Background: RhoGDI is a key regulator and a chaperon of Rho GTPases.
Results: RhoGDI strongly discriminates between GDP- and GTP-bound forms of prenylated RhoA, although both complexes are of high affinity.
Conclusion: We provide direct evidence for the existence of two populations of the RhoGDI·RhoA complexes in the cell, characterized by different lifetimes.
Significance: The obtained data allows us to formulate the model for membrane delivery and extraction of Rho GTPases.
Keywords: GTPase, Membrane Transport, Molecular Chaperone, Protein Isoprenylation, Rho GTPases
Abstract
Small GTPases of the Rho family regulate cytoskeleton remodeling, cell polarity, and transcription, as well as the cell cycle, in eukaryotic cells. Membrane delivery and recycling of the Rho GTPases is mediated by Rho GDP dissociation inhibitor (RhoGDI), which forms a stable complex with prenylated Rho GTPases. We analyzed the interaction of RhoGDI with the active and inactive forms of prenylated and unprenylated RhoA. We demonstrate that RhoGDI binds the prenylated form of RhoA·GDP with unexpectedly high affinity (Kd = 5 pm). The very long half-life of the complex is reduced 25-fold on RhoA activation, with a concomitant reduction in affinity (Kd = 3 nm). The 2.8-Å structure of the RhoA·guanosine 5′-[β,γ-imido] triphosphate (GMPPNP)·RhoGDI complex demonstrated that complex formation forces the activated RhoA into a GDP-bound conformation in the absence of nucleotide hydrolysis. We demonstrate that membrane extraction of Rho GTPase by RhoGDI is a thermodynamically favored passive process that operates through a series of progressively tighter intermediates, much like the one that is mediated by RabGDI.
Introduction
The Rho family of GTPases plays important roles in a plethora of cellular functions. The defining members of this family, RhoA, Rac1, and Cdc42, regulate cytoskeleton remodeling, cell polarity (1, 2), and transcription (3), as well as the cell cycle (4). Similar to other Ras family GTPases, the Rho proteins cycle between the inactive GDP-bound and the active GTP-bound effector binding competent states. This cycle is tightly controlled by guanine nucleotide exchange factors (GEFs)2 that facilitate loading Rho with guanosine triphosphate (5) and GTPase-activating proteins that dramatically accelerate the intrinsic rate of GTP hydrolysis (6).
Similar to the majority of other Ras superfamily members, the Rho proteins are posttranslationally prenylated on their C termini. Rho GTPases are predominantly singly geranylgeranylated, although farnesylation has also been reported (7). These modifications enable small GTPases to reversibly and dynamically associate with intracellular membranes. In a number of cases, membrane delivery was shown to be functionally coupled to GTPase activation. For instance, GTP loading of the Arf GTPases exposes their otherwise buried myristoylated N-terminal helix and promotes association of Arf proteins with the plasma membrane (8). Similarly, GEF-mediated loading of Rab GTPases with GTP reduces its affinity for its chaperone, Rab GDP dissociation inhibitor (RabGDI), by 3 orders of magnitude and promotes association of GTP-bound RabGTPases with their target membrane (9).
As in the case of Rab proteins, membrane/cytosol cycling of geranylgeranylated Rho GTPases is regulated by functionally similar but structurally unrelated GDP dissociation inhibitors (RhoGDIs). In contrast to the RabGDI, RhoGDIs were reported to interact with both GDP- and GTP-bound Rho proteins with similar efficiency (10–13). However, the exact affinities of these interactions are still disputed (14–16).
The lack of clear evidence for the role of nucleotide exchange in the dissociation of Rho·RhoGDI complexes has led to a search for alternative mechanisms of the membrane recruitment of Rho. A number of putative GDI displacement factors were proposed to promote dissociation of the RhoGDI·Rho complexes in vivo. These include receptors, such as p75 NTR (17) and integrins (18), and associated proteins, such as ERM (19) and merlin (20). However, no thermodynamically sound model has been proposed that would explain how GDI displacement factors could reduce the high affinity of the RhoGDI/Rho interaction. One alternative explanation is that these molecules act as scaffold proteins that recruit RhoGEFs (21–23) and membrane phosphoinositides (21), which have been proposed to facilitate Rho·RhoGDI complex dissociation (22). Phosphorylation of RhoGDI or Rho proteins was also shown to influence the Rho/RhoGDI interaction, strongly affecting their association in the cell (23–27). Phosphorylation of Rho GTPases is believed to promote their extraction from the membranes by RhoGDI, whereas phosphorylation of RhoGDI diminishes its ability to bind Rho.
