Background: Endothelial cell (EC) Toll-like receptor 2 (TLR2) signaling induces inflammatory events.
Results: NF-κB, p38-MAPK, JNK, and ERK5 promote, whereas MEK1 suppresses, EC TLR2 signaling.
Conclusion: ERK5 is a newly identified mediator of TLR2 signaling, and TLR2 signaling pathways differ in ECs and monocytes.
Significance: TLR2 signaling differences can be exploited therapeutically for endothelial-specific or leukocyte-specific inflammatory responses.
Keywords: Endothelium, Inflammation, MAP Kinases (MAPKs), NF-κB, Toll-like Receptors (TLR), TLR2
Abstract
Endothelial cell (EC) Toll-like receptor 2 (TLR2) activation up-regulates the expression of inflammatory mediators and of TLR2 itself and modulates important endothelial functions, including coagulation and permeability. We defined TLR2 signaling pathways in EC and tested the hypothesis that TLR2 signaling differs in EC and monocytes. We found that ERK5, heretofore unrecognized as mediating TLR2 activation in any cell type, is a central mediator of TLR2-dependent inflammatory signaling in human umbilical vein endothelial cells, primary human lung microvascular EC, and human monocytes. Additionally, we observed that, although MEK1 negatively regulates TLR2 signaling in EC, MEK1 promotes TLR2 signaling in monocytes. We also noted that activation of TLR2 led to the up-regulation of intracellularly expressed TLR2 and inflammatory mediators via NF-κB, JNK, and p38-MAPK. Finally, we found that p38-MAPK, JNK, ERK5, and NF-κB promote the attachment of human neutrophils to lung microvascular EC that were pretreated with TLR2 agonists. This study newly identifies ERK5 as a key regulator of TLR2 signaling in EC and monocytes and indicates that there are fundamental differences in TLR signaling pathways between EC and monocytes.
Introduction
The endothelium is an integral component of host innate immune response. Microbial infections and tissue injury induce nearby endothelial cells (ECs)2 to up-regulate proteins that recruit and activate leukocytes at sites of inflammation (1, 2). Although the leukocyte adhesion cascade ultimately helps to clear the host of infectious agents and to repair damaged tissues, during disseminated infections or inflammatory disorders the activation of the endothelium at sites remote from the inciting source can lead to the dysregulation of a variety of microvascular functions, causing organ failure and subsequent death (3–5). Vascular ECs are uniquely situated to detect the presence of pathogens within the vasculature as they are in direct and constant contact with the circulating blood. Although not classically viewed as central regulators of innate immune responses, vascular ECs contain inflammatory signaling pathways and express several Toll-like receptors (TLRs) that bind components of microorganisms and certain endogenous molecules released by damaged or necrotic cells (2, 6–8).
The TLRs are expressed in intracellular compartments or at the cell surface in several different cell types. Each member binds a conserved subset of pathogen-associated molecule patterns such as triacylated and diacylated lipoproteins (TLR2/1 and TLR2/6), LPS (TLR4), nucleic acids (TLR3, TLR7, TLR8, and TLR9), and flagellin (TLR5) or endogenous damage-associated molecule patterns such as HMGB1 (TLR4) or mitochondrial DNA (TLR9) to induce activation of TLR intracellular signaling pathways (8, 9). In particular, TLR2 recognizes a diverse set of triacylated and diacylated microbial lipoproteins present on both Gram-negative and Gram-positive bacteria and forms heterodimers with either TLR1 or TLR6, respectively (10–12). TLR2 is expressed by leukocytes and ECs, and multiple factors have been found to induce EC expression of TLR2, including bacterial lipoproteins, LPS, histamine, HMGB1, TNFα, IFNγ, and IL-1β (13–19). We previously reported that the TLR2-dependent activation of ECs leads to a variety of outcomes, including altered endothelial permeability, EC apoptosis, a shift in the expression of coagulation pathway factors to reflect a pro-coagulatory state, and up-regulation of proteins necessary for neutrophil recruitment, adherence, and activation at sites of inflammation (16, 20). Many of the TLR2-dependent outcomes observed in ECs differ from those of mononuclear cells, most notably the lack of TNFα expression, the expression and up-regulation of E-selectin and plasminogen activator inhibitor-1 (PAI-1) activity, and the modulation of endothelial permeability. In fact, the expression of TLR2 itself is regulated differently in human ECs and mononuclear cells; in ECs, TLR2 lipoproteins induce the expression of TLR2, whereas in mononuclear cells they do not (16, 21). Thus we hypothesized that different signaling pathways might be utilized in ECs and monocytes and performed studies to analyze the expression profiles for TLR1, TLR2, and TLR6 and assess the role of the mitogen-activated protein kinases (MAPKs) and nuclear factor κB (NF-κB) in TLR2-activated inflammatory pathways in primary human vascular ECs.
The NF-κB and the MAPK pathways have been extensively studied downstream of inflammatory stimuli. The NF-κB family of transcription factors consists of RelA (p65), c-Rel, RelB, NF-κB1, and NF-κB2 (22). Activation of the canonical NF-κB pathway results in the degradation of bound IκBα, IκBβ, or IκBϵ in the cytoplasm, which leads to the translocation of NF-κB to the nucleus to mediate transcriptional events (22, 23). Proinflammatory stimuli can also induce the transactivation of p65 through phosphorylation, such as occurs after the TLR2-dependent activation of macrophages (24, 25). The MAPK family members p38-MAPK, JNK, and ERK1/2 participate in the inflammatory response and are induced by TLR2 activation in macrophages (26–28). There is some indication that another MAPK family member, ERK5, also participates in inflammatory signaling (29). The MAPKs can induce proinflammatory gene expression by directly activating transcription factors and by regulating proteins involved in transcriptional and translational control; however, ERK1/2 and p38-MAPK have also been reported to negatively regulate inflammatory gene expression (26, 30–32, 50, 70).
Leukocyte inflammatory signaling cascades have been more extensively studied than those of ECs, but there is evidence suggesting that the signaling pathways utilized by ECs and monocytes overlap considerably (33–35). Guided by known TLR2 signaling pathways in leukocytes and by known inflammation pathways in ECs, in the current report we defined several inflammatory signaling pathways downstream of TLR2 in ECs and tested the hypothesis that TLR2 signaling differs in ECs and monocytes. We observe that the p65 subunit of NF-κB and the MAPK family members p38-MAPK, JNK, and ERK5 are all required for the TLR2-induced up-regulation of inflammatory proteins and PAI-1 in ECs. We also provide evidence that MEK1 negatively regulates TLR2-activated inflammatory responses in ECs, whereas MEK1-positive regulates them in human monocytes. Thus divergent TLR2 signaling pathways exist in human ECs and monocytes.
EXPERIMENTAL PROCEDURES
Cell Lines
Human umbilical vein endothelial cells (HUVEC, passage 2–6) (Lonza, Walkersville, MD) and human lung microvascular endothelial cells (HMVEC-L, passage 4–6) (Lonza) were incubated at 37 °C under humidified 5% CO2. HMVEC-L were prepared from both male and female cadaver donors. HUVEC were grown in EGM-2 and HMVEC-L in EGM-2MV (Lonza). Endothelial growth medium was supplemented with 2% FCS. Peripheral blood mononuclear cells (PBMCs) and the monocytic cell line THP-1 were maintained in RPMI 1640 supplemented with 10% FBS, l-glutamine, and antibiotics. THP-1 cells were differentiated into macrophages with phorbol 12-myristate 13-acetate (PMA) as described previously (36).
Agonist and Inhibitor Treatments
Adherent cells were grown to 100% confluence before treatment, whereas suspension cells were seeded at 2.5 × 105 cells/cm2 and were agonist-treated the next day. Cells were treated with either the TLR2 agonist Pam3Cys-SKKK or FSL-1 (EMC Microcollections, Tubingen, Germany) at 1 or 10 μg/ml, recombinant human TNFα (Peprotech, Inc., Rocky Hill, NJ) at 0.05 or 0.10 μg/ml, recombinant human EGF (Sigma) at 200 ng/ml or hydrogen peroxide at 3 mm unless otherwise noted for the indicated times. Preparations of Pam3Cys, FSL-1, and TNFα contained <5 pg of LPS/μg of protein based on the Limulus-amebocyte lysate assay (Associates of Cape Cod, Inc., East Falmouth, MA). Cells were preincubated with the MAPK inhibitors SB203580 (10 μm) and SP600125 (10 μm) (Calbiochem) and BIRB 796 (10 μm), AEG3482 (10 μm), PD184352 (1 μm), and XMD8–92 (5 μm) (Axon Medchem; Netherlands) for 1 h before and continuously during agonist treatment.
PBMC and Neutrophil Isolation
Heparinized whole blood was collected by venipuncture from healthy human volunteers. Human PBMCs and neutrophils were isolated by gradient centrifugation using LymphoprepTM and PolymorphprepTM (Axis-Shield, Oslo, Norway), respectively. PBMCs were isolated according to the manufacturer's supplied instructions and neutrophils as described previously (37).
siRNA Transfection of HUVEC
NF-κB p65 (RELA) (L-003533-00-0005), TLR2 (D-001810-10-05), ERK5 (L-003513-00-0005), and control (L-005120-01-0005) siRNA ON-TARGET plus SMARTPools (Thermo Scientific, Rockford, IL) were used in accordance with the manufacturer's suggested protocol for HUVEC transfections using DharmaFECT 4 transfection reagent. Protein cell lysates (see below) were prepared 72 h post-siRNA transfection, and cell culture supernatants were collected 68 h post-siRNA transfection (48 h + 20 h agonist treatment).
NF-κB Electrophoretic Mobility Shift Assays (EMSA)
Cells lysates were prepared using a nuclear extraction kit (Panomics, Fremont, CA). NF-κB(1) EMSA Probe Set (AY1030P) and NF-κB(1) EMSA kit (AY1030; Panomics) were used to detect the presence of active NF-κB according to the manufacturer's supplied protocol. Protocol specifics were as follows. 10 μg of the nuclear fraction was used per reaction, and samples were run in non-denaturing 6% polyacrylamide gels. Oligos were transferred to Ball Biodyne B nylon membrane using a semidry transfer apparatus (Bio-Rad), and oligos were immobilized by placing the membrane in a UV Stratalinker® (Stratagene) for 3 min at 120 mJ and then placed in foil and baked for 1 h. After incubation with substrate detection solution, the membrane was exposed to film for 20–24 h before development. Direct densitometry measurements were performed on all EMSAs using ImageJ.
