposed within the optimization procedure for added efficiency (Lasker et al., 2010a).
MultiFit Interface in Chimera
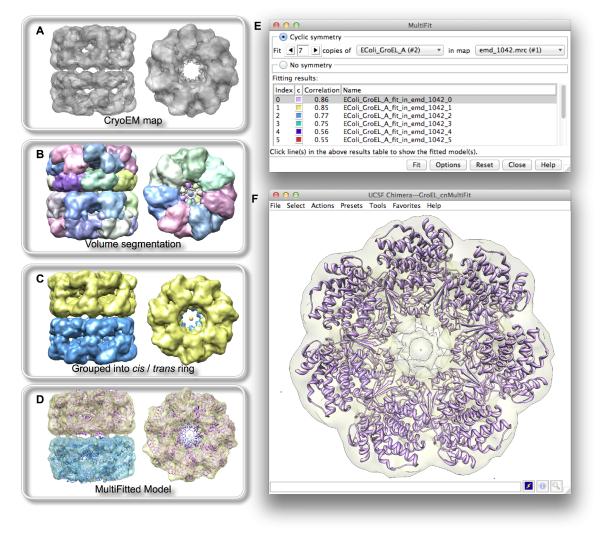
Chimera’s MultiFit GUI (under Tools -> Volume Data in the menu) takes as input one or more protein structures and an EM density map. Chimera allows editing structures and maps to generate the desired inputs: structures can be combined and subsets of their atoms selected or deleted, and specific regions of maps can be extracted. The user can further specify the level and resolution of the EM map, as well as the number of copies of subunits. Multiple copies of a structure can be fit assuming cyclic symmetry, or multiple different structures fit without using symmetry. The results are returned as a list of possible configurations with correlation scores indicating their goodness of fit to the density (Figures 2 and 5). Choosing a row in the list displays the corresponding set of structures in the Chimera graphics window. More than one row can be chosen to display multiple sets of results simultaneously for comparison; the different solutions are shown with different colors. A typical run takes a few minutes and can be performed locally or remotely via a web service.
Figure 2.
MultiFit GUI and multiple fitting of GroEL subunits into EM maps. (A) Cryo-EM map of the apo form of GroEL. (B) Cryo-EM map separated and colored by Chimeras Segment Map tool. (C) Segmentation regions grouped and merged into cis and trans rings. (D) Multiple subunits placed within the segmented map according to one of the solutions from MultiFit. (E) Chimera’s MultiFit interface, showing fitting with cyclic symmetry. The table lists solutions to the multiple fitting problem and their correlation coefficient values. (F) The EM map and top-scoring solution (index 0 in the table) are shown in Chimeras 3D graphics window.
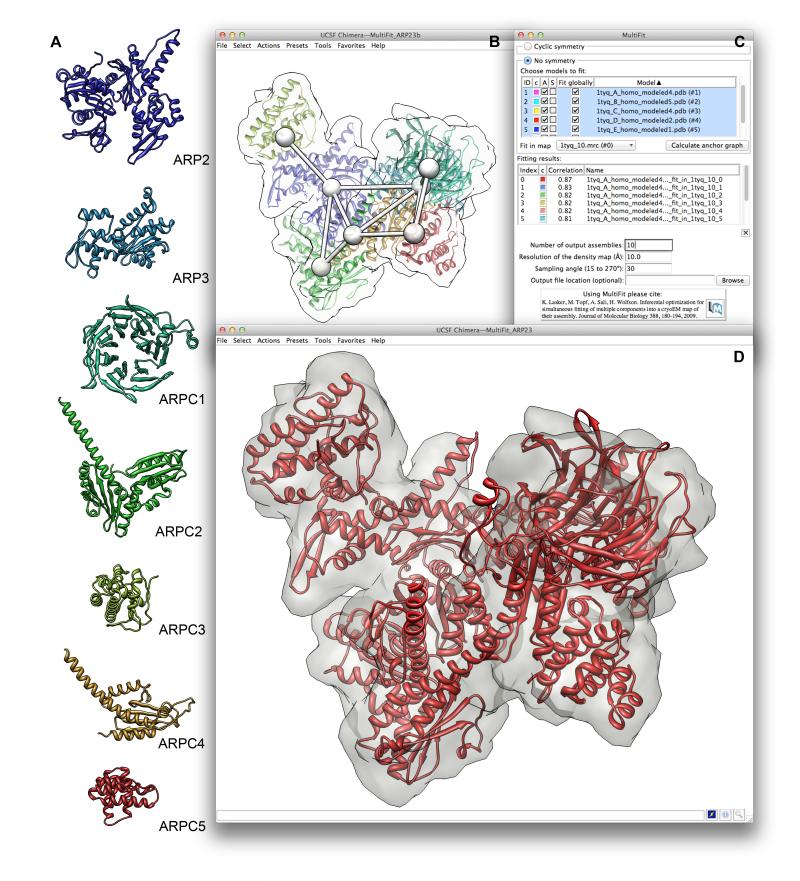
Figure 5.