Despite numerous reports on the possible mechanisms that govern membrane delivery and recycling of Rho GTPases, a kinetic and thermodynamic description of such mechanisms is still outstanding. Therefore, this study reports on the quantitative biophysical analysis of RhoA interaction with RhoGDI with the objective of determining the kinetic and thermodynamic framework of the Rho functional cycle. In contrast to previous findings, we have demonstrated that RhoGDI discriminates strongly between different nucleotide-bound forms of prenylated and unprenylated RhoA. We provide direct evidence for the existence of two populations of the RhoGDI·RhoA complex in the cell, characterized by different lifetimes. We also propose a thermodynamic model of RhoGDI-mediated RhoGTPase recycling.
MATERIALS AND METHODS
Protein Expression and Purification
All proteins used in this study were expressed in the Escherichia coli BL21(DE3) Codon plus RIL strain (Stratagene). Expression and purification of FTase, GGTase-I, and RhoAF25N were carried out as described previously (28). RhoGDI was cloned in-frame with maltose-binding protein using the pOPINM vector (29), expressed in E. coli, and purified by nickel-nitrilotriacetic acid chromatography. The tag was removed by proteolysis with Precision protease, and the protein was further purified by gel filtration on a Superdex 200 26/60 column (GE Healthcare). The L193A mutant of RhoAF25N was constructed by PCR mutagenesis of the pGATEV-RhoAF25N plasmid using the forward 5′-GATGGGTATCCATGGCTGATG-3′, and reverse primer 5′-GGATCCCTCGAGTCATGCGACAAGGC-3′, and subcloning the resulting PCR product into the NcoI and XhoI sites of the pGATEV vector (30). The C-terminal citrine-RhoGDI was cloned using the in-fusion protocol into the pOPINE vector (29). The CFP N-terminal fusion of RhoA was cloned into the pGATEV vector using the forward 5′-CATCACAACACTAGTATGGTGAGCAAGGGCGAGGAGCTG-3′, and reverse 5′-ATTCTCATCAGCC-ATGGACTTGTACAGCTCGTCCATGCCG-3′, primers and cloning of the PCR fragment into SpeI and NcoI sites.
Preparation of GMPPNP-, mantGDP-, and mantGMPPNP-bound forms of the RhoA
Nucleotide exchange on the RhoA molecule was performed as described earlier (31).
Enzymatic Prenylation of the RhoA
Purified RhoA was enzymatically prenylated in vitro by GGTase-I using geranylgeranyl or NBD-geranyl pyrophosphates as substrates. RhoAL193A was farnesylated by FTase using FPP as a substrate. The prenylation reaction was carried out in 2–4 ml of prenylation buffer containing 25 mm Hepes-NaOH, pH 7.2, 40 mm NaCl, 2 mm MgCl2, 2 mm dithioerythritol, 20 μm GDP or 20 μm GMPPNP, and 20 μm ZnCl2. A typical reaction contained 60 nmol of the prenyltransferase, mixed with 2-fold excess of RhoA and 260 nmol of GGPP (Sigma) or 550 nmol of NDB-GPP (28). The RhoA farnesylation reaction contained 10 nmol of FTase, 100 nmol of RhoA, and 400 nmol of FPP (Sigma).
After incubation for 4 h at room temperature, the reaction mixture was chromatographed on a Superdex 200 10/30 gel filtration column (GE Healthcare) equilibrated with prenylation buffer without ZnCl2. The flow rate was 0.35 ml/min, and fractions of 0.3 ml were collected and analyzed by 15% SDS-PAGE. When the fluorescent analog was used as a substrate, the gel was scanned using a Typhoon Trio fluorescent scanner (GE Healthcare) (excitation 488 nm; cutoff filter 520 nm), and the gel was subsequently stained with Coomassie Blue. Isoprenoid conjugation was confirmed by electrospray ionization-mass spectrometry of the selected fractions. The eluted fractions containing the protein complex were combined, concentrated, flash-frozen in liquid nitrogen, and stored at −80 °C.
The complex of geranylgeranylated RhoA and GGTase-I was obtained by size exclusion chromatography on a Superdex 200 column of the in vitro prenylation reaction containing equimolar amounts of GGTase-I and RhoA. The fractions containing the protein complex were collected, concentrated to 10 mg/ml, and stored as described above.