Immunoblots
Cells were lysed with PLC lysis buffer (50 mm Hepes, pH 7.5, 150 mm sodium chloride, 10% glycerol, 1% Triton X-100, 1.5 mm MgCl2, 1 mm EGTA, 100 mm sodium fluoride, 0.5 mm sodium vanadate) plus protease inhibitor mixture (Sigma), and protein concentrations of the lysates were estimated using the RCDC protein assay kit (Bio-Rad). Total proteins were separated by SDS-PAGE and then transferred to PVDF membranes (Pall Corp, Ann Arbor, MI). Membranes were blocked in 3% BSA for 45 min at room temperature (RT) and then incubated with primary antibody solution overnight at 4 °C. Membranes were the washed and then incubated with suitable peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA). Immunoblots were developed using SuperSignal West Dura Extended Duration Substrate (34076; Thermo Scientific), and the signal was detected using a Gel Logic 2200 Imaging System (Eastman Kodak Co.) run on Carestream Imaging Software (Carestream Health, Rochester, NY). Antibodies were: TLR2 (0.5 μg/ml, AF2616, R&D Systems, Minneapolis, MN), E-selectin (2.5 μg/ml, BBA18, R&D Systems), phospho-JNK (0.5 μg/ml, AF1205, R&D Systems), phospho-p38MAPK (1:1000, 4631, Cell Signaling, Danvers, MA), phospho-ERK1/2 (1:1000, 9106, Cell Signaling), phospho-ERK5 (1:1000, 3371, Cell Signaling), JNK (1:1000, 9252, Cell Signaling), p38MAPK (1:1000, 9212, Cell Signaling), ERK1/2 (1:2000, 9107, Cell Signaling), ERK5 (1:1000, 3372, Cell Signaling), phospho-p65 Ser-536 (1:1000, 3033, Cell Signaling), NF-κB p65 (1 μg/ml, SC-8008X, Santa Cruz Biotechnologies Inc., Santa Cruz, CA), IκBα (0.1 μg/ml, MAB4299, R&D Systems), IκBβ (1 μg/ml, MAB3425, R&D Systems), IκBϵ (1 μg/ml, MAB4300, R&D Systems), and actin (0.1 μg/ml, A2066, Sigma).
Cell-based ELISAs
HUVEC were grown in 48-well plates. The cells were gently washed once with PBS (w/Ca2+ and Mg2+) and incubated with either 2.5 μg/ml E-selectin (BBA18; R&D Systems), ICAM-1 (BBA3; R&D Systems), goat IgG (NI02; Calbiochem) or mouse IgG1 (MAB002; R&D Systems) diluted in Iscove's modified Dulbecco's medium containing 1% FBS for 1 h at 37 °C. After 4 washes, the cells were then incubated with either a 1:2000 dilution of bovine anti-goat-HRP or goat anti-mouse-HRP (Jackson ImmunoResearch) diluted in Iscove's modified Dulbecco's medium containing 1% FBS for 1 h at 37 °C. After 4 washes, bound HRP substrate was detected using a TMB reagent (KPL, Gaithersburg, MD), and the absorbance was read at 450 nm, and values were corrected at 570 nm. A crystal violet assay was performed to assess the relative cell density in each well (38). If significant differences were observed between treatments, the absorbance values for the particular assay were corrected.
Sandwich ELISAs
Supernatant concentrations of IL-6, CSF-2, and CSF-3 were detected using Human DuoSet kits (R&D Systems). IL-8 levels were detected using a human IL-8 ELISA kit (BD Biosciences). Active human PAI-1 was detected using the PAI-1 functional assay ELISA kit (Molecular Innovations, Novi, MI). All assays were performed according to the manufacturer's supplied protocol. Values were corrected when a significant difference in cell density was observed between treatments based on crystal violet assay.
Immunofluorescence
HUVEC were grown on collagen-I-coated coverslips. The cells were washed once with PBS (w/Ca2+ and Mg2+), then fixed with 4% paraformaldehyde for 10 min at RT, washed, and permeabilized with PBS containing 0.5% Triton X-100 5 min at RT. HUVEC were then blocked with 3% normal goat serum (TLR2) or 2% BSA (p65) containing 1 μg/ml human IgG (R&D Systems) in PBS-Tween for 30 min at RT. For TLR2, HUVEC were incubated with either FITC-conjugated mouse anti-human TLR2 (5 μg/ml; mab-mtlr2f, Invivogen, San Diego, CA) or FITC-conjugated mouse IgG1 (1:10; 556649, BD Pharmingen) diluted in 4% normal goat serum in PBS-Tween overnight at 4 °C. Coverslips were then washed and stained and mounted to microscope slides with ProLong® Gold antifade reagent with DAPI (Invitrogen) according to the manufacturer's protocol. For P65, HUVEC were preincubated in the presence of 100 ng/ml leptomycin B (Sigma) for 1 h, then incubated with either mouse anti-human p65 (10 μg/ml; SC-8008 X; Santa Cruz) or mouse IgG1 (10 μg/ml; MAB002; R&D Systems) diluted in 2% BSA in PBS for 1 h at RT. The cells were then washed and incubated with goat anti-mouse-FITC (5 μg/ml; AP180F; Millipore; MA) diluted in 2% BSA in PBS for 30 min at RT. Nuclei were stained with DAPI (0.1 μg/ml; VWR) for 10 s before mounting on microscope slides with Vectashield (Fisher).
Flow Cytometry
HUVEC were detached using Accutase Cell Detachment Solution (Innovative Cell Technologies, San Diego, CA). HUVEC and THP-1 cells were passed through a 40 μm filter, counted, and aliquoted at 1 × 106 cells per sample. The cells were then washed using flow cytometry staining buffer (R&D Systems) and incubated with 10 μg/ml human IgG (R&D Systems) in flow cytometry staining buffer for 15 min at 4 °C. After washing twice with flow cytometry staining buffer, the cells were incubated for 1 h at 4 °C with either FITC-conjugated mouse anti-human TLR2 (5 μg/ml; mab-mtlr2f, Invivogen), E-selectin (2.5 μg/ml; BBA21; R&D Systems), ICAM-1 (2.5 μg/ml; BBA20; R&D Systems), or mouse IgG1 (1:10; BD Pharmingen). All samples were washed 2 more times with flow cytometry staining buffer and then were analyzed by flow cytometry (BD LSRII Flow Cytometer; BD Biosciences).
Quantitative Real-time PCR
TaqMan human TOLL-like receptor pathway arrays (4418838), specific gene expression assays (HPRT1 (Hs01003267_m1), TLR1 (Hs00413978_m1), TLR2 (Hs00152932_m1), TLR6 (Hs00271977_s1), TRAF6 (Hs00371512_g1), and the manufacturer suggested assay reagents were purchased from Applied Biosystems (Foster City, CA). HUVEC monolayers, HMVEC-L monolayers, and THP-1 cells were lysed, and mRNA was isolated using TRIzol according to the manufacturer's supplied protocol (Invitrogen). mRNA concentrations were determined with a ND-1000 (NanoDrop®/Thermo Fisher Scientific), and mRNA was reverse-transcribed to cDNA using the High Capacity RNA-to-cDNA kit using 2 μg of mRNA per reaction (Invitrogen). An input of 2.5 ng of cDNA in 10-μl total reaction volume per well containing TaqMan® Fast Advanced Master Mix (Applied Biosystems) was used in all quantitative real-time PCR experiments, and quantitative real-time PCR was performed using the StepOnePlusTM System (Applied Biosystems). PCR activation at 95 °C for 20 s was followed by 40 cycles of 1 s at 95 °C and 20 s at 60 °C. The average Ct value of two technical replicates was used in all calculations with specific gene expression assays. The average Ct value of the internal controls HPRT1 and 18S were used to calculate ΔCt values for the array samples as this combination of reference genes displayed the lowest standard deviation among groups. HPRT1 alone was used to calculate ΔCt values for specific gene expression assays. For the human Toll-like receptors pathway arrays, the initial data analysis was performed using the 2−ΔΔCt method (39), and the data were corrected using log transformation, mean centering, and auto scaling as described by Willems et al. (40). For the specific gene expression assays, statistical analyses were calculated from the uncorrected ΔCt values. The methods of calculation utilized assume an amplification efficiency of 100% between successive cycles. The quantitative real-time PCR array data discussed in this publication have been deposited in NCBI Gene Expression Omnibus (41).
Neutrophil Adhesion Assay
RPMI 1640 medium without phenol red was used in all steps. After isolation, neutrophils were resuspended in RPMI 1640 at 5 × 106 cells/ml and then incubated with 3 μm calcein-AM (Invitrogen) for 30 min at 37 °C in the dark. The neutrophils were then washed 2× with prewarmed RPMI 1640 and resuspended in RPMI 1640 at 2.5 × 106 cells/ml. Just before adding neutrophils, 48-well plates containing HUVEC or HMVEC-L monolayers were washed 3× with RPMI 1640 containing 3% BSA. Neutrophils were then added at 5 × 105 cells/200 μl/well and allowed to incubate for 60 min at 37 °C in the dark. 200 μl of PBS (w/Ca2+ and Mg2+) was then added to each well, and the fluorescence was read at an excitation of 485 nm and an emission of 520 nm in a FLUOstar OPTIMA fluorescent plate reader (BMG Labtech, Cary, NC) (prereading). The plate was then washed 5× with PBS (w/Ca2+ and Mg2+), and fluorescence intensity was read again (post reading). The % adherence was calculated by first subtracting the background fluorescence of the ECs, then dividing the post reading by the prereading and multiplying by 100%. Of note, some variation in the percent adherence was observed between experiments, which were directly correlated to the degree of confluency of the HMVEC-L and HUVEC monolayers at the start of the adhesion assay (data not shown). Neutrophil adhesion was visualized using a Zeiss Axio Imager.D1 microscope.
Statistics
Data are expressed as the mean ± S.D. The data were analyzed using t tests (unequal variance, one-tail) or analysis of variance using a Dunnett's post-test when analyzing the Pam3Cys time course data. GraphPad Prism or Excel® 2008 were used for statistical analyses. p values ≤0.05 were considered significant. All data are represented as the mean ± S.D.
Online Supplemental Material
Supplemental Table S1 displays the complete data set for the human Toll-like receptors pathway arrays. Supplemental Table S2 shows the RQ and ΔCt Pam3Cys time course data for TRAF6, whereas supplemental Table S3 shows the RQ and ΔCt Pam3Cys time course data for TLR1, TLR2, and TLR6 expression in HUVEC and HMVEC-L. Supplemental Table S4 describes the MAPK/K inhibitors used in experiments. Supplemental Fig. S1 shows that ERK1/2 are constitutively active when HUVEC are grown in EGM-2 media and the Pam3Cys-induced activation of p38-MAPK and JNK is dependent on TLR2.
RESULTS
HUVEC Express TLR signaling-associated Genes
We have previously observed that the activation of endothelial TLR2 induces the expression of genes specifically involved in the neutrophil response (i.e. IL6, IL8, CSF2, CSF3, ICAM1, and SELE) and also induces the expression of TLR2 itself (16, 20). This suggests that ECs contain inflammatory signaling pathways similar to those utilized by leukocytes. We, therefore, performed gene expression analysis of human umbilical vein ECs (HUVEC) as a model EC type and THP-1 as a model human monocytic cell type using Toll-like receptor pathway arrays (supplemental Tables S1 and S2). The results show that the vast majority of TLR signaling-associated genes analyzed were detected in both HUVEC and THP1.