(A) Comparative models of the ARP2/3 subunits. The seven subunits of the ARP2/3 complex are modeled using MODELLER based on multiple templates. (B) Anchor graph of the ARP2/3 complex as generated by MultiFit for the density map simulated from crystal structure 1TYQ. 1TYQ is also shown, with subunits colored uniquely. (C) Chimera’s MultiFit interface, showing fitting without symmetry. The table lists solutions to the multiple fitting problem and their correlation coefficient values. (D) The top-scoring MultiFit model within the density map simulated from 1TYQ.
IMP: FoXS
Fast X-Ray Scattering (FoXS), another module of IMP, is a rapid and accurate method for calculating a theoretical SAXS profile given an atomic structure (Schneidman-Duhovny et al., 2010). The method explicitly computes all interatomic distances and models the first solvation layer based on atomic solvent-accessible surface areas. Alternatively, a fast coarse-grained profile can be calculated based on protein Cα positions only. The theoretical SAXS profile can be fitted to an experimental SAXS profile by minimizing a penalty (χ2) function.
FoXS interface in Chimera
The FoXS interface (in the Chimera menu, Tools -> Higher-Order Structure -> Small-Angle X-Ray Profile) takes as input one or more entire structures or the currently selected atoms and, optionally, an experimental SAXS profile. Results are returned as a two-dimensional plot (Figure 3). If an experimental profile is provided, the theoretical profile will be scaled to fit the experimental profile and the χ value representing their quality of fit reported in the legend. Advanced options include whether or not to adjust excluded volume and hydration parameters to improve the fit, whether or not to apply a background adjustment to the experimental data, and whether or not to perform coarse-graining. The typical running time is less than a second for a system of a thousand atoms, and can extend to a few minutes for tens of thousands of atoms. Users can modify the input structures and recalculate the SAXS profile, and multiple results can be shown on the same plot.
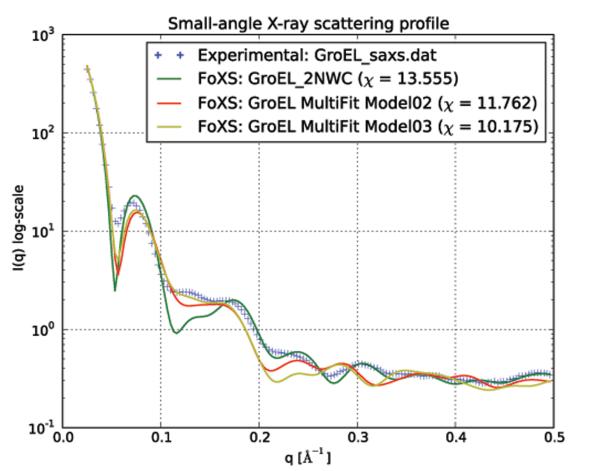
Figure 3.
SAXS profiles of GroEL. The Chimera-FoXS interface takes as input structures in Chimera and, optionally, an experimental SAXS profile. The IMP FoXS module calculates theoretical SAXS profiles from the structures and plots the results with the experimental data, if provided. Shown here are theoretical SAXS profiles calculated from the apo form structure of GroEL (PDB: 2NWC; green line) and the structures with the two best correlations (turquoise and red lines, respectively) from 11 MultiFit models, along with the experimental profile (blue crosses). A smaller value of χ indicates a theoretical profile more similar to the experimental profile.
Rotamers
Further structural refinement can employ the Chimera Rotamers tool. This tool allows viewing and evaluating likely conformations of amino acid sidechains and incorporating them into structures. A residue can be updated to a different conformation of the same type of amino acid residue or mutated into a different type.
Rotamer libraries are catalogs of distinct conformations of amino acid sidechains and their probabilities, usually extracted from a sample of high-quality structures (Dunbrack, 2002; Lovell et al., 2000). The probabilities can reflect not only residue type but also other information, such as the backbone of the residue, defined by the φ and ψ dihedral angles. The Rotamers tool in Chimera further combines data from a rotamer library with the evaluation of local non-bonded interactions to facilitate identifying likely sidechain conformations in the context of an entire structure.
Rotamers interface in Chimera
In Chimera, one or more amino acid residues can be selected to indicate positions of interest within a protein structure. The Rotamers tool (under Tools -> Structure Editing in the menu) allows specifying an amino acid residue type, which could differ from that in the structure, and the rotamer library to use. Three library options are provided: the Dunbrack backbone-dependent rotamer library (Dunbrack, 2002), the Richardson backbone-independent library (Lovell et al., 2000) with the author-recommended common-atom values, and the Richardson library with modal (peak) values instead of common-atom. At each selected position, a “bouquet” of rotamers is displayed (Figure 4). The rotamer sidechain torsion (χ) angles and probabilities from the library are listed in a separate window. Choosing one or more rows in the list with the mouse displays only the corresponding rotamers in the main window and hides the others. Importantly, the probabilities in the list are taken from the rotamer library and are not affected by the structural environment, except by φ and ψ angles when the Dunbrack library is used. The rotamer list Columns menu allows calculating the number of clashes (bad contacts) and hydrogen bonds formed by each rotamer with its surroundings, and showing these results as additional columns in the list. If a suitable density map is present, the rotamers can also be evaluated for their fit to the density. The list can be sorted by the values in any column by clicking the column header. The library and local environment information together with interactive viewing of specific rotamers facilitate identifying the best conformation given the entire context. A single rotamer can be chosen and used to replace the pre-existing sidechain coordinates. The Rotamers tool is also useful for revealing cases where an experimental structure has a nonrotameric conformation, possibly due to other local constraints, or suggesting how well a structure might accommodate a particular mutation and what effect that mutation might have on its function.