Labeling of the RhoGDI with Maleimide Dyes
The tetramethylrhodamine-maleimide dye was obtained from Molecular Probes, and RhoGDI labeling was performed according to the manufacturer's protocol. The degree of labeling, based on spectroscopic observation, was typically 50%.
Fluorescence Measurements
Fluorescence spectra and long-time base fluorescence measurements were performed at 25 °C in 1-ml quartz cuvettes (Hellma, Germany) with continuous stirring on a Spex Fluoromax-3 or Fluoromax-4 spectrofluorometer (Jobin Yvon Inc., USA). Fluorescence titrations were carried out in buffer containing 25 mm Hepes-NaOH, pH 7.2, 40 mm NaCl, 2 mm MgCl2, 5 mm dithioerythritol, and 20 μm guanosine nucleotide (GDP or GMPPNP). NBD-Geranyl was excited at 479 nm (slit 4 nm), and the emission was monitored at 560 nm (slit 8 nm). The dissociation of the fluorescent CFP-RhoA-GG·RhoGDI-citrine complex was monitored with extinction at 436 nm (4 nm slits), and emission spectra at 475–530 nm were read. The FRET efficiency (EFRET) was defined as intensity of the citrine (acceptor, 530 nm) divided by the sum of the intensity of the CFP (donor, 475 nm) and acceptor.
 |
Primary data analysis was performed with the programs Grafit 5.0 (Erithacus software) and “fluorescence” implementation (Jobin Yvon Inc., USA) of Origin 7.0 (Originlab Corp., USA).
Fluorescence Cross-correlation Measurements
Measurements were done on an inverted laser scanning confocal microscope Zeiss LSM 710 FCS, equipped with a Confocor 3 module (FCCS detector head and APD detectors). Normally, the excitation laser beams were focused on 20 μl of the sample, placed on the well of an in-house made silicon plate mounted on an optically clear coverslip. Emitted light from the 458-nm and 561-nm lasers passed through an adjustable pinhole and a 458/561 main beam splitter and was separated on a 565-nm diachronic mirror. The 470–540-nm spectra segment was used to quantify fluorescence from CFP, whereas a long pass 580-nm filter was set for measuring the tetramethylrhodamine dye signal. The laser power was set up to achieve a count rate (CPM) that was not giving detectable cross-talk. Typically, CPM was kept at 8 to 12 kHz. The data acquisition time was at least 100 s and included 10 runs of 10 s each. The collected data were analyzed using the ZEN 2011 Zeiss software package. The theoretical basics of autocorrelation and cross-correlation analysis are described everywhere (32). Experimental autocorrelation functions were fitted in a three-dimensional diffusion model (33) for a 2-component model. The Kd for the interaction of geranylgeranylated CFP-RhoA and tetramethylrhodamine-labeled RhoGDI (RhoGDI-TMR) was quantified based on the assumption that the amplitude of the cross-correlation function (CCF) corresponds linearly to the concentration of the CFP-RhoA-GG·RhoGDI-TMR complex. The CCF values obtained by titration of the 50 nm CFP-RhoA-GG·RhoGDI-TMR solution with farnesylated RhoA were fitted to the 3-component competitive model in Dynafit 4.0 (Biokin Ltd.) (34).
Analysis of Titrations of NBD-geranylated RhoA Versus Increasing Concentration of RhoGDI or GGTase-I
Titrations were fitted to the explicit solution of the quadratic equation describing the P + L ⇔ PL binding equilibrium, where Kd is defined as Kd = [P][L]/[PL]. For titrations performed in this study, the concentration of RhoA-geranyl-NBD (RhoA-GNBD) was fixed [P0], and increasing concentrations of GGTase-I or RhoGDI [L0] were added. Total concentrations of [P0] and [L0] are the sum of free and bound species. Under these conditions, the fluorescence is described as,
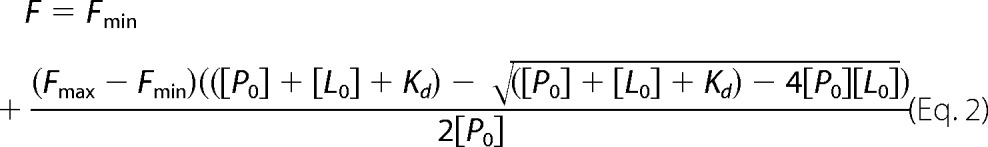 |
where F is the measured fluorescence intensity and Fmax and Fmin refer to the maximal and minimal of its value. The parameters [P0], Fmax, Fmin, and Kd were allowed to vary during the fit. The nonlinear least-square regression was performed in Grafit 5.0 (Erithacus software).