HUVEC Express Low Levels of TLR2 in Comparison to Monocytes, Whereas Bacterial Lipopeptides Induce the Intracellular Expression of EC TLR2
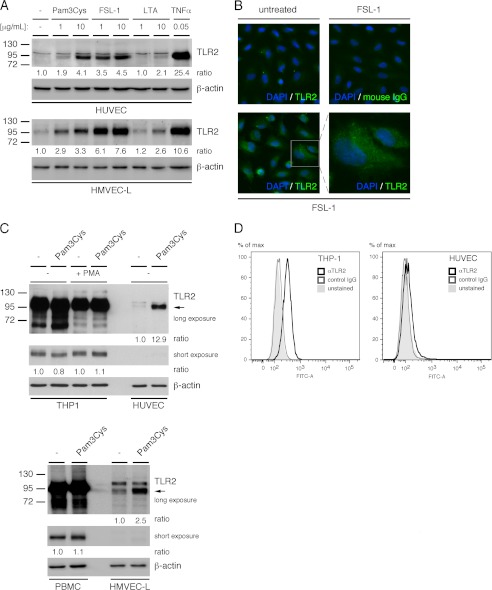
To extend our previous finding that the triacylated bacterial lipopeptide TLR1/2 agonist, Pam3Cys, induces the expression of TLR2, we analyzed the gene expression profiles for TLR2 and its dimerization partners TLR1 and TLR6 in both HUVEC and human lung microvascular EC (HMVEC-L) treated with Pam3Cys (16). We observe that Pam3Cys induces TLR2 but neither TLR1 nor TLR6 gene expression in both EC types (supplemental Table S3). In addition to Pam3Cys, we also observe that the diacylated bacterial lipopeptide TLR2/6 agonist, FSL-1, but not lipoteichoic acid, induces a robust expression of TLR2 in HUVEC and HMVEC-L (Fig. 1A). The minimal response induced by lipoteichoic acid may be explained by recent studies that suggest that optimal TLR2 activation may not occur after binding lipoteichoic acid (10, 12, 42). Using immunofluorescence microscopy, we observed that a majority of the TLR2 up-regulation after FSL-1 treatment is intracellular rather than at the cell surface (Fig. 1B). This is consistent with our previous results indicating that Pam3Cys induces only a minor increase in EC TLR2 surface expression (16). In comparison to THP-1 and PBMC, HUVEC and HMVEC-L, respectively, express relatively low levels of TLR2 before and after treatment with Pam3Cys (Fig. 1C). However, using flow cytometry, we verified that TLR2 is expressed on the surface of HUVEC before agonist treatment, albeit at much lower levels than observed for THP1 cells (Fig. 1D). Taken together, the data indicate that TLR2 is expressed at low levels on the cell surface of human vascular ECs, that treatment with bacterial lipopeptides up-regulates EC TLR2 mRNA and protein expression, and that the majority of the TLR2 protein expressed after FSL-1 treatment is located intracellularly rather than on the cell surface.
FIGURE 1.
TLR2 bacterial lipopeptide agonists promote endothelial cell expression of TLR2. A, HUVEC and HMVEC-L monolayers were treated with either medium alone, Pam3Cys, FSL-1, lipoteichoic acid, or TNFα for 20 h after which TLR2 protein levels were assessed by immunoblot. B, TLR2 expression in HUVEC after treatment with FSL-1 (10 μg/ml) for 20 h was assessed by immunofluorescence microscopy. C, THP-1 monocytes, phorbol 12-myristate 13-acetate (PMA) macrophage-differentiated THP-1 cells, PBMCs, HMVEC-L, or HUVEC monolayers were left untreated or treated with Pam3Cys (10 μg/ml) for 20 h. TLR2 protein levels (upper and middle panels) were assessed by immunoblot. D, flow cytometry was performed to assess basal surface expression of TLR2 on THP-1 monocytes and HUVEC. IgG-treated and unstained cells were used as antibody specificity controls. The densitometry values shown are the ratio of TLR2 to actin relative to the untreated sample. All experiments were performed at minimum three times.
TLR2 Induces NF-κB Activation in HUVEC
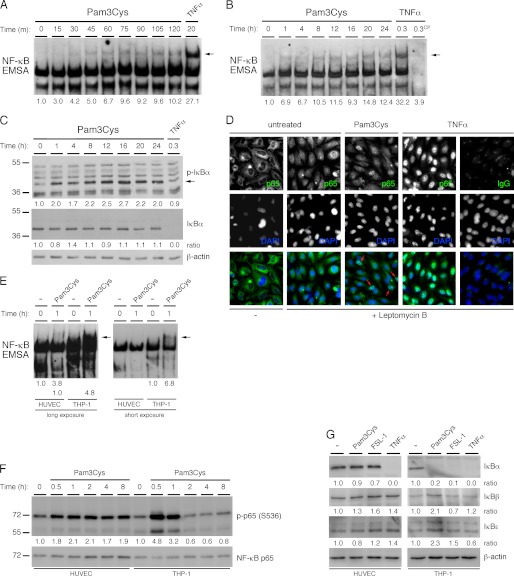
We hypothesized that NF-κB may be activated in ECs in response to TLR2 agonists and, therefore, analyzed the activation of NF-κB at intervals up to 24 h in HUVEC treated with Pam3Cys. The results show that NF-κB is activated beginning between 15 and 30 min after the addition of Pam3Cys and that the activation is sustained out to 24 h (Fig. 2, A and B). However, we did not observe the same degree of NF-κB activation in response to Pam3Cys as that observed with TNFα treatment. Consistent with the less robust activation of NF-κB observed in the EMSAs, we observed only a slight degradation of IκBα over a 24-h time course of treatment with Pam3Cys, although we did detect considerable IκBα phosphorylation, indicating active degradation of IκBα (Fig. 2C). We used immunofluorescence microscopy to analyze the extent of nuclear localization and thus activation of NF-κB in HUVEC treated with Pam3Cys, again using TNFα as a positive control. HUVEC were pretreated with leptomycin B, a nuclear export inhibitor, to confine any active NF-κB within the nucleus. Although there was little or no NF-κB within the nuclei of untreated HUVEC, the nuclei of leptomycin B treatment cells displayed significant p65 staining, indicating that there is a constant shuttling of NF-κB to and from the nucleus under our growth conditions (Fig. 2D). In leptomycin B-treated cells, TNFα treatment led to nuclear localization of almost all the cytoplasmic NF-κB. In contrast, Pam3Cys treatment caused the accumulation of NF-κB within the nuclei of only some HUVEC, indicating TLR2 activation induces a pleiotropic pattern of NF-κB activation in HUVEC (Fig. 2D). Next, a side-by-side comparison of NF-κB activation in HUVEC and THP-1 cells was performed. The results show that although NF-κB is active downstream of TLR2 in both cells types, the activation was much more robust in the THP-1 monocytes (Fig. 2E). Because phosphorylation of Ser-536 on the p65 subunit of NF-κB can induce its transactivation potential, we analyzed the kinetics of p65 Ser-536 phosphorylation at intervals up to 8 h in both HUVEC and THP-1 cells treated with Pam3Cys (22, 24). The results show that in THP-1 cells, Pam3Cys induces a robust, but transient phosphorylation of p65, whereas in HUVEC, the phosphorylation of p65 is less robust but is sustained over 8 h (Fig. 1F). This indicates that the duration of p65 transcriptional activity is more protracted in ECs. Because IκBα is not the only IκB family member able to retain NF-κB within the cytoplasm, we assessed for the involvement of two additional members, IκBβ and IκBϵ. We analyzed the degradation of the three classical IκBs in HUVEC and THP1 cells for comparison. Although TNFα induces detectable degradation of IκBα in both HUVEC and THP1, Pam3Cys and FSL-1 only induced the degradation of IκBα in THP1 cells but not in HUVEC (Fig. 2G). We did not detect significant degradation of either IκBβ or IκBϵ, suggesting that they may not be involved in the TLR2-dependent activation of NF-κB in either HUVEC or THP1 cells. In conclusion, the data indicate that bacterial lipopeptides induce a less robust activation of NF-κB in HUVEC in comparison monocytes or that induced in HUVEC by TNFα; however, the transactivation potential of NF-κB in ECs appears more prolonged.
FIGURE 2.
TLR2 agonists induce NF-κB activation in endothelial cells. A and B, HUVEC monolayers were either treated at varying time points with Pam3Cys (10 μg/ml) or 20 min with TNFα (100 ng/ml). Activation of NF-κB was assessed by EMSAs. A cold probe was used to show NF-κB binding specificity. C, HUVEC monolayers were treated with either Pam3Cys (10 μg/ml) for intervals through 24 h or with TNFα (100 ng/ml) for 20 min. IkBα phosphorylation levels as well as protein levels were assessed by immunoblot. D, the localization of the p65 subunit of NF-κB was assessed by immunofluorescence microscopy in untreated HUVEC monolayers in the presence or absence of leptomycin B (100 ng/ml; 1 h preincubation), and cells were treated for 1 h with Pam3Cys (10 μg/ml) or TNFα (100 ng/ml) in the presence leptomycin B. Arrows indicate Pam3Cys-treated cells showing more nuclear than cytoplasmic staining of p65. E, HUVEC monolayers and THP-1cells were either left untreated or treated for 1 h with Pam3Cys (10 μg/ml). Activation of NF-κB was assessed by EMSAs. F, HUVEC monolayers and THP-1 cells were treated at varying time points with Pam3Cys (10 μg/ml). NF-κB p65 Ser-536 phosphorylation levels as well as p65 protein levels were assessed by immunoblot. G, HUVEC monolayers and THP-1 cells were treated for 20 min with either Pam3Cys (10 μg/ml), FSL-1 (10 μg/ml), or TNFα (100 ng/ml). IκBα, IκBβ, and IκBϵ protein levels were assessed by immunoblot. Direct densitometry measurements were performed on phosphoprotein blots and EMSAs, whereas the ratio to actin was calculated for protein blots. For each cell type, the densitometry values are relative to the untreated sample. EMSA and immunoblotting experiments were performed at minimum 3 times, whereas the immunofluorescence experiment was performed 2 times.
NF-κB Is Essential for TLR2-dependent Outcomes in HUVEC
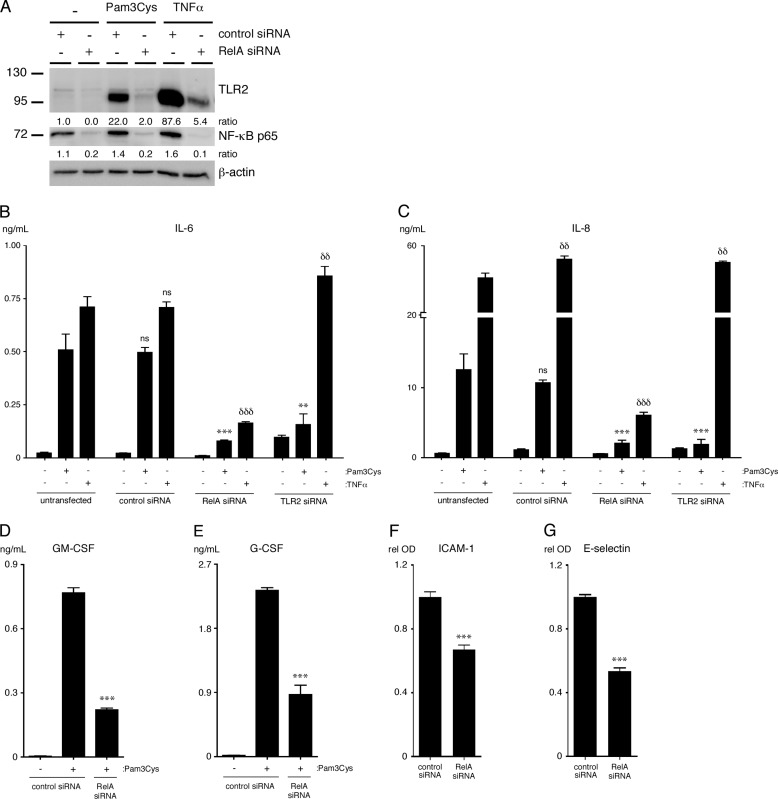
We used siRNA knockdown of the p65 subunit of NF-κB to determine the requirement of NF-κB in the up-regulation of IL-6, IL-8, GM-CSF, G-CSF, ICAM-1, E-selectin, and TLR2 expression in ECs treated with TLR2 agonists. The results show that the p65 subunit of NF-κB is required for the up-regulation of TLR2, IL-6, IL-8, GM-CSF, G-CSF, ICAM-1, and E-selectin by treatment with Pam3Cys (Fig. 3, A–G). As a control, specific siRNA knockdown of TLR2 expression confirmed that TLR2 expression is required for induction of IL-6 and IL-8 by Pam3Cys but not by TNFα (Fig. 3, B and C).