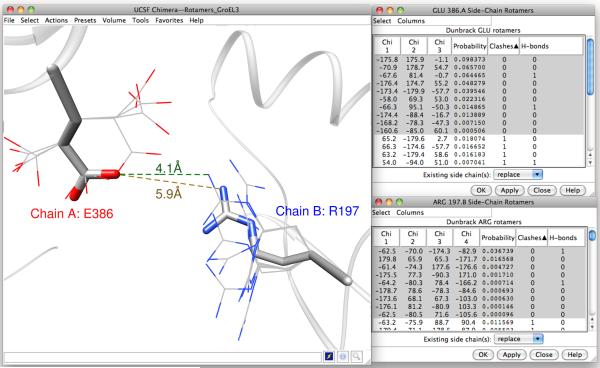
Figure 4.
Intersubunit salt bridge and Chimera Rotamers. Residues E386 from chain A and R197 from chain B in the optimal model from MultiFit are shown as thick “sticks.” Shown as thin lines are sidechain conformations for these residues from the Dunbrack backbone-dependent rotamer library. The Chimera Rotamers tool lists the sidechain torsion angles, backbone-dependent probabilities, clashes, and number of H-bonds for each rotamer. While the backbone positions of these residues are too far apart to allow formation of the salt bridge thought to exist in the apo form of GroEL, a closer interaction (4.1 Å as compared to 5.9 Å in the MultiFit result) can be obtained using favorable, zero-clash rotamers of these residues.
Modeling the GroEL complex
Bacterial chaperonin GroEL is a widely studied ATP-regulated molecular machine, composed of two back-to-back stacked rings (cis and trans), each containing seven 60 kDa subunits of the same sequence (Horwich et al., 2007; Ranson et al., 2001; Sigler et al., 1998; Zeilstraryalls et al., 1991). In this example, we model the structure of the E. coli GroEL structure based on an EM density map (EMDB: 1042) (Ranson et al., 2001) and structural homologs of the subunit. We then assess models by a SAXS profile and examine sidechain rotamers of a pair of residues that form a salt bridge.
Homology modeling of GroEL monomer
The homology modeling input is the amino acid sequence of the target, E. coli GroEL (UniProt id: P0A6F5) (Blattner et al., 1997; Burland et al., 1995; Hemmingsen et al., 1988). BLAST was launched via a Chimera web service to search the PDB database for known protein structures with sequence similarity to the target (the E. coli structures were filtered out from the search results). The following GroEL structures were chosen as the templates: chains A and B from Mycobacterium tuberculosis chaperonin 60.2 (PDB: 1SJP) (Qamra and Mande, 2004), chain H from Thermus thermophilus chaperonin (PDB: 1WE3) (Shimamura et al., 2004) and chain A from Paracoccus denitrificans chaperonin-60 (PDB: 1IOK) (Fukami et al., 2001) The pairwise identities between the target and template sequences are 59.3, 57.1, 63.2, and 68.0%, respectively (Figure 1).
The homology modeling process was launched as a web service from the Chimera-MODELLER interface. After 20 minutes on the Linux cluster running the service, ten MODELLER models were returned along with statistical measures of model accuracy, GA341 (Melo et al., 2002) and Discrete Optimized Protein Energy (DOPE and zDOPE, raw and normalized, respectively) (Shen and Sali, 2006). A zDOPE score below -1 indicates that the distribution of atom pair distances in the model resembles that found in a large sample of known protein structures. The “Fetch Scores” option on the results dialog was used to calculate two additional scores, Estimated RMSD and Estimated Overlap relative to the true structure (Eramian et al., 2008).
These models can also be assessed by comparison to any of the several previously solved structures of E. coli GroEL. The highest-resolution structure available from the PDB for the intact complex in the apo form is 2NWC (3.02Å) (Kiser et al., 2007). We chose to compare the monomer models to chain A of 2NWC, or 2NWC_A. The ten models were superimposed on 2NWC_A using the Chimera MatchMaker tool, with resulting Cα RMSDs ranging from 1.19 to 1.73Å for all residues present in the structure (2NWC_A contains 524 of the 548 residues found within the UniProt sequence file).
The result so far is a model of a single subunit. While GroEL consists of two back-to-back stacked 7-subunit rings, we modeled one ring at a time using MultiFit’s cyclic symmetry mode.