Global Fitting Analysis of Competitive Titrations
Competitive titration was performed by titrating the mixture of fluorescent RhoA-GNBD and prenylated RhoA·GGTase-I complex with increasing concentrations of RhoGDI. The global fitting procedure of the obtained data were performed in Dynafit 4.0 (Biokin Ltd.) (34).
Stopped-flow Measurements
The kinetics of the prenylated RhoA interaction with RhoGDI was analyzed using an SF-61MX stopped flow apparatus (High Tech Scientific). Measurements were carried out at 25 °C in degassed buffer containing 25 mm Hepes-NaOH, pH 7.2, 40 mm NaCl, 2 mm MgCl2, and 5 mm dithioerythritol. Fluorescence of the NBD group was excited at 479 nm, and the data were collected with a 515-nm cut-off filter. Transient kinetics of the RhoGDI interaction with RhoA bound to mant-labeled guanine nucleotides was measured under the same conditions, except that fluorescence of the mant group was excited via FRET from tryptophan at 289 nm and detected through a 395-nm cut-off filter. To improve the signal to noise ratio, data from 5 to 10 measurements were averaged and used for further analysis. Data collection and primary analysis of the rate constants were performed with the High Tech Scientific package, and the secondary analysis was performed using Grafit 5.0 (Erithacus software).
Isothermal Titration Calorimetry Measurements
Binding affinities of the unprenylated GDP- and GMPPNP-bound forms of RhoA to RhoGDI were determined using ITC200 (MicroCal). All proteins were kept in buffer containing 25 mm Hepes-NaOH, pH 7.2, 40 mm NaCl, 2 mm MgCl2, and 1 mm DTT. The concentration of RhoA protein in the syringe was at least 10-fold higher than the concentration of protein in the cell (typically 10 μm). In control experiments, the respective protein was injected from the syringe into the buffer solution, and the recorded background signals were subtracted from the titration experiment data. The titration experiments were carried out at 25 °C in triplicate. The data obtained were fitted using the MicroCal-ITC implementation of the Origin 7 software package.
Crystallization of RhoA·GMPPNP-GG·RhoGDI Complex, Data Collection and Structure Determination
Initial crystallization conditions were determined at room temperature using the PEG Ion kit from Hampton Research in 200-nl sitting drops, set up against a 50-μl reservoir with a Mosquito nanoliter dispensing robot (TTP LabTech). Promising conditions were optimized with respect to precipitant composition, pH, temperature, and protein concentration and were transferred to hanging drops prepared by mixing 1 μl of protein complex solution with 1 μl of precipitant mixture. Crystals for data collection were obtained at 20 °C with a reservoir consisting of 20% (w/v) PEG 3350, 0.2 m MgCl2, and 0.1 m Hepes-NaOH, pH 7.4. The protein complex was used at 13 mg/ml in buffer containing 25 mm Hepes-NaOH, pH 7.2, 40 mm NaCl, 2 mm MgCl2, 10 μm GMPPNP, and 1 mm Tris(2-carboxyethyl)phosphine.
Prior to flash-cooling in liquid nitrogen, the crystals were washed briefly in 25% (w/v) PEG 3350, 5% (v/v) glycerol, 0.2 m MgCl2, and 0.1 m Hepes, pH 7.4. Diffraction data were collected at 100 K at station MX2 of the Australia Synchrotron (Melbourne, Australia). The data were processed with XDS (55). The crystals belonged to the orthorhombic space group P212121 and contained 1 RhoA-GG·RhoGDI complex in the asymmetric unit.
Initial phases were determined by molecular replacement with PHASER (56) of the CCP4 suite, using coordinates of Cdc42·GDP-GG·RhoGDI (PDB code 1DOA) from which the nucleotide, Mg2+, and prenylated C terminus of Cdc24 had been deleted. The model was then corrected by alternating rounds of refinement in REFMAC5 (57) and manual adjustment in COOT (58). Restraint libraries were generated with PRODRG (59). Interface analysis was performed with PISA at the EBI website (35). Structure figures were generated using PyMOL.