FIGURE 3.
Pam3Cys-induced cytokine production and surface adhesion molecule up-regulation is dependent on NF-κB p65 expression in HUVEC. A, HUVEC were transfected with RelA (p65) siRNA or control siRNA and then treated with either Pam3Cys (10 μg/ml) or TNFα (50 ng/ml) for 20 h. TLR2 and p65 protein levels were assessed by immunoblot. The densitometry values shown are the ratio of either TLR2 or p65 to actin relative to the untreated control siRNA sample. B and C, levels of IL-6 and IL-8 were quantified in supernatants of HUVEC that were transfected with either RelA, TLR2, or control siRNA and then treated with either Pam3Cys (10 μg/ml) or TNFα (50 ng/ml) for 20 h (n = 4). **, p < 0.01; ***, p < 0.001, Pam3Cys-treated untransfected versus Pam3Cys-treated siRNA transfected; δδ, p < 0.01; δδδ, p < 0.001, TNFα-treated untransfected versus TNFα-treated siRNA transfected. ns, not significant. D and E, levels of GM-CSF and G-CSF were quantified in supernatants of HUVEC that were transfected with RelA siRNA or control siRNA and then left either untreated or treated with Pam3Cys (10 μg/ml) for 20 h (n = 4). F and G, surface expression of the adhesion molecules ICAM-1 and E-selectin was assessed using a cell-based ELISA on HUVEC that were transfected with RelA siRNA or control siRNA and then treated with Pam3Cys (10 μg/ml) for 20 h (n = 4). ***, p < 0.001, Pam3Cys-treated control siRNA versus Pam3Cys-treated RelA siRNA. All experiments were performed a minimum of 3 times. rel OD, relative optical density.
TLR2 Induces p38-MAPK, JNK, and ERK5 Activation in HUVEC
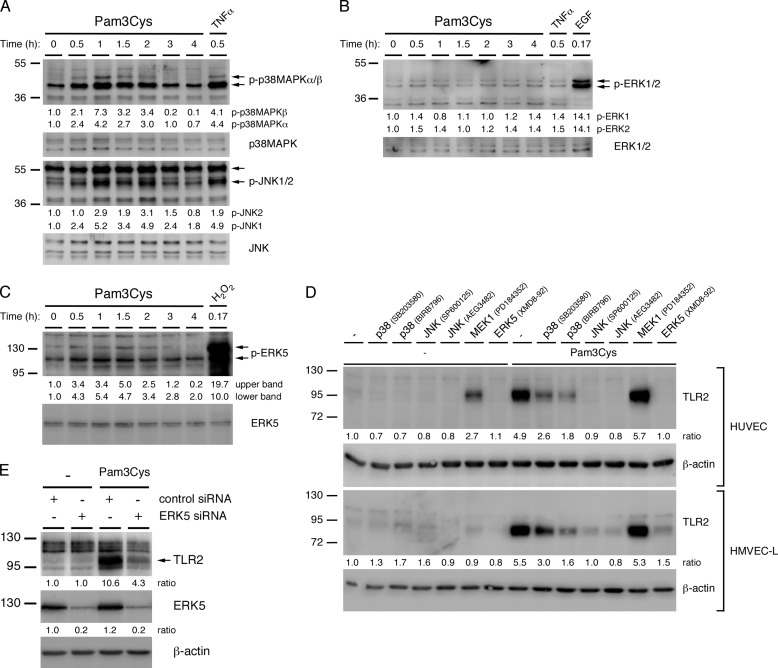
The MAPK signaling pathways have been extensively investigated as mediators of inflammatory gene expression in mononuclear cells. We analyzed the activity of the four conventional MAPK families, p38-MAPK, JNK, ERK1/2, and ERK5, in HUVEC at intervals of up to 4 h of Pam3Cys treatment. We found that p38-MAPK, JNK, and ERK5 are transiently activated between 30 min and 2 h of Pam3Cys treatment (Fig. 4, A and C). Although we were unable to consistently detect an increase or decrease in ERK1/2 activity after Pam3Cys treatment, inhibitor studies showed that there is a basal level of ERK1/2 activity present under our culture conditions (Fig. 4B and supplemental Fig. S1A). In the MAPK experiments, TNFα was used as a positive control for p38-MAPK and JNK activation, EGF was used for ERK1/2 activation, and hydrogen peroxide was used for ERK5 activation. Using TLR2 siRNA knockdown, we also showed that the activation of p38-MAPK and JNK in HUVEC is TLR2-dependent (supplemental Fig. S1B). Additionally, we found that the activity of p38-MAPK increases incrementally in response to higher concentrations of Pam3Cys (supplemental Fig. S1C).
FIGURE 4.
p38-MAPK, JNK, and ERK5 are activated by Pam3Cys and promote the TLR2-mediated expression of TLR2 in ECs. HUVEC monolayers were treated with Pam3Cys (10 μg/ml) for intervals through 4 h. A, p38-MAPKα/β phosphorylation levels and p38-MAPK protein levels, JNK1/2 phosphorylation levels, and JNK protein levels were assessed by immunoblot. TNFα (100 ng/ml) was used for a positive control. B, ERK1/2 phosphorylation levels and protein levels were assessed by immunoblot. EGF (200 ng/ml) was used for a positive control. C, ERK5 phosphorylation levels and protein levels were assessed by immunoblot. Hydrogen Peroxide (3 mm) was used for a positive control. A faint upper band observed in the phospho-ERK5 immunoblot that did not co-migrate with the majority of ERK5 protein may represent a more highly phosphorylated, smaller ERK5 protein fraction. D, expression of TLR2 protein was assessed by immunoblotting lysates of HUVEC or HMVEC-L monolayers that were pretreated for 1 h with the indicated MAPK inhibitors or with medium and then with Pam3Cys (10 μg/ml) or medium for an additional 20 h while in the continuous presence of inhibitor. E, expression of TLR2 and ERK5 proteins was assessed by immunoblotting lysates of HUVEC that were transfected with either ERK5 or control siRNA and then with Pam3Cys (10 μg/ml) or medium for 20 h. Direct densitometry measurements were performed on phospho-protein blots, whereas the ratio to actin was calculated for protein blots. All densitometry values are relative to the untreated sample. All experiments were performed at a minimum three times.
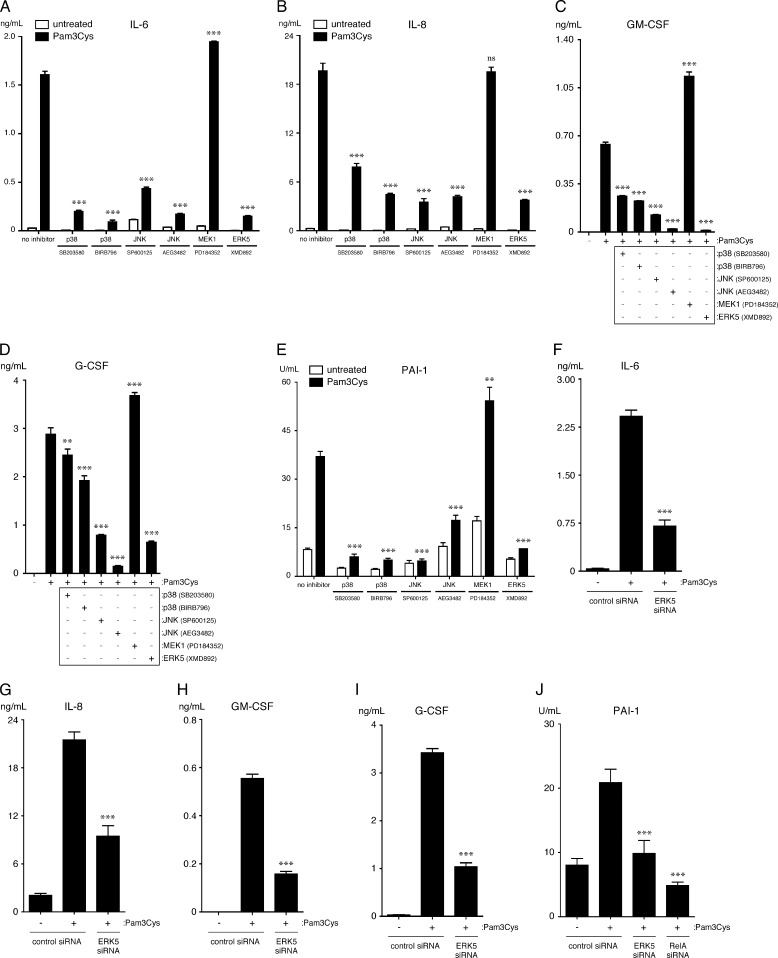
p38-MAPK, JNK, and ERK5 Promote, Whereas MEK1 Suppresses TLR2-dependent Up-regulation of TLR2, Cytokines, and PAI-1 Activity in Endothelial Cells
To determine the contribution of the various MAPK family members to the up-regulation of the aforementioned inflammatory proteins, TLR2 and PAI-1, we utilized a variety of MAPK inhibitors. These include the p38-MAPK inhibitors SB203580 and BIRB 796, the JNK inhibitors SP600125 and AEG3482, the MEK1 inhibitor PD184352 (1 μm), and the ERK5 inhibitor XMD8–92 (supplemental Table S4). Because to our knowledge no suitable ERK1/2 inhibitors are available, we utilized an inhibitor of MEK1, the upstream kinase of ERK1/2, to investigate their contribution to EC TLR2 signaling. We did not use the commonly used MEK inhibitors PD98059 and U0126 or higher concentrations of PD184352, as these inhibit both MEK1 and MEK5, thereby affecting the activity of both ERK1/2 and ERK5 (43, 44). We first analyzed the effects of the inhibitors on the Pam3Cys-induced up-regulation of TLR2 in HUVEC and HMVEC-L. We found that TLR2 up-regulation is dependent on JNK and ERK5 activity and to a lesser degree on p38-MAPK activity but not MEK1 activity (Fig. 4D). Interestingly, TLR2 expression is augmented after the inhibition of MEK1 in the absence of TLR2 agonist in HUVEC, suggesting that constitutive activation of MEK1 may be necessary to suppress TLR2 expression. Because ERK5 has not previously been reported to be involved in TLR2 expression, we verified its role using siRNA knockdown in transiently transfected HUVEC. Consistent with the inhibitor studies, knockdown of ERK5 prevents TLR2 up-regulation in HUVEC, indicating it is essential for the Pam3Cys-induced up-regulation of TLR2 in ECs (Fig. 4E). Importantly, knockdown of ERK5 only minimally affected HUVEC viability and/or growth under our culture conditions, which was accounted for in subsequent ELISA experiments (data not shown).