Multiple fitting GroEL cis and trans rings
Next, we used MultiFit to simultaneously fit seven copies of the subunit into the cryo-EM density map of the GroEL cis ring complex and then repeated the procedure for the trans ring (Figure 2). A cryo-EM map of the ATP-bound state of GroEL is available from the EM Data Bank (EMDB: 1042; Ranson et al., 2001). The cis ring can be extracted using the “region bounds” option in the Chimera volume viewer or using Chimera’s segmentation tool (Pintilie et al., 2010). Through the Chimera-MultiFit GUI, the model of the monomer from MODELLER previously identified as the best match to 2NWC_A (lowest-RMSD, see above) and the segmented cryo-EM map were chosen and submitted to the MultiFit web service. The top eleven solutions were returned. Similarly, the trans ring density was extracted and MultiFit was used to model the second heptameric ring. Subsequently, the two ring models were combined into a single atomic structure (Figure 2D) using their positions relative to the original map. The highest-correlation structures of the two rings were combined to give one atomic model, the second-highest-correlation structures of the two rings to give a second model, and so on for a total of six models of the apo form of GroEL.
Superposition of the highest-correlation model with the known structure of E. coli GroEL (PDB: 2NWC) using Chimera’s MatchMaker tool revealed a Cα RMSD of 2.66 Å for 7336 Cα atom pairs.
SAXS Profiles of GroEL
To further assess the results from previous steps, SAXS profiles of the six model structures and 2NWC were calculated and fit to the experimental profile provided by Chiu and Ludtke (Ludtke et al., 2001). We chose the coarse-grained SAXS profile option (Cα atoms only) because the complexes are large (52,668 atoms), allowing calculating and fitting a theoretical profile in approximately 2 minutes. The best match between the experimental and theoretical profiles is for Model03, with a mediocre χ of 10.2 (Figure 3). The relatively high χ value could be a consequence of several factors. First, the solution conformation of GroEL may differ from the crystal structure, either within subunits or between subunits. Second, it is also possible that multiple conformations are present in solution. Third, the SAXS profile may not have been determined or computed accurately.
GroEL salt bridge examined with Rotamers
Intersubunit salt bridges play a crucial role in the cooperativity of GroEL (Ranson et al., 2001) and its transitions between functional states (Hyeon et al., 2006; Yang et al., 2009). In the T state, residue E386 at the end of helix M in the intermediate domain forms salt bridges with R284, R285 and R197 on adjacent subunits. These salt bridges stabilize the apo form of GroEL’s T state. During the transition from the T state to the R state that occurs with ATP binding and hydration, these contacts are disrupted and E386 forms an intersubunit salt bridge with K80 instead. It is desirable to study the sidechains of these residues, but the resolution of the EM density map is too low for a generating a precise model based on the map alone. Instead, we employ the Chimera Rotamers tool. The Dunbrack rotamer library was used in this example. Figure 4 shows the rotamers of E386 and R197 that have zero clashes (bad atomic contacts) with their surroundings (thin wires), so these are the rotamers that “fit” into the structure. The sidechain conformations in the model (which were determined by MODELLER at the monomer-modeling stage) are shown with sticks. For E386, the highest-probability zero-clash rotamer matches the conformation in the model. The probabilities are based only on backbone conformation, so clashes are the more important criterion in the context of the structure. For R197, none of the zero-clash rotamers closely matches the conformation in the model, but these rotamers are no less reasonable, and in fact the rotamer listed third in Figure 4 (third-highest-probability of the zero-clash rotamers) is better positioned to form a salt bridge with E386 than the conformation in the model (4.1Å between NH2 and OE1 vs. 5.9Å). 4.1Å is still too far for a good salt bridge interaction, but as close as possible given the backbone geometry.
Modeling the ARP2/3 complex
The actin-related protein-2/3 (ARP2/3), a seven-protein asymmetric complex, plays a major role in the formation of branched actin-filament networks during diverse processes ranging from cell motility to endocytosis (Goley and Welch, 2006). The bovine ARP2/3 complex structure has been determined by X-ray crystallography (Robinson et al., 2001), revealing its molecular organization. However, a structural understanding of how the ARP2/3 complex mediates actin filament formation is still limited. EM studies have been useful for investigating ARP2/3 function (Egile et al., 2005). To demonstrate the potential application of the Chimera MultiFit tool, we simultaneously fit comparative models of the ARP2/3 subunits into a simulated 15Å density map of ATP-bound ARP2/3 (PDB id: 1TYQ)(Nolen et al., 2004) created using Chimera’s “molmap” command (Figure 5).
Comparative modeling of ARP2/3 subunits
Comparative models were calculated separately for each of the seven target subunits. BLAST was launched via Chimeras web service to search the PDB database for homologous protein structures. Twelve structures were used to model each of the seven target subunits (Table A.1); 1TYQ subunits were filtered out since these were used to generate the assembly density map.
Table A.1.