RESULTS
Interaction Analysis of RhoGDI with Unprenylated RhoA
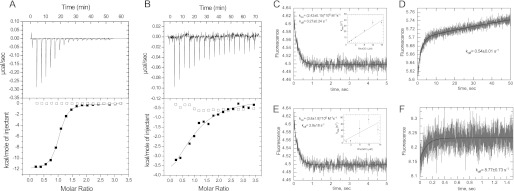
To gain insight into the mechanism of the interaction of RhoGDI with RhoA GTPase and the role of the nucleotide-bound state in this process, we assessed the thermodynamics of this interaction. We measured the affinity of the interaction between RhoGDI and recombinant unprenylated RhoA, loaded with GDP- or the unhydrolyzable GTP analog GMPPNP, using isothermal titration calorimetry (ITC). We found that RhoGDI forms a complex with GDP- and GMPPNP-bound RhoA with Kd values of 0.17 and 2.5 μm, respectively (Fig. 1, A and B). To confirm the obtained affinity values using an independent method, we decided to characterize the kinetics of this interaction. We loaded RhoA molecules with the fluorescent nucleotide mant-GDP or mant-GMPPNP, a nonhydrolyzable fluorescent analog of GTP. Rapid mixing of the mant nucleotide-loaded RhoA with RhoGDI in a stopped-flow apparatus resulted in time-dependent fluorescence signal changes. The plots in Fig. 1, C and E, represent a typical fit to the experimental data. The insets show the dependence of the pseudo-first order rate constants on the concentration of RhoGDI. Although the data give a good estimate of the kon values, the koff cannot be extracted from the intercept with y axis reliably, due to its low value in the case of GDP-bound RhoA and low signal amplitudes in the case of the GMPPNP-bound form. Therefore, we determined koff values experimentally by displacing mant nucleotide-labeled RhoA from RhoA·RhoGDI complexes with an excess of nonlabeled GDP-bound RhoA (Fig. 1, D and F). Analysis of the kinetic parameters of the RhoGDI interaction with both nucleotide-bound forms of RhoA (Table 1) shows that the affinity differences stem from a 15-fold faster off-rate of the RhoGDI complex with the activated RhoA. The affinities that were calculated from the kinetic constants were 0.2 μm for GDP-bound and 2.3 μm for GMPPNP-bound RhoA and are in good agreement with the Kd values obtained in the ITC measurements.
FIGURE 1.
Interaction analysis of RhoGDI with unprenylated RhoA. A, representative ITC data for titration of 10 μm RhoGDI with either 160 μm RhoA·GDP (filled squares) or buffer (open squares). Solid curve represents a fit of the data to a 1:1 stoichiometry binding model with a calculated Kd value 213 nm; B as in A except that RhoGDI was titrated with 130 μm RhoA·GMPPNP (filled squares) or buffer (open squares). The fit resulted in a Kd value of 5.7 μm. C, kinetic analysis of the RhoGDI interaction with unprenylated RhoA·mGDP. A typical fluorescence change observed upon mixing 0.4 μm RhoAmGDP with 8 μm RhoGDI. The inset represents a plot of the observed rates as function of RhoGDI concentration. D, fluorescence change upon mixing of 0.4 μm RhoA·mGDP·RhoGDI complex with 5 μm RhoA·GDP. E, fluorescence change resulting from mixing of 0.4 μm RhoA·mGMPPNP with 12 μm RhoGDI. The inset represents the plot of observed rates as a function of RhoGDI concentration. F, fluorescence change upon mixing of 1.2 μm RhoA·mGMPPNP·RhoGDI with 20 μm RhoA·GMPPNP.
TABLE 1.
Summary of thermodynamic and kinetic parameters of the RhoGDI interaction with GDP- and GMPPNP-associated RhoA
| Protein | kon | koff | Kd |
|---|---|---|---|
| μm−1 s−1 | s−1 | m | |
| RhoA·GDP | 2.42 ± 0.15 | 0.54 ± 0.01 | 0.17 × 10−6 |
| RhoA·GMPPNP | 3.8 ± 1.9 | 8.77 ± 0.73 | 2.5 × 10−6 |
| CFP-RhoA·GDP-GG | 37a | (2.0 ± 0.02) × 10−4 | (5.4 ± 2.5) × 10−12 |
| CFP-RhoA·GMPPNP-GG | 1.6a | (4.6 ± 0.03) × 10−3 | (2.9 ± 0.65) × 10−9 |
| RhoA·GDP-GNBD | 10.7 ± 0.7 | (5.8 ± 0.3) × 10−2 | (2.0 ± 0.15) × 10−9 |
| RhoA·GDP-F | NDb | ND | (2.5 ± 1.1) × 10−9 |
a Calculated from kon = koff /Kd.
b ND, not determined.