We next analyzed the effects of the MAPK inhibitors on the Pam3Cys-dependent expression of IL-6, IL-8, GM-CSF, and G-CSF and up-regulation of PAI-1 activity. We observed that inhibition of p38-MAPK, JNK, or ERK5 activity significantly reduced the levels of IL-6, IL-8, GM-CSF, G-CSF, and PAI-1 activity in culture supernatants during a 20-h treatment of HUVEC with Pam3Cys (Fig. 5, A–E). We again verified the role of ERK5 using siRNA knockdown in transiently transfected HUVEC. Consistent with the inhibitor studies, knockdown of ERK5 prevents the expression and/or secretion of IL-6, IL-8, GM-CSF, G-CSF, and PAI-1 (Fig. 5, F–J). Surprisingly, inhibition of MEK1 augmented Pam3Cys-induced secretion of IL-6, GM-CSF, G-CSF, but not IL-8, and up-regulation of PAI-1 activity, suggesting that MEK1 negatively regulates the expression and/or secretion of these proteins and PAI-1 activity (Fig. 5, A–E).
FIGURE 5.
p38-MAPK, JNK, and ERK5 facilitate, whereas MEK1 suppresses, TLR2-mediated up-regulation of cytokine expression and PAI-1 activity by HUVEC. A–D and I, levels of IL-6, IL-8, GM-CSF, G-CSF, and PAI-1 activity were quantified in supernatants of HUVEC that were pretreated for 1 h with the indicated MAPK inhibitors or medium and then treated with Pam3Cys (10 μg/ml) or medium for an additional 20 h while in the continuous presence of inhibitor (n = 3). GM-CSF and G-CSF were not detected in the untreated samples. **, p < 0.01; ***, p < 0.001, Pam3Cys versus Pam3Cys plus inhibitor; ns, not significant. E–H, levels of IL-6, IL-8, G-CSF, and GM-CSF were quantified in supernatants of HUVEC that were transfected with either ERK5 or control siRNA and then treated with Pam3Cys (10 μg/ml) or medium for 20 h (n = 3). ***, p < 0.001, Pam3Cys-treated control siRNA versus Pam3Cys-treated ERK5 siRNA. Inhibitor experiments were performed three times, whereas ERK5 siRNA experiments were performed two times.
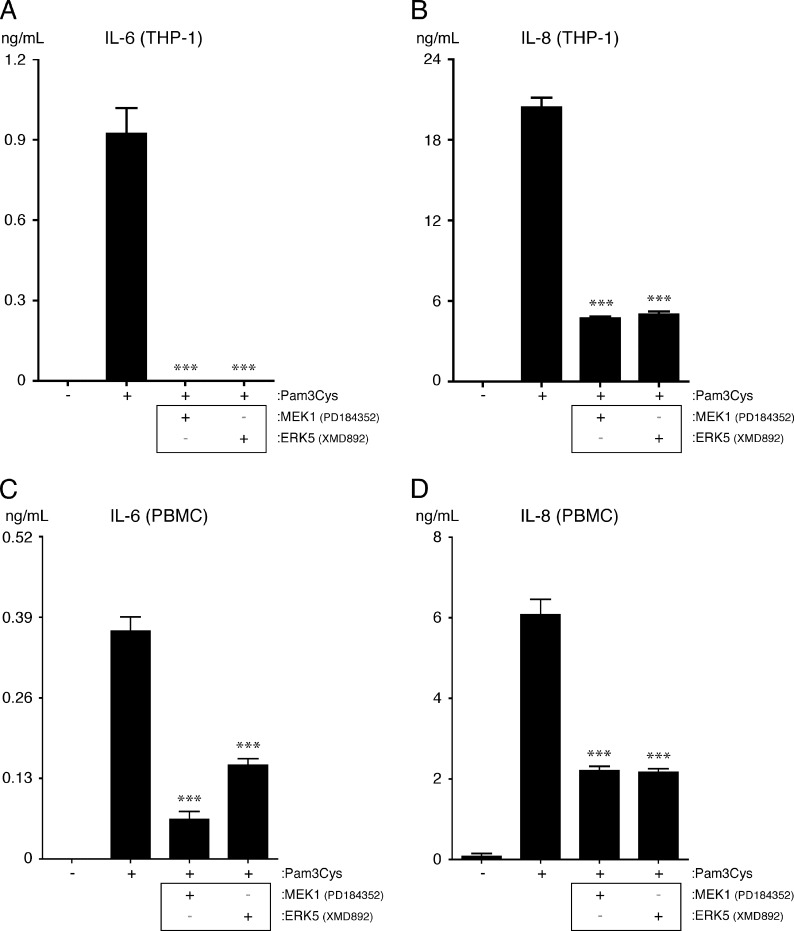
ERK5 and MEK1 Promote the TLR2-dependent Up-regulation of Cytokines in Monocytes
Because a proinflammatory role for ERK5 downstream of TLR2 in leukocytes has also not been reported previously, we tested its contribution to cytokine expression in both THP-1 cells and human PBMCs. The results show that inhibition of ERK5 reduces the expression of IL-6 and IL-8 by THP-1 cells and PBMCs, indicating that ERK5 promotes cytokine expression downstream of TLR2 in both ECs and monocytes (Fig. 6, A–D). Importantly, we found that inhibition of MEK1 also reduces the expression of IL-6 and IL-8 in THP-1 cells and PBMCs, which is opposite to what we observed in ECs (Fig. 6, A–D). Thus divergent signaling pathways exist downstream of TLR2 in ECs and monocytes.
FIGURE 6.
MEK1 and ERK5 promote cytokine expression from THP-1 cells and PBMCs. Equal numbers of THP-1 cells or PBMCs were seeded the day before to inhibitor and agonist treatment. The cells were either left in medium or pretreated for 1 h with the MAPK inhibitors PD184352 (1 μm) or XMD8–92 (5 μm). The cells were then either left untreated, treated with Pam3Cys (10 μg/ml), or treated with Pam3Cys (10 μg/ml) while in the continuous presence of inhibitor for 20 h. A and B, IL-6 and IL-8 levels were quantified in THP-1 culture supernatants by ELISA (n = 4). IL-6 and IL-8 were not detected in the absence of Pam3Cys. C and D, IL-6 and IL-8 levels were quantified in PBMC culture supernatants by ELISA (n = 4). IL-6 was not detected in the absence of Pam3Cys. ***, p < 0.001, Pam3Cys versus Pam3Cys plus inhibitor. Of note, we did not observe either THP-1 or PBMCs to express PAI-1 after Pam3Cys treatment (data not shown). All experiments were performed two times.
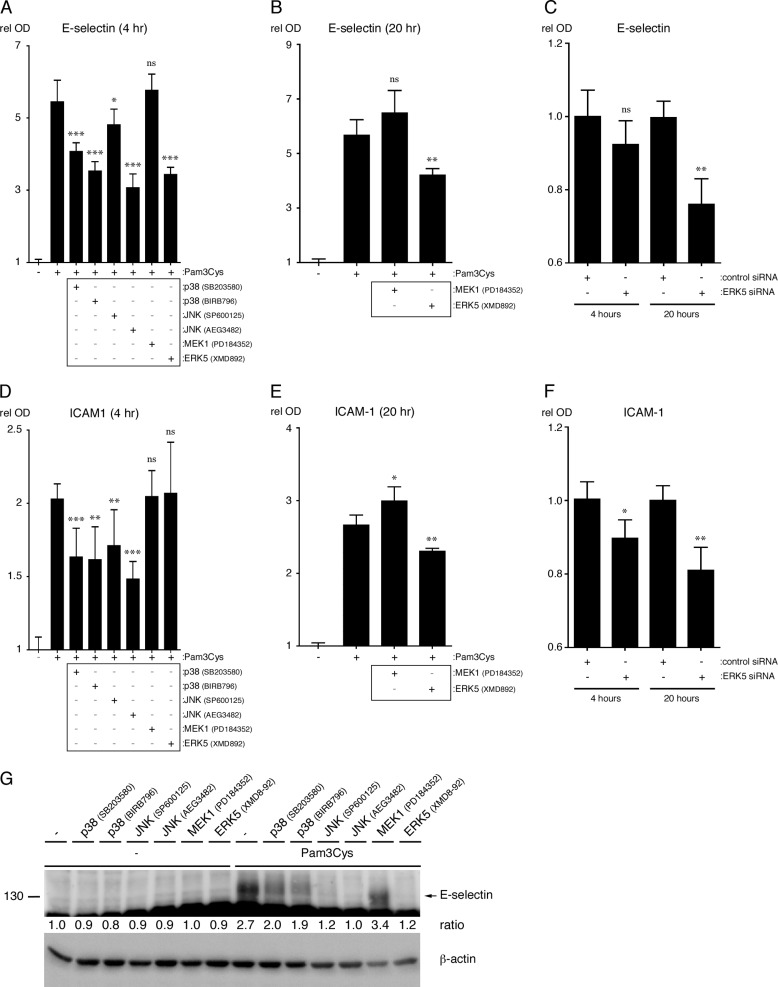
p38-MAPK, JNK, and ERK5 Promote TLR2-dependent Up-regulation of Adhesion Molecules in Endothelial Cells
We next analyzed the contribution of the MAPKs to the TLR2-dependent surface expression of E-selectin and ICAM-1 in HUVEC. After 4 h of Pam3Cys treatment in the presence of MAPK inhibitors, p38-MAPK, JNK, and ERK5 activity were shown to be necessary for maximal surface expression of E-selectin, whereas only p38-MAPK and JNK appear to be necessary for maximal ICAM-1 surface expression at the 4 h time point (Fig. 7, A and D). Flow cytometry analysis performed on HUVEC treated with FSL-1 for 4 h in the presence of the MAPK inhibitors yielded similar results as the E-selectin and ICAM-1 cell ELISAs (data not shown). To test the involvement of MEK1 and ERK5 at later time points, HUVEC were treated for 20 h in the presence or absence of the relevant inhibitors. The results show that ERK5 is required for the maximal surface expression of both adhesion molecules, whereas MEK1 inhibition slightly increases ICAM-1 surface expression (Fig. 7, B and E). To verify the results of the ERK5 inhibitor studies, we again knocked down the expression of ERK5 in HUVEC using siRNA. Like the inhibitor studies, the results show that ERK5 expression is necessary for the maximal expression of both E-selectin and ICAM-1 after 20 h of Pam3Cys treatment (Fig. 7, C and F). Finally, we assessed the total protein levels for E-selectin after 20 h of Pam3Cys treatment in the presence of MAPK inhibitors. The results show that p38-MAPK, JNK, and ERK5 promote E-selectin protein expression (Fig. 7G). Taken together, the patterns suggest that p38-MAPK, JNK, and ERK5 all promote ICAM-1 and E-selectin surface expression, whereas MEK1 has only a minor effect on their expression.
FIGURE 7.
p38-MAPK, JNK, and ERK5 promote TLR2 agonist driven surface expression of adhesion molecules in HUVEC. A, B, D, and E, HUVEC were pretreated for 1 h with the indicated MAPK inhibitors or with medium and then treated with Pam3Cys (10 μg/ml) in the continuous presence of inhibitor. The surface expression of E-selectin and ICAM-1 was quantified at 4 and 20 h of Pam3Cys treatment (n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001, Pam3Cys versus Pam3Cys plus inhibitor; ns, not significant. C and F, HUVEC were transfected with ERK5 or control siRNA and then treated with Pam3Cys (10 μg/ml) for either 4 or 20 h (n = 3). *, p < 0.05; **, p < 0.01, Pam3Cys-treated control siRNA versus Pam3Cys-treated ERK5 siRNA. G, E-selectin protein levels after 20 h of Pam3Cys treatment in the presence or absence of inhibitors were assessed by immunoblot. The densitometry values shown are the ratio of E-selectin to actin, relative to the untreated control sample. Cell ELISA experiments were performed three times, whereas the E-selectin immunoblot experiment was performed two times. rel OD, relative optical density.