Homology modeling templates of Arp2/3 Complex
| PDBID | Name | Species | Resolution (Å) |
|---|---|---|---|
| 1K8K | Crystal Structure of Arp2/3 Complex | Bos taurus | 2.00 |
| 1U2V | Crystal structure of Arp2/3 complex with bound ADP and calcium | Bos taurus | 2.55 |
| 2P9I | Crystal Structure of bovine Arp2/3 Complex co-crystallized with ADP and crosslinked with gluteraldehyde |
Bos taurus | 2.46 |
| 2P9K | Crystal structure of bovine Arp2/3 complex co-crystallized with ATP and crosslinked with glutaraldehyde |
Bos taurus | 2.59 |
| 2P9L | Crystal Structure of bovine Arp2/3 complex | Bos taurus | 2.65 |
| 2P9N | Crystal Structure of bovine Arp2/3 complex co-crystallized with ADP | Bos taurus | 2.85 |
| 2P9P | Crystal Structure of bovine Arp2/3 complex co-crystallized with ADP | Bos taurus | 2.90 |
| 2P9S | Structure of bovine Arp2/3 complex co-crystallized with ATP/Mg2 + | Bos taurus | 2.68 |
| 2P9U | Crystal structure of bovine Arp2/3 complex co-crystallized with AMP- PNP and calcium |
Bos taurus | 2.75 |
| 3DXK | Structure of Bos Taurus Arp2/3 Complex with Bound Inhibitor CK0944636 |
Bos taurus | 2.70 |
| 3DXM | Structure of Bos taurus Arp2/3 Complex with Bound Inhibitor CK0993548 |
Bos taurus | 2.85 |
| 3RSE | Structural and biochemical characterization of two binding sites for nucleation promoting factor WASp-VCA on Arp2/3 complex |
Bos taurus | 2.65 |
A MODELLER comparative modeling process was then launched from the Chimera-MODELLER interface separately for each target subunit. Modeling a single subunit took 4-7 minutes on the Linux cluster running the MODELLER web service and produced five comparative models and associated quality scores, as described above. The “Fetch Scores” option in the results dialog was used to calculate two additional scores, Estimated Cα-RMSD and Estimated Overlap relative to the hypothetical true structure (Eramian et al., 2008). The model with the best Estimated Cα-RMSD was selected for each subunit (Table A.2).
Table A.2.
Comparative models used for the ARP2/3 subunits
| Subunit (UNIPROT code, name: #aa) |
Top Estimated RMSD (Å) |
RMSD between 1TYQ and top model (Å) |
Shared region in 1TYQ and top model (#aa) |
Covered region (start-end, %) |
Native Overlap (%) |
|---|---|---|---|---|---|
| P61157,ARP2:418 | 0.949 | 0.808 | 399 | 1-418,100% 140-364,58% 1-372,100% 1-281,94% | 95% |
| A7MB62,ARP3:394 | 6.844 | 0.411 | 208 | 92% | |
| Q58CQ2,ARPC1:372 | 1.177 | 0.252 | 342 | 92% | |
|
Q3MHR7, ARPC2:300 |
2.836 | 0.714 | 274 | 96% | |
| Q3T035,ARPC3:178 | 0.280 | 0.248 | 169 | 1-178,100% 1-168,100% 36-151,77% | 95% |
| Q148J6,ARPC4:168 | 4.217 | 0.493 | 166 | 97% | |
| Q3SYX9,ARPC5:151 | 2.421 | 3.470 | 134 | 86% |
Multiple fitting
The space sampled in simultaneous asymmetric fitting is generally larger than that sampled in symmetric fitting, as fewer geometric restraints are provided. Thus, the user has the option of performing either global or local searches via MultiFit. With the global search option, no prior information regarding subunit positions and orientations is used, so no initial placement of the structures is required. Alternatively, the user can approximately place the subunits within the density map and perform local searching, in which case MultiFit will sample possible solutions for each subunit only within a bounding box surrounding the initial placement. These initial placements can come from individual subunit fitting (using, for example, the Chimera Fit in Map tool), manual visual fitting, or other methods based on prior knowledge of the complex. For the ARP2/3 system, running MultiFit with global sampling gave correlation scores for the top ten solutions ranging from 0.8 to 0.86 and took approximately 5 minutes. Comparison of the top-scoring model to the reference crystal structure gave a Cα-RMSD value of 6.09 Å and 31% of Cα atoms superposing within 3.5 Å of their native positions. For local sampling Chimera is first used to launch a MultiFit anchor graph calculation. The anchor graph suggests approximate centroid positions for the seven subunits (Figure 5C). The user can then use the anchor graph as a guide for placing the individual subunits within the density. Once initial positions are set, a MultiFit run with local sampling can be launched from Chimera. With this approach, the top ten solutions for the ARP2/3 assembly had correlation scores ranging from 0.93 to 0.96. The local sampling run took about twice as long as global; although both calculations are parallelized on the web server (otherwise global sampling could take hours), the local search employs finer sampling. Comparison of the top ten local fitting solutions to the reference crystal structure gave Ca-RMSD values of 3.9-4.7 Å and 72-80% of Cα atoms superposing within 3.5Å of their native positions.