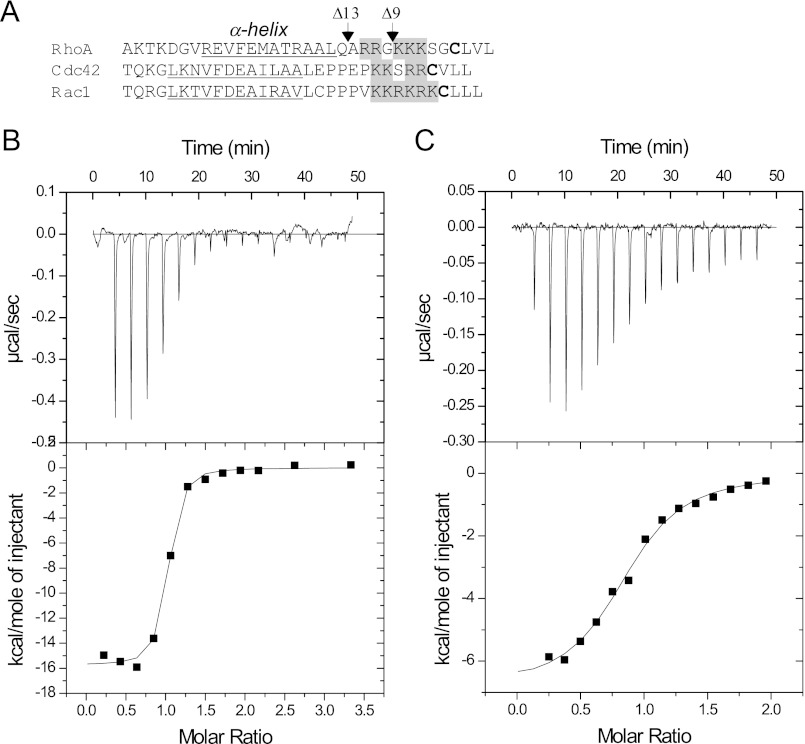
To assess the role of the RhoA C terminus in RhoGDI·Rho complex assembly, we prepared C-terminal truncated Δ9 and Δ13 RhoA mutants that lacked the C-terminal stretch lysines or in addition arginine residues, respectively (Fig. 2A). Interaction of the truncation mutants with RhoGDI was assessed by ITC as described for the wild-type protein. The titration experiments showed that deletion of 9 C-terminal residues led to a 15-fold decrease in the affinity of the RhoA/RhoGDI interaction (Fig. 2, B and C). Further deletion of C-terminal residues did not significantly decrease the affinity (supplemental Fig. S1, A and B). These data suggest that similar to the Rab GTPases, the C terminus of the RhoA significantly contributes to the affinity of the RhoGDI·Rho complex (36). The presence of a lysine- and arginine-rich patch in the C terminus of other Rho GTPases suggests that this is a generic feature that may be important for RhoGDI-mediated membrane recycling of Rho GTPases, because it is also involved in the interaction with negatively charged lipids (22, 37).
FIGURE 2.
Thermodynamic analysis of RhoGDI interaction with C-terminal-truncated RhoA mutants. A, the alignment of human RhoA, Cdc42, and Rac1 protein sequences. The prenylatable cysteine is highlighted in bold, the gray background represents conservative sequence of positively charged amino acid residues, and the C-terminal α-helix is underlined. The C-terminal truncation of RhoA are indicated by arrows at corresponding positions. Titration of 10 μm solution of wild-type RhoA (B) or RhoA with a C-terminal deletion of 13 residues (C) with RhoGDI.