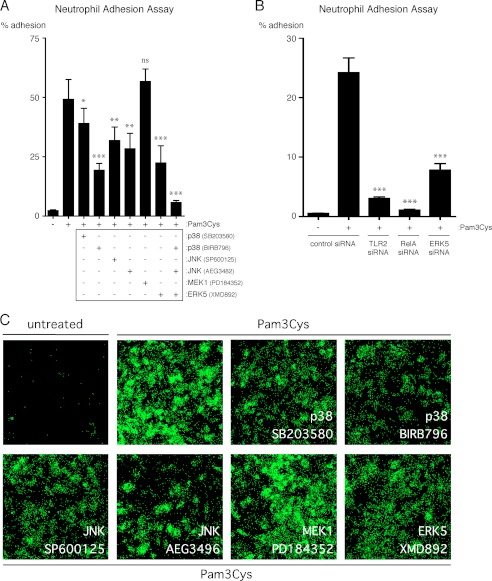
p38-MAPK, JNK, ERK5, and NF-κB Promote the Adhesion of Neutrophils to Lung Microvascular Endothelial Cells Activated by TLR2 Lipopeptide Agonist
To determine the contribution of p38-MAPK, JNK, MEK1, ERK5, and NF-κB to the Pam3Cys-dependent up-regulation of adhesion molecules on ECs in a more functional assay, we performed adhesion assays with primary human neutrophils and HMVEC-L. After a 4 h treatment of HMVEC-L monolayers with Pam3Cys in the presence of MAPK inhibitors, p38-MAPK, JNK, and ERK5 activity were shown to be necessary for maximal neutrophil adhesion to HMVEC-L (Fig. 8, A and C). We also observe that MEK1 does not significantly regulate neutrophil adhesion to HMVEC-L after 4 h of Pam3Cys treatment, which is consistent with the results obtained from the E-selectin and ICAM-1 cell ELISAs (Figs. 7, A and D, and 8A). To verify the results of the ERK5 inhibitor studies and to confirm a role for NF-κB, we knocked down the expression of ERK5 and p65 in HMVEC-L and performed a neutrophil adhesion assay after a 4-h treatment with Pam3Cys. Consistent with the inhibitor studies, the knockdown studies indicate that ERK5 expression is necessary for the maximal adhesion of neutrophils and that NF-κB is also critical for neutrophil adhesion to Pam3Cys-treated HMVEC-L (Fig. 8B). As a control, we also knocked down the expression of TLR2 in HMVEC-L to confirm that the effects of Pam3Cys on neutrophil adhesion are specific (Fig. 8B). Similar results were obtained using HUVEC (data not shown). Taken together, these results indicate that the TLR2-dependent activation of human lung microvascular ECs results in the up-regulation of adhesion molecules at the cell surface that are able to retain neutrophils and that this process is dependent on p38-MAPK, JNK, ERK5, and NF-κB activity.
FIGURE 8.
p38-MAPK, JNK, ERK5, and NF-κB promote TLR2-dependent adhesion of primary human neutrophils to HMVEC-L. A, HMVEC-L were pretreated for 1 h with the indicated MAPK inhibitors or with medium and then treated with Pam3Cys (10 μg/ml) in the continuous presence of inhibitor for 4 h. The HMVEC-L were then washed, and calcein AM-labeled primary human neutrophils were then allowed to adhere for 1 h. Non-adherent neutrophils were subsequently removed, and the percentage of remaining, adherent neutrophils was calculated (n = 5). *, p < 0.05; **, p < 0.01; ***, p < 0.001, Pam3Cys versus Pam3Cys plus inhibitor. ns, not significant. B, HMVEC-L were transfected with either TLR2, RelA (p65), ERK5, or control siRNA and then treated with Pam3Cys (10 μg/ml) for 4 h. The percentage of adherent neutrophils was determined as in A (n = 4). ***, p < 0.001, Pam3Cys-treated control siRNA versus Pam3Cys-treated TLR2, RelA, or ERK5 siRNA. C, HMVEC-L were pretreated for 1 h with the indicated MAPK inhibitors or with medium and then treated with Pam3Cys (10 μg/ml) in the continuous presence of inhibitor for 4 h. The HMVEC-L were then washed, and calcein AM-labeled primary human neutrophils were then allowed to adhere for 1 h. Non-adherent neutrophils were subsequently removed, and the adherent calcein AM-labeled neutrophils were then imaged using fluorescence microscopy (5× magnification). All neutrophil adhesion assays were performed two times.
DISCUSSION
Microvascular circulatory abnormalities resulting from endothelial activation and dysfunction are believed to lie at the heart of sepsis-induced respiratory failure (3–5). The endothelium participates in the septic response through the production of inflammatory mediators, up-regulation of adhesion molecules, modulation of coagulation pathways, and alteration of barrier functions (1, 2). We previously found that bacterial lipoprotein TLR2 agonists, which are ubiquitously expressed by all bacteria that cause sepsis, induce endothelial inflammation and neutrophil adhesion to EC monolayers, increase lung neutrophil trafficking and respiratory dysfunction, and modulate the activity of several coagulation pathway intermediaries (16, 20, 45). The present studies delved into the signaling pathways involved in the TLR2-dependent activation of endothelial inflammation and modulation of coagulation pathway intermediaries.
There are several novel and important aspects of these studies. First, ERK5 promotes the TLR2-mediated expression of TLR2 and inflammatory proteins in primary human ECs and monocytes. A role for ERK5 in these TLR2-mediated processes has not been described previously in any cell type. Additionally, our neutrophil adhesion assays suggest that ERK5 plays an important role in the signaling pathways that lead to the adherence of neutrophils to the human lung endothelium during a bacterial infection. Second, MEK1, the upstream kinase of ERK1/2, negatively regulates TLR2-mediated EC expression of inflammatory proteins and of TLR2 itself. This is opposite to the role that has been reported for MEK1 in leukocytes, which we also confirmed in our current studies using human monocytes. We speculate that the opposite effects of MEK1 inhibition in these cells may reflect a fundamental difference between inflammatory responses of the endothelium and monocytes/macrophages. Third, we have extended our prior work by showing that bacterial lipopeptides up-regulate EC expression of TLR2, but not of TLR1 or TLR6, and demonstrated that the newly synthesized TLR2 protein is primarily located in intracellular compartments rather than at the cell surface. Importantly, this is in sharp contrast to monocytes, whereby Pam3Cys does not influence TLR2 expression, and TLR2 is primarily expressed on the surface. An additional important aspect of this work is that we have demonstrated that TLR2-activated signaling occurs in primary human lung microvascular endothelial cells. This strongly supports our hypothesis that endothelial TLR2 signaling pathways are important in the lung response to systemic infections. Table 1 summarizes our findings on the effects of the different MAPK inhibitors and of RNAi on the TLR2-mediated responses.
TABLE 1.
Summary of MAPK and NF-κB results in endothelial cells
+, positive regulation; −, negative regulation; NS, not significant.
| Output | p38-MAPKa | JNKb | MEK1c | ERK5d | NF-κBe |
|---|---|---|---|---|---|
| IL-6f | +/+ | +/+ | − | +/+ | + |
| IL-8f | +/+ | +/+ | NS | +/+ | + |
| GM-CSFf | +/+ | +/+ | − | +/+ | + |
| G-CSFf | +/+ | +/+ | − | +/+ | + |
| PAI-1f | +/+ | +/+ | − | +/+ | + |
| E-selecting | +/+ | +/+ | NS | +/NS | |
| E-selectinh | NS | +/+ | + | ||
| E-selectinj | +/+ | +/+ | − | +/? | |
| ICAM-1g | +/+ | +/+ | NS | NS/+ | |
| ICAM-1h | − | +/+ | + | ||
| Neutrophil Adhesioni | +/+ | +/+ | NS | +/+ | + |
| TLR2j | +/+ | +/+ | − | +/+ | + |
| TLR2k | +/+ | +/+ | +l | +/? |
a SB203580/BIRB796.
b SP600125/AEG3482.
c PD184352, 1 μm.
d XMD8–92/ERK5 siRNA.
e RelA siRNA.
f Sandwich ELISA: HUVEC treated for 20 h with 10 μg/ml Pam3Cys.
g Cell ELISA: HUVEC treated for 4 h with 10 μg/ml Pam3Cys.
h Cell ELISA: HUVEC treated 20 h with 10 μg/ml Pam3Cys.
i Neutrophil adhesion assay: HMVEC-L treated for 4 h with 10 μg/ml Pam3Cys.
j Immunoblot: HUVEC treated for 20 h with 10 μg/ml Pam3Cys.
k Immunoblot: HMVEC-L treated 20 hr with with 10 μg/ml Pam3Cys.
l The densitrometry ratio for Pam3Cys + PD184352 (1 μm) was only slightly less than that of the Pam3Cys without the inhibitor ratio in this experiment and may not be significant.
In comparison to the monocytic cell line THP-1 and PBMCs, we observed that resting ECs express relatively low levels of TLR2 protein. We suspect that this lower base-line TLR2 expression may serve to prevent the systemic activation of endothelium during minor infections or transient bacteremia that occurs regularly during normal daily life, such as when there is a break in mucosal or skin integrity. We also observed that endothelial TLR2 is up-regulated by exposure to TLR2 agonists, which perhaps allows a more robust response in ECs during prolonged or repeated exposure to microbial threats. The primarily intracellular localization of TLR2 in ECs suggests that TLR2 agonists may have to be first internalized to activate ECs. Intracellularly localized TLR2 could also be necessary to recognize intracellular pathogens (46–48). Of note and consistent with previously published data, we also observe that TLR2 expression is not induced in monocytes after Pam3Cys treatment, emphasizing yet another profound difference between EC and monocyte biology (21).
NF-κB activity is centrally involved in inflammatory gene expression in ECs (33, 34). Although NF-κB activation was less robust in HUVEC in response to TLR2 agonist, we found that NF-κB is necessary for the maximal TLR2-induced up-regulation of inflammatory mediators, PAI-1 and TLR2. In contrast to THP-1 cells, Pam3Cys-induced phosphorylation of p65 Ser-536 was sustained out to 8 h after Pam3Cys treatment in EC. The protracted activation of p65 in the nucleus of ECs may compensate for relatively lower amounts of NF-κB heterodimers present there. This may in fact partially explain our observations that comparable amounts of cytokines are produced in monocytes and ECs after 20 h of Pam3Cys treatment. Of note, unlike for ECs, in human monocytes/macrophages neither LPS nor the TLR2 agonist MALP-2 induced the expression of TLR2 despite robustly activating NF-κB (21). This indicates that the transcription of TLR2 is regulated differently in human ECs and macrophages.