Discussion
UCSF Chimera is an extensible program for interactive visualization and analysis of molecular structures and related data, including density maps, sequence alignments, docking results, and molecular dynamics trajectories. MODELLER (http://salilab.org/modeller) implements comparative protein structure modeling by satisfaction of spatial restraints and can perform many additional tasks, including de novo modeling of loops and optimization of protein structure with respect to a flexibly defined objective function. IMP (http://salilab.org/imp) is a suite of modules, including MultiFit (http://modbase.compbio.ucsf.edu/multifit/) and FoXS (http://modbase.compbio.ucsf.edu/foxs/), for the integrative structural characterization of macromolecular assemblies.
The current work integrates comparative modeling with MODELLER, multiple-structure fitting into EM density maps with MultiFit, and SAXS profile calculation and comparison with FoXS into the Chimera system, with easy-to-use graphical user interfaces. The calculations can be run locally or via web services hosted by the UCSF Resource for Biocomputing, Visualization, and Informatics.
Chimera includes complete documentation and can be downloaded free of charge for noncommercial use, with versions available for Mac, Windows, and Linux (http://www.rbvi.ucsf.edu/chimera). The interfaces and features described here are available in Chimera daily builds dated September 1, 2011 and later, and will be available in Chimera production releases version 1.6 and higher. In addition, video tutorials illustrating use of the tools described in this paper are available at http://www.rbvi.ucsf.edu/chimera/videodoc/JSB-Yang/index.html.
Limitations should be noted. MultiFit performs rigid-body rather than flexible fitting, and the symmetric fitting currently handles only cyclic symmetries. Furthermore, the Chimera interfaces do not provide access to all options that would be available from running MODELLER or IMP modules directly. There is a trade-off between keeping the interfaces simple and easy to use, versus accommodating more controls and more types of calculations. For example, the Chimera-MODELLER interface allows including water and/or ligand molecules from template structures, but only a single protein chain or subunit can be modeled at a time.
However, the integration described here brings several advantages. The accessibility of MODELLER, MultiFit, and FoXS calculations is enhanced by simple graphical interfaces and the provision of web services, so that local installations are not required. Chimera can be used to search the PDB for modeling templates and to prepare structures, sequence alignments, and density maps as inputs to the modeling and fitting calculations. Results can be analyzed in several ways, including measuring distances and angles, identifying hydrogen bonds and other contacts, coloring to show properties such as residue hydrophobicity and sequence conservation, and superimposing structures.
Chimera’s task manager monitors web service data transfers and execution progress. Advanced users/developers could potentially use their own web services to run calculations launched from Chimera. Chimera’s web services are implemented using the Opal Toolkit (nbcr.net/software/opal) (Krishnan et al., 2009; Krishnan et al., 2006).
Conclusions
To date, structure determination of challenging macromolecular assemblies requires integration of different data types obtained by multiple methods (Alber et al., 2007). An integrated visualization-based platform can greatly facilitate modeling tasks and lower the barrier to their use. We have illustrated the modeling of two multi-protein complexes from sequence to 3D structure using Chimera, MODELLER, and IMP. We expect that integrative modeling protocols, coupled with a user-friendly visualization tool such as Chimera, will become increasingly useful and facilitate maximizing the coverage, accuracy, resolution and efficiency of the structural characterization of macromolecular assemblies.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Prof. Wah Chiu and Dr. Steven Ludtke for providing the SAXS experimental data for the GroEL example. The research of KL was supported by continuous mentorship from Prof. Haim J. Wolfson as well as a fellowship from the Clore Foundation Ph.D Scholars program. KL’s research was carried out in partial fulfillment of the requirements for a Ph.D. degree from TAU. This work was funded by NIH grants R01 GM083960, U54 RR022220, and PN2 EY016525 to AS and P41 RR001081 to TEF.