Fluorescent Prenylated RhoA as a Sensor of RhoGDI/Rho Interaction
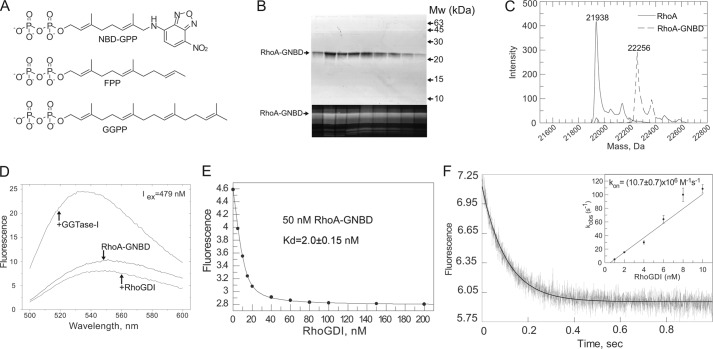
To elucidate the mechanism underlying RhoGDI-mediated delivery and extraction of Rho GTPases, it is important to characterize RhoGDI interactions with prenylated Rho proteins. Hence, we sought to develop a fluorescent sensor that would report on the interaction of RhoGDI with prenylated RhoGTPases. This is a challenging task, because prenylated proteins display low water solubility, which largely precludes their direct biophysical analysis. All previous experiments relied on detergent-solubilized prenylated proteins, a situation that is known to strongly change the properties of prenylated proteins (41, 42). To alleviate this problem, we took advantage of synthetic phosphoisoprenoids modified with the fluorescent NBD group. These analogs of FPP and GGPP can transfer fluorescent lipid moieties onto proteins by protein prenyltransferases. These are less hydrophobic than the native lipids due to the shorter isoprenoid chain (28) (Fig. 3A). We enzymatically prenylated the RhoA protein using recombinant GGTase-I and NBD-geranyl pyrophosphate as a lipid donor. The reaction mixture was resolved on a size exclusion column, and the fluorescently prenylated RhoA was eluted at a position corresponding to a 20-kDa monomer. The protein was homogeneous (Fig. 3B) and remained soluble in the absence of detergent, even at concentrations above 10 mg/ml. ESI-MS analysis of the fractions revealed that the majority of protein was modified with geranyl-NBD (Fig. 3C). Purified RhoA-GNBD was also correctly folded, because it was able to form a stoichiometric complex with RhoGDI, as determined by gel filtration analysis (supplemental Fig. S2A).
FIGURE 3.
Construction and analysis of prenylated fluorescent RhoA sensor. A, chemical structure of NBD-GPP in comparison with structures of FPP and GGPP. B, SDS-PAGE analysis of RhoA-GNBD eluted from Superdex 200 size exclusion column. The upper panel shows a gel stained with Coomassie Blue, whereas the lower panel shows a fluorescent scan of the same gel (excitation laser, 473 nm; cutoff filter, 510 nm). C, ESI-MS analysis of RhoA-GNBD. D, emission spectra of 50 nm solution of RhoA-GNBD before or after addition of 1 μm RhoGDI or GGTase-I. E, titration of RhoGDI to 50 nm solution of RhoA-GNBD. The fluorescence of the NBD group was excited at 479 nm, and the emission was collected at 560 nm. The Kd value of 2 nm was calculated by fitting data to the quadratic equation using Grafit 5.0. F, kinetic analysis of RhoGDI interaction with the RhoA-GNBD. The graph represents a typical time course of fluorescence signal changes upon rapid mixing of 100 nm RhoA-GNBD and 1.5 μm RhoGDI at 25 °C. The solid curve shows the single-exponential fit to the data. The inset represents a plot of the observed pseudo-first order rate constant kobs versus concentration of RhoGDI.
Addition of saturating concentrations of RhoGDI to a solution of RhoA-GNBD resulted in an ∼1.5-fold decrease in NBD fluorescence that could be used for titration experiments (Fig. 3D). Fitting the data to a quadratic equation led to a Kd value of 2 nm (Fig. 3E). To confirm this affinity estimate by an independent method and gain insight into the kinetics of the interaction, we rapidly mixed both proteins in a stopped-flow apparatus. The observed trace could be fitted using a single exponential function (Fig. 3F). A linear fit of the observed first-order association rate constants at a range of concentrations allowed us to determine a kon value of 10.7 × 106 m−1 s−1 for this reaction. To obtain the koff value for this interaction, we mixed 100 nm RhoGDI·Rho-GNBD complex with 1 μm farnesylated RhoA. This resulted in a time-dependent increase in fluorescence, reflecting dissociation of RhoGDI·RhoA-GNBD and formation of the RhoGDI·Rho-F complex. The fit of the data resulted in a dissociation rate constant of 0.06 s−1 (supplemental Fig. S2B). From the obtained kinetic parameters, we calculated a Kd value of 5.6 nm, which is in good agreement with the equilibrium titration experiments. These data demonstrate that RhoA-GNBD binds to RhoGDI with an affinity higher than that corresponding to Kd of 7, 30, or 180 nm, respectively, estimated for RhoGDI interactions with the native geranylgeranylated Rho protein Cdc42 (15, 16, 38). Taken together, these results suggest that NBD-G mimics the native isoprenoid and that the developed sensor can be used to analyze RhoA/RhoGDI interactions.
Interaction of Farnesylated RhoA with RhoGDI
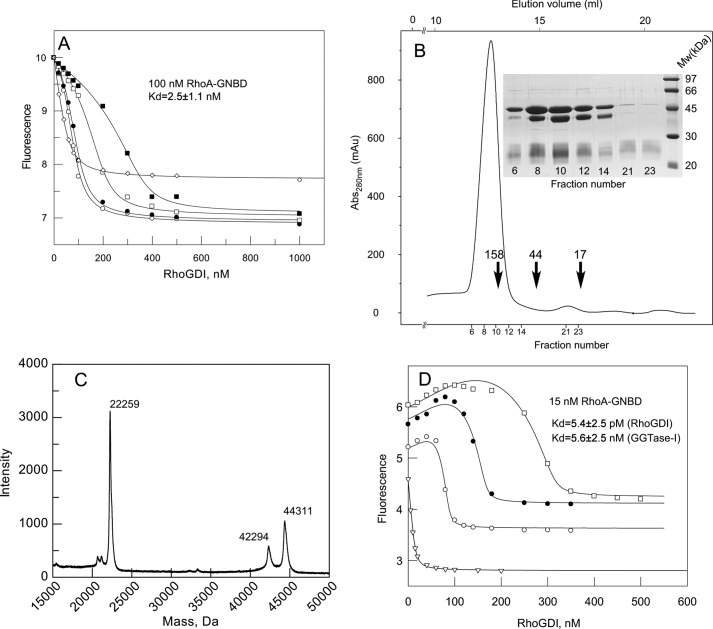
Several Rho proteins can exist in both farnesylated and geranylgeranylated forms in vivo (39–41, 45, 46). Thus, it is expected that RhoGDI will govern the interaction of both prenylated forms with membranes, but the biophysical parameters may differ significantly. To analyze the interaction of farnesylated RhoA with RhoGDI, we generated the RhoAL193A mutant, which is a more efficient FTase substrate than wild-type of RhoA. The protein was in vitro farnesylated by recombinant mammalian FTase and purified by gel filtration (supplemental Fig. S3). As expected, farnesylated RhoAL193A (RhoA-F) was soluble at concentrations above 1 mg/ml. To determine the affinity of RhoA-F for RhoGDI, we titrated 100 nm RhoA-GNBD with increasing concentrations of RhoGDI in the presence of 50, 100, 200, or 400 nm GDP bound RhoA-F (Fig. 4A). The titration curves showed an initial “lag” that was most prominent at high concentrations of RhoA-F. This suggests that RhoGDI preferentially forms a complex with the fluorescently silent RhoA-F. Data analysis was performed via least square fitting using a 3-component competitive model (42). The fit to the data shown in Fig. 4A yielded a Kd value of 2.5 nm for the RhoGDI interaction with farnesylated RhoA. These data suggest that farnesylated RhoA has the capacity to form a tight complex with RhoGDI in vivo.
FIGURE 4.
Interaction analysis of prenylated RhoA·GDP with RhoGDI. A, titration of the RhoGDI into a mixture of fluorescently labeled 100 nm RhoA-GNBD (λex/em 479/560 nm) in the absence (open diamonds) and presence of increasing concentrations of RhoA-F: 50 (open circles), 100 (filled circles), 200 (open squares), and 400 nm (filled squares). The data were fitted to a competitive model, resulting in a Kd value of 2.5 nm. B, purification of the RhoA-GG·GGTase-I complex by size exclusion chromatography and SDS-PAGE analysis of complex containing fractions. Arrows indicate elution volumes of molecular weight standards. C, the MALDI-MS analysis of the purified RhoA-GG·GGTase-I complex. D, titration of RhoGDI to a 15 nm solution of RhoA-GNBD in the absence (open triangles) or presence of 100 (open circles), 200 (filled circles), or 400 nm (open squares) RhoA-GG·GGTase-I complex. The data were fitted globally by numerical simulation to a competitive binding model in the program Dynafit 4.0.
Preparation of Solution-stabilized Geranylgeranylated RhoA and Analysis of Its Interaction with RhoGDI
The data described above demonstrate that prenylation of RhoA significantly increases its affinity for RhoGDI. However, so far, it is unclear to what extent an increase in the length of the isoprenoid chain would influence the strengths of this interaction. This is particularly important because Rho proteins are predominantly geranylgeranylated. In contrast to RhoA-GNBD and farnesylated RhoA, geranylgeranylated RhoA is insoluble in water and rapidly aggregates. To overcome this problem, we took advantage of the observation that prenyltransferases form stable complexes with their reaction products in the absence of and excess of lipid donor (43). Hence, we geranylgeranylated RhoA in vitro with