We observed that inhibition of MEK1 augments the TLR2-dependent activation of ECs, whereas inhibition or siRNA knockdown of ERK5 reduces TLR2-dependent activation of ECs. Additionally, MEK1 inhibition induces the up-regulation of endothelial TLR2 even in the absence of TLR2 agonists. These data suggest that MEK1 negatively regulates, whereas ERK5 promotes TLR2 signaling pathways in ECs. Because TLR2 activation did not modulate ERK1/2 activity, the only reported substrates of MEK1, we suspect that the basal activity of ERK1/2 observed under our growth conditions is sufficient to constitutively repress the transcription of some inflammatory genes, including TLR2 (49). We do not, however, rule out the possibility that Pam3Cys induces a change in ERK1/2 activity at later time points to promote negative feedback pathways or that unidentified substrates for MEK1 may exist. Nonetheless, the mechanism by which MEK1 negatively regulates TLR2 signaling in ECs is currently not known, but one possibility is through the ERK1/2-dependent up-regulation of MAPK phosphatase DUSP1/MKP-1 (50–52).
Because few substrates of ERK5 have been conclusively identified, it is unclear how ERK5 participates in EC TLR2 signaling. The few published reports on the role of ERK5 in TLR-mediated signaling show conflicting results. For example, LPS has been reported to activate ERK5 in RAW264.7 macrophages but has also been reported to not activate ERK5 in BAC1.2F5 macrophages (29, 53). However, ERK5 does have a critical role in EC biology (54). Members of the MEF2 family of transcription factors (i.e. MEF2A, -2C, and -2D) have been identified as ERK5 substrates (55). MEF2D binds to the promoter of, and induces the transcription of c-Jun, an AP-1 transcription factor, suggesting a role of ERK5 in AP-1 regulation (56, 57). ERK5 also induces the expression of lung Krüppel-like factor (lKLF or KLF2) through MEF2 transcription factors (58). In ECs, overexpression of KLF2 represses a variety of inflammatory outcomes (59–62). Our experiments suggest that ERK5 activity promotes TLR2-induced inflammatory protein expression, indicating that an ERK5-MEF2-KLF2 signaling axis may not be active downstream of TLR2 in ECs. The ubiquitously expressed gap junction protein connexin 43 is also a substrate of ERK5, and its phosphorylation by ERK5 induces gap junction uncoupling between adjacent cells (63, 64). We have reported that TLR2 activation augments EC monolayer permeability, which we hypothesize may be related to ERK5 activation and connexin 43-associated gap junction uncoupling (16).
We observed that JNK and p38-MAPK also have central roles in endothelial TLR2 signaling to inflammatory and coagulation (PAI-1) endpoints as well as expression of TLR2 itself. In addition to acting through AP-1 transcription factors, p38-MAPK promotes mRNA stabilization, influences NF-κB activity, modulates gene expression by regulating chromatin modifiers, and directly influences protein translation downstream of inflammatory stimuli (25, 31, 65–69). Further studies will be required to define which of these many different p38-MAPK-dependent mechanisms are responsible for TLR2 signaling in ECs.
This study deepens the understanding of microvascular responses to infection by delineating pathways involved in TLR2-mediated endothelial signaling. The microvasculature is centrally involved in the host response to infections, including those that are necessary for controlling bacterial burden and repairing damaged tissue. Dysregulated endothelial responses can contribute to microvascular dysfunction leading to organ injury and failure during inflammatory disorders such as sepsis (4, 5). We have identified novel aspects of TLR2 signaling in human ECs, including that ERK5 activity is required for many of the EC effects of TLR2 activation and that MEK1 activity down-regulates TLR2-mediated activation of EC inflammatory responses. We have also demonstrated that signal transduction pathways downstream of TLR2 differ between human leukocytes and ECs. Furthermore, our study confirms functional relevance by showing that TLR2 activation in human lung microvascular ECs causes the robust adherence of neutrophils to the endothelium, suggesting that TLR2 plays a central role in lung responses to disseminated bacterial infections. The identification of endothelial-specific TLR2 signaling pathways and the identification of ERK5 as a central regulator of TLR2-mediated inflammatory signaling could have important implications for the design of therapies targeting endothelial-specific or leukocyte-specific inflammatory responses.
Acknowledgments
We thank Drs. Arun Prakash and Arnoud Sonnenberg for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01AI058106 (NIAID; to J. Hellman). This work was also supported by the University of California San Francisco Department of Anesthesia and Perioperative Care.

This article contains supplemental Tables S1–S4 and Fig. S1.
The quantitative real-time PCR array data have been deposited in the NCBI Gene Expression Omnibus (GEO Series accession no. GSE33449).
- EC
- endothelial cell
- TLR
- Toll-like receptor
- PAI-1
- plasminogen activator inhibitor-1
- HUVEC
- human umbilical vein endothelial cell
- HMVEC-L
- human lung microvascular endothelial cell
- RT
- room temperature
- PBMC
- peripheral blood mononuclear cell
- G-CSF
- granulocyte colony-stimulating factor
- ICAM-1
- intercellular adhesion molecule 1.
REFERENCES
- 1. Ley K., Laudanna C., Cybulsky M. I., Nourshargh S. (2007) Getting to the site of inflammation. The leukocyte adhesion cascade updated. Nat. Rev. Immunol. 7, 678–689 [DOI] [PubMed] [Google Scholar]
- 2. Hickey M. J., Kubes P. (2009) Intravascular immunity, The host-pathogen encounter in blood vessels. Nat. Rev. Immunol. 9, 364–375 [DOI] [PubMed] [Google Scholar]
- 3. Hotchkiss R. S., Karl I. E. (2003) The pathophysiology and treatment of sepsis. N. Engl. J. Med. 348, 138–150 [DOI] [PubMed] [Google Scholar]
- 4. Aird W. C. (2003) The role of the endothelium in severe sepsis and multiple organ dysfunction syndrome. Blood 101, 3765–3777 [DOI] [PubMed] [Google Scholar]
- 5. Rittirsch D., Flierl M. A., Ward P. A. (2008) Harmful molecular mechanisms in sepsis. Nat. Rev. Immunol. 8, 776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pober J. S., Sessa W. C. (2007) Evolving functions of endothelial cells in inflammation. Nat. Rev. Immunol. 7, 803–815 [DOI] [PubMed] [Google Scholar]
- 7. Takeuchi O., Akira S. (2010) Pattern recognition receptors and inflammation. Cell 140, 805–820 [DOI] [PubMed] [Google Scholar]
- 8. Lin Q., Li M., Fang D., Fang J., Su S. B. (2011) The essential roles of Toll-like receptor signaling pathways in sterile inflammatory diseases. Int. Immunopharmacol. 11, 1422–1432 [DOI] [PubMed] [Google Scholar]
- 9. Kang J. Y., Lee J. O. (2011) Structural biology of the Toll-like receptor family. Annu. Rev. Biochem. 80, 917–941 [DOI] [PubMed] [Google Scholar]
- 10. Zähringer U., Lindner B., Inamura S., Heine H., Alexander C. (2008) TLR2- promiscuous or specific? A critical re-evaluation of a receptor expressing apparent broad specificity. Immunobiology 213, 205–224 [DOI] [PubMed] [Google Scholar]
- 11. Jin M. S., Kim S. E., Heo J. Y., Lee M. E., Kim H. M., Paik S. G., Lee H., Lee J. O. (2007) Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a triacylated lipopeptide. Cell 130, 1071–1082 [DOI] [PubMed] [Google Scholar]
- 12. Kang J. Y., Nan X., Jin M. S., Youn S. J., Ryu Y. H., Mah S., Han S. H., Lee H., Paik S. G., Lee J. O. (2009) Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity 31, 873–884 [DOI] [PubMed] [Google Scholar]
- 13. Talreja J., Kabir M. H., B Filla M., Stechschulte D. J., Dileepan K. N. (2004) Histamine induces Toll-like receptor 2 and 4 expression in endothelial cells and enhances sensitivity to Gram-positive and Gram-negative bacterial cell wall components. Immunology 113, 224–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li Y., Xiang M., Yuan Y., Xiao G., Zhang J., Jiang Y., Vodovotz Y., Billiar T. R., Wilson M. A., Fan J. (2009) Hemorrhagic shock augments lung endothelial cell activation. Role of temporal alterations of TLR4 and TLR2. Am. J. Physiol. Regul. Integr. Comp. Physiol. 297, R1670–R1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grote K., Schuett H., Salguero G., Grothusen C., Jagielska J., Drexler H., Mühlradt P. F., Schieffer B. (2010) Toll-like receptor 2/6 stimulation promotes angiogenesis via GM-CSF as a potential strategy for immune defense and tissue regeneration. Blood 115, 2543–2552 [DOI] [PubMed] [Google Scholar]
- 16. Shin H. S., Xu F., Bagchi A., Herrup E., Prakash A., Valentine C., Kulkarni H., Wilhelmsen K., Warren S., Hellman J. (2011) Bacterial lipoprotein TLR2 agonists broadly modulate endothelial function and coagulation pathways in vitro and in vivo. J. Immunol. 186, 1119–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Satta N., Kruithof E. K., Reber G., de Moerloose P. (2008) Induction of TLR2 expression by inflammatory stimuli is required for endothelial cell responses to lipopeptides. Mol. Immunol. 46, 145–157 [DOI] [PubMed] [Google Scholar]
- 18. Fan J., Frey R. S., Malik A. B. (2003) TLR4 signaling induces TLR2 expression in endothelial cells via neutrophil NADPH oxidase. J. Clin. Invest. 112, 1234–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Faure E., Thomas L., Xu H., Medvedev A., Equils O., Arditi M. (2001) Bacterial lipopolysaccharide and IFNγ induce Toll-like receptor 2 and Toll-like receptor 4 expression in human endothelial cells. Role of NF-κB activation. J. Immunol. 166, 2018–2024 [DOI] [PubMed] [Google Scholar]
- 20. Wilhelmsen K., Mesa K. R., Prakash A., Xu F., Hellman J. (2011) Activation of endothelial TLR2 by bacterial lipoprotein upregulates proteins specific for the neutrophil response. Innate Immun, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haehnel V., Schwarzfischer L., Fenton M. J., Rehli M. (2002) Transcriptional regulation of the human toll-like receptor 2 gene in monocytes and macrophages. J. Immunol. 168, 5629–5637 [DOI] [PubMed] [Google Scholar]
- 22. Vallabhapurapu S., Karin M. (2009) Regulation and function of NF-κB transcription factors in the immune system. Annu. Rev. Immunol. 27, 693–733 [DOI] [PubMed] [Google Scholar]
- 23. Scheidereit C. (2006) IκB kinase complexes. Gateways to NF-κB activation and transcription. Oncogene 25, 6685–6705 [DOI] [PubMed] [Google Scholar]
- 24. Perkins N. D. (2006) Post-translational modifications regulating the activity and function of the nuclear factor κB pathway. Oncogene 25, 6717–6730 [DOI] [PubMed] [Google Scholar]
- 25. Verstak B., Nagpal K., Bottomley S. P., Golenbock D. T., Hertzog P. J., Mansell A. (2009) MyD88 adapter-like (Mal)/TIRAP interaction with TRAF6 is critical for TLR2- and TLR4-mediated NF-κB proinflammatory responses. J. Biol. Chem. 284, 24192–24203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cargnello M., Roux P. P. (2011) Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 75, 50–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Horng T., Barton G. M., Flavell R. A., Medzhitov R. (2002) The adaptor molecule TIRAP provides signaling specificity for Toll-like receptors. Nature 420, 329–333 [DOI] [PubMed] [Google Scholar]
- 28. Farhat K., Riekenberg S., Heine H., Debarry J., Lang R., Mages J., Buwitt-Beckmann U., Röschmann K., Jung G., Wiesmüller K. H., Ulmer A. J. (2008) Heterodimerization of TLR2 with TLR1 or TLR6 expands the ligand spectrum but does not lead to differential signaling. J. Leukoc. Biol. 83, 692–701 [DOI] [PubMed] [Google Scholar]
- 29. Zhu W., Downey J. S., Gu J., Di Padova F., Gram H., Han J. (2000) Regulation of TNF expression by multiple mitogen-activated protein kinase pathways. J. Immunol. 164, 6349–6358 [DOI] [PubMed] [Google Scholar]
- 30. Miller M. (2009) The importance of being flexible. The case of basic region leucine zipper transcriptional regulators. Curr. Protein Pept. Sci. 10, 244–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cuadrado A., Nebreda A. R. (2010) Mechanisms and functions of p38 MAPK signaling. Biochem. J. 429, 403–417 [DOI] [PubMed] [Google Scholar]
- 32. Anderson P. (2008) Post-transcriptional control of cytokine production. Nat. Immunol. 9, 353–359 [DOI] [PubMed] [Google Scholar]
- 33. Collins T., Read M. A., Neish A. S., Whitley M. Z., Thanos D., Maniatis T. (1995) Transcriptional regulation of endothelial cell adhesion molecules. NF-κB and cytokine-inducible enhancers. FASEB J. 9, 899–909 [PubMed] [Google Scholar]
- 34. Ye X., Ding J., Zhou X., Chen G., Liu S. F. (2008) Divergent roles of endothelial NF-κB in multiple organ injury and bacterial clearance in mouse models of sepsis. J. Exp. Med. 205, 1303–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Read M. A., Whitley M. Z., Gupta S., Pierce J. W., Best J., Davis R. J., Collins T. (1997) Tumor necrosis factor α-induced E-selectin expression is activated by the nuclear factor-κB and c-JUN N-terminal kinase/p38 mitogen-activated protein kinase pathways. J. Biol. Chem. 272, 2753–2761 [DOI] [PubMed] [Google Scholar]
- 36. Daigneault M., Preston J. A., Marriott H. M., Whyte M. K., Dockrell D. H. (2010) The identification of markers of macrophage differentiation in PMA-stimulated THP-1 cells and monocyte-derived macrophages. PLoS One 5, e8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nuzzi P. A., Lokuta M. A., Huttenlocher A. (2007) Analysis of neutrophil chemotaxis. Methods Mol. Biol. 370, 23–36 [DOI] [PubMed] [Google Scholar]
- 38. Yen J., Kramer S. M. (1991) A rapid in vitro cytotoxicity assay for the detection of tumor necrosis factor on human BT-20 cells. J. Immunother. 10, 174–181 [DOI] [PubMed] [Google Scholar]
- 39. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 40. Willems E., Leyns L., Vandesompele J. (2008) Standardization of real-time PCR gene expression data from independent biological replicates. Anal. Biochem. 379, 127–129 [DOI] [PubMed] [Google Scholar]
- 41. Edgar R., Domrachev M., Lash A. E. (2002) Gene Expression Omnibus. NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schmidt R. R., Pedersen C. M., Qiao Y., Zähringer U. (2011) Chemical synthesis of bacterial lipoteichoic acids. An insight on its biological significance. Org. Biomol. Chem. 9, 2040–2052 [DOI] [PubMed] [Google Scholar]
- 43. Mody N., Leitch J., Armstrong C., Dixon J., Cohen P. (2001) Effects of MAP kinase cascade inhibitors on the MKK5/ERK5 pathway. FEBS Lett. 502, 21–24 [DOI] [PubMed] [Google Scholar]
- 44. Kamakura S., Moriguchi T., Nishida E. (1999) Activation of the protein kinase ERK5/BMK1 by receptor-tyrosine kinases. Identification and characterization of a signaling pathway to the nucleus. J. Biol. Chem. 274, 26563–26571 [DOI] [PubMed] [Google Scholar]
- 45. Petersen B., Bloch K. D., Ichinose F., Shin H. S., Shigematsu M., Bagchi A., Zapol W. M., Hellman J. (2008) Activation of Toll-like receptor 2 impairs hypoxic pulmonary vasoconstriction in mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 294, L300–L308 [DOI] [PubMed] [Google Scholar]
- 46. Takeuchi O., Hoshino K., Akira S. (2000) Cutting edge. TLR2-deficient and MyD88-deficient mice are highly susceptible to Staphylococcus aureus infection. J. Immunol. 165, 5392–5396 [DOI] [PubMed] [Google Scholar]
- 47. Menzies B. E., Kourteva I. (1998) Internalization of Staphylococcus aureus by endothelial cells induces apoptosis. Infect Immun 66, 5994–5998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Beekhuizen H., van de Gevel J. S., Olsson B., van Benten I. J., van Furth R. (1997) Infection of human vascular endothelial cells with Staphylococcus aureus induces hyperadhesiveness for human monocytes and granulocytes. J. Immunol. 158, 774–782 [PubMed] [Google Scholar]
- 49. Shaul Y. D., Seger R. (2007) The MEK/ERK cascade. From signaling specificity to diverse functions. Biochim. Biophys. Acta 1773, 1213–1226 [DOI] [PubMed] [Google Scholar]
- 50. Chen P., Li J., Barnes J., Kokkonen G. C., Lee J. C., Liu Y. (2002) Restraint of proinflammatory cytokine biosynthesis by mitogen-activated protein kinase phosphatase-1 in lipopolysaccharide-stimulated macrophages. J. Immunol. 169, 6408–6416 [DOI] [PubMed] [Google Scholar]
- 51. Bertelsen T., Iversen L., Riis J. L., Arthur J. S., Bibby B. M., Kragballe K., Johansen C. (2011) The role of mitogen- and stress-activated protein kinase 1 and 2 in chronic skin inflammation in mice. Exp. Dermatol. 20, 140–145 [DOI] [PubMed] [Google Scholar]
- 52. Salojin K. V., Owusu I. B., Millerchip K. A., Potter M., Platt K. A., Oravecz T. (2006) Essential role of MAPK phosphatase-1 in the negative control of innate immune responses. J. Immunol. 176, 1899–1907 [DOI] [PubMed] [Google Scholar]
- 53. Rovida E., Spinelli E., Sdelci S., Barbetti V., Morandi A., Giuntoli S., Dello Sbarba P. (2008) ERK5/BMK1 is indispensable for optimal colony-stimulating factor 1 (CSF-1)-induced proliferation in macrophages in a Src-dependent fashion. J. Immunol. 180, 4166–4172 [DOI] [PubMed] [Google Scholar]
- 54. Roberts O. L., Holmes K., Müller J., Cross D. A., Cross M. J. (2009) ERK5 and the regulation of endothelial cell function. Biochem. Soc. Trans. 37, 1254–1259 [DOI] [PubMed] [Google Scholar]
- 55. Kato Y., Zhao M., Morikawa A., Sugiyama T., Chakravortty D., Koide N., Yoshida T., Tapping R. I., Yang Y., Yokochi T., Lee J. D. (2000) Big mitogen-activated kinase regulates multiple members of the MEF2 protein family. J. Biol. Chem. 275, 18534–18540 [DOI] [PubMed] [Google Scholar]
- 56. Kato Y., Kravchenko V. V., Tapping R. I., Han J., Ulevitch R. J., Lee J. D. (1997) BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. EMBO J. 16, 7054–7066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Han T. H., Prywes R. (1995) Regulatory role of MEF2D in serum induction of the c-jun promoter. Mol. Cell. Biol. 15, 2907–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sohn S. J., Li D., Lee L. K., Winoto A. (2005) Transcriptional regulation of tissue-specific genes by the ERK5 mitogen-activated protein kinase. Mol. Cell. Biol. 25, 8553–8566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. SenBanerjee S., Lin Z., Atkins G. B., Greif D. M., Rao R. M., Kumar A., Feinberg M. W., Chen Z., Simon D. I., Luscinskas F. W., Michel T. M., Gimbrone M. A., Jr., García-Cardeña G., Jain M. K. (2004) KLF2 Is a novel transcriptional regulator of endothelial proinflammatory activation. J. Exp. Med. 199, 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lin Z., Kumar A., SenBanerjee S., Staniszewski K., Parmar K., Vaughan D. E., Gimbrone M. A., Jr., Balasubramanian V., García-Cardeña G., Jain M. K. (2005) Kruppel-like factor 2 (KLF2) regulates endothelial thrombotic function. Circ. Res. 96, e48–57 [DOI] [PubMed] [Google Scholar]
- 61. Bhattacharya R., Senbanerjee S., Lin Z., Mir S., Hamik A., Wang P., Mukherjee P., Mukhopadhyay D., Jain M. K. (2005) Inhibition of vascular permeability factor/vascular endothelial growth factor-mediated angiogenesis by the Kruppel-like factor KLF2. J. Biol. Chem. 280, 28848–28851 [DOI] [PubMed] [Google Scholar]
- 62. Atkins G. B., Jain M. K. (2007) Role of Krüppel-like transcription factors in endothelial biology. Circ. Res. 100, 1686–1695 [DOI] [PubMed] [Google Scholar]
- 63. Solan J. L., Lampe P. D. (2009) Connexin43 phosphorylation. Structural changes and biological effects. Biochem. J. 419, 261–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cameron S. J., Malik S., Akaike M., Lerner-Marmarosh N., Yan C., Lee J. D., Abe J., Yang J. (2003) Regulation of epidermal growth factor-induced connexin 43 gap junction communication by big mitogen-activated protein kinase1/ERK5 but not ERK1/2 kinase activation. J. Biol. Chem. 278, 18682–18688 [DOI] [PubMed] [Google Scholar]
- 65. Winzen R., Kracht M., Ritter B., Wilhelm A., Chen C. Y., Shyu A. B., Müller M., Gaestel M., Resch K., Holtmann H. (1999) The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 18, 4969–4980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sandler H., Stoecklin G. (2008) Control of mRNA decay by phosphorylation of tristetraprolin. Biochem. Soc. Trans. 36, 491–496 [DOI] [PubMed] [Google Scholar]
- 67. Vermeulen L., De Wilde G., Van Damme P., Vanden Berghe W., Haegeman G. (2003) Transcriptional activation of the NF-κB p65 subunit by mitogen- and stress-activated protein kinase-1 (MSK1). EMBO J. 22, 1313–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Saccani S., Pantano S., Natoli G. (2002) p38-Dependent marking of inflammatory genes for increased NF-κB recruitment. Nat. Immunol. 3, 69–75 [DOI] [PubMed] [Google Scholar]
- 69. Wang X., Flynn A., Waskiewicz A. J., Webb B. L., Vries R. G., Baines I. A., Cooper J. A., Proud C. G. (1998) The phosphorylation of eukaryotic initiation factor eIF4E in response to phorbol esters, cell stresses, and cytokines is mediated by distinct MAP kinase pathways. J. Biol. Chem. 273, 9373–9377 [DOI] [PubMed] [Google Scholar]
- 70. Ananieva O., Darragh J., Johansen C., Carr J. M., McIlrath J., Park J. M., Wingate A., Monk C. E., Toth R., Santos S. G., Iversen L., Arthur J. S. (2008) The kinases MSK1 and MSK2 act as negative regulators of Toll-like receptor signaling. Nat. Immunol. 9, 1028–1036 [DOI] [PubMed] [Google Scholar]