References
- Alber F, Forster F, Korkin D, Topf M, Sali A. Integrating diverse data for structure determination of macromolecular assemblies. Annu Rev Biochem. 2008;77:443–477. doi: 10.1146/annurev.biochem.77.060407.135530. [DOI] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper, et al. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Burland V, Plunkett G, Sofia HJ, Daniels DL, Blattner FR. Analysis of the Escherichia-Coli Genome VI: DNA-Sequence of the Region from 92.8 through 100 Minutes. Nucleic Acids Res. 1995;23:2105–2119. doi: 10.1093/nar/23.12.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch GS, Hendrix DK, Ferrin TE. Nucleic acid visualization with UCSF Chimera. Nucleic Acids Res. 2006;34:e29. doi: 10.1093/nar/gnj031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbrack RL., Jr. Rotamer libraries in the 21st century. Curr Opin Struct Biol. 2002;12:431–440. doi: 10.1016/s0959-440x(02)00344-5. [DOI] [PubMed] [Google Scholar]
- Egile C, Rouiller I, Xu XP, Volkmann N, Li R, et al. Mechanism of filament nucleation and branch stability revealed by the structure of the Arp2/3 complex at actin branch junctions. PLoS Biol. 2005;3:e383. doi: 10.1371/journal.pbio.0030383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eramian D, Eswar N, Shen MY, Sali A. How well can the accuracy of comparative protein structure models be predicted? Protein Sci. 2008;17:1881–1893. doi: 10.1110/ps.036061.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, et al. Comparative Protein Structure Modeling Using MODELLER, Current Protocols in Protein Science. John Wiley & Sons, Inc; 2001. [Google Scholar]
- Fabiola F, Chapman MS. Fitting of high-resolution structures into electron microscopy reconstruction images. Structure. 2005;13:389–400. doi: 10.1016/j.str.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Fiser A, Sali A. ModLoop: automated modeling of loops in protein structures. Bioinformatics. 2003;19:2500–2501. doi: 10.1093/bioinformatics/btg362. [DOI] [PubMed] [Google Scholar]
- Fiser A, Do RKG, Sali A. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster F, Webb B, Krukenberg KA, Tsuruta H, Agard, et al. Integration of small-angle X-ray scattering data into structural modeling of proteins and their assemblies. J Mol Biol. 2008;382:1089–1106. doi: 10.1016/j.jmb.2008.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami TA, Yohda M, Taguchi H, Yoshida M, Miki K. Crystal structure of chaperonin-60 from Paracoccus denitrificans. J Mol Biol. 2001;312:501–509. doi: 10.1006/jmbi.2001.4961. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure. 2005;13:473–482. doi: 10.1016/j.str.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- Guharoy M, Janin J, Robert CH. Side-chain rotamer transitions at protein-protein interfaces. Proteins. 2010;78:3219–3225. doi: 10.1002/prot.22821. [DOI] [PubMed] [Google Scholar]
- Hemmingsen SM, Woolford C, van der Vies SM, Tilly K, Dennis DT, et al. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature. 1988;333:330–334. doi: 10.1038/333330a0. [DOI] [PubMed] [Google Scholar]
- Horwich AL, Fenton WA, Chapman E, Farr GW. Two families of chaperonin: physiology and mechanism. Annu Rev Cell Dev Biol. 2007;23:115–145. doi: 10.1146/annurev.cellbio.23.090506.123555. [DOI] [PubMed] [Google Scholar]
- Hyeon C, Lorimer GH, Thirumalai D. Dynamics of allosteric transitions in GroEL. Proc Natl Acad Sci U S A. 2006;103:18939–18944. doi: 10.1073/pnas.0608759103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiser PD, Lodowski DT, Palczewski K. Purification, crystallization and structure determination of native GroEL from Escherichia coli lacking bound potassium ions. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:457–461. doi: 10.1107/S1744309107020295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan S, Clementi L, Ren J, Papadopoulos P, Li W. Design and Evaluation of Opal2: A Toolkit for Scientific Software as a Service; Proceedings of the 2009 Congress on Services - I. IEEE Computer Society; 2009.pp. 709–716. [Google Scholar]
- Krishnan S, Stearn B, Bhatia K, Baldridge KK, Li W, et al. Opal: SimpleWeb Services Wrappers for Scientific Applications; International Conference on Web Services (ICWS); 2006.2006. pp. 823–832. [Google Scholar]
- Lasker K, Sali A, Wolfson HJ. Determining macromolecular assembly structures by molecular docking and fitting into an electron density map. Proteins: Structure, Function, and Bioinformatics. 2010a;78:3205–3211. doi: 10.1002/prot.22845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasker K, Topf M, Sali A, Wolfson HJ. Inferential Optimization for Simultaneous Fitting of Multiple Components into a CryoEM Map of Their Assembly. J Mol Biol. 2009;388:180–194. doi: 10.1016/j.jmb.2009.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasker K, Phillips JL, Russel D, Velazquez-Muriel J, Schneidman-Duhovny D, et al. Integrative structure modeling of macromolecular assemblies from proteomics data. Molecular & cellular proteomics : MCP. 2010b;9:1689–1702. doi: 10.1074/mcp.R110.000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell SC, Word JM, Richardson JS, Richardson DC. The penultimate rotamer library. Proteins. 2000;40:389–408. [PubMed] [Google Scholar]
- Ludtke SJ, Jakana J, Song JL, Chuang DT, Chiu W. A 11.5 angstrom single particle reconstruction of GroEL using EMAN. J Mol Biol. 2001;314:253–262. doi: 10.1006/jmbi.2001.5133. [DOI] [PubMed] [Google Scholar]
- Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, et al. Comparative protein structure modeling of genes and genomes. Annu Rev Bioph Biom. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- Melo F, Sánchez R, Sali A. Statistical potentials for fold assessment. Protein Sci. 2002;11:430–448. doi: 10.1002/pro.110430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng EC, Pettersen EF, Couch GS, Huang CC, Ferrin TE. Tools for integrated sequence-structure analysis with UCSF Chimera. Bmc Bioinformatics. 2006;7:339. doi: 10.1186/1471-2105-7-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JH, Huang CC, Babbitt PC, Ferrin TE. structureViz: linking Cytoscape and UCSF Chimera. Bioinformatics. 2007;23:2345–2347. doi: 10.1093/bioinformatics/btm329. [DOI] [PubMed] [Google Scholar]
- Navaza J, Lepault J, Rey FA, Alvarez-Rua C, Borge J. On the fitting of model electron densities into EM reconstructions: a reciprocal-space formulation. Acta Crystallogr D. 2002;58:1820–1825. doi: 10.1107/s0907444902013707. [DOI] [PubMed] [Google Scholar]
- Nolen BJ, Littlefield RS, Pollard TD. Crystal structures of actin-related protein 2/3 complex with bound ATP or ADP. Proc Natl Acad Sci U S A. 2004;101:15627–15632. doi: 10.1073/pnas.0407149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov MV, Svergun DI. Analysis of X-ray and neutron scattering from biomacromolecular solutions. Curr Opin Struc Biol. 2007;17:562–571. doi: 10.1016/j.sbi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pieper U, Webb BM, Barkan DT, Schneidman-Duhovny D, Schlessinger A, et al. ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 2011;39:D465–474. doi: 10.1093/nar/gkq1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J Struct Biol. 2010;170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons C, D’Abramo M, Svergun DI, Orozco M, Bernado P, et al. Structural characterization of protein-protein complexes by integrating computational docking with small-angle scattering data. J Mol Biol. 2010;403:217–230. doi: 10.1016/j.jmb.2010.08.029. [DOI] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- Qamra R, Mande SC. Crystal Structure of the 65-Kilodalton Heat Shock Protein, Chaperonin 60.2, of Mycobacterium tuberculosis. J. Bacteriol. 2004;186:8105–8113. doi: 10.1128/JB.186.23.8105-8113.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranson NA, Farr GW, Roseman AM, Gowen B, Fenton WA, et al. ATP-bound states of GroEL captured by cryo-electron microscopy. Cell. 2001;107:869–879. doi: 10.1016/s0092-8674(01)00617-1. [DOI] [PubMed] [Google Scholar]
- Robinson CV, Sali A, Baumeister W. The molecular sociology of the cell. Nature. 2007;450:973–982. doi: 10.1038/nature06523. [DOI] [PubMed] [Google Scholar]
- Robinson RC, Turbedsky K, Kaiser DA, Marchand JB, Higgs HN, et al. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Bernal R, Pletnev SV. Combining electron microscopic with X-ray crystallographic structures. Journal of Structural Biology. 2001;136:190–200. doi: 10.1006/jsbi.2002.4435. [DOI] [PubMed] [Google Scholar]
- Russel D, Webb B, Lasker K, Velazquez-Muriel J, Tjioe E, et al. Putting the pieces together: Integrative Modeling Platform for structure determination of macromolecular assemblies. PLoS Biol. 2011 doi: 10.1371/journal.pbio.1001244. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative Protein Modeling by Satisfaction of Spatial Restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010;38:W540–W544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M.-y., Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura T, Koike-Takeshita A, Yokoyama K, Masui R, Murai N, et al. Crystal structure of the native chaperonin complex from Thermus thermophilus revealed unexpected asymmetry at the cis-cavity. Structure. 2004;12:1471–1480. doi: 10.1016/j.str.2004.05.020. [DOI] [PubMed] [Google Scholar]
- Sigler PB, Xu Z, Rye HS, Burston SG, Fenton WA, et al. Structure and function in GroEL-mediated protein folding. Annu Rev Biochem. 1998;67:581–608. doi: 10.1146/annurev.biochem.67.1.581. [DOI] [PubMed] [Google Scholar]
- Svergun D, Barberato C, Koch MHJ. CRYSOL - a Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates} Journal of Applied Crystallography. 1995;28:768–773. [Google Scholar]
- Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing (vol 76, pg 2879,1999) Biophys J. 1999;77:2896–2896. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tama F, Miyashita O, Brooks CL. Normal mode based flexible fitting of high-resolution structure into low-resolution experimental data from cryo-EM. Journal of Structural Biology. 2004;147:315–326. doi: 10.1016/j.jsb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Topf M, Lasker K, Webb B, Wolfson H, Chiu W, et al. Protein structure fitting and refinement guided by cryo-EM density. Structure. 2008;16:295–307. doi: 10.1016/j.str.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wriggers W. Using Situs for the integration of multi-resolution structures. Biophys Rev. 2010;2:21–27. doi: 10.1007/s12551-009-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wriggers W, Chacón P. Modeling Tricks and Fitting Techniques for Multiresolution Structures. Structure. 2001;9:779–788. doi: 10.1016/s0969-2126(01)00648-7. [DOI] [PubMed] [Google Scholar]
- Yang Z, Majek P, Bahar I. Allosteric transitions of supramolecular systems explored by network models: application to chaperonin GroEL. PLoS Comput Biol. 2009;5:e1000360. doi: 10.1371/journal.pcbi.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilstraryalls J, Fayet O, Georgopoulos C. The Universally Conserved Groe (Hsp60) Chaperonins. Annu Rev Microbiol. 1991;45:301–325. doi: 10.1146/annurev.mi.45.100191.001505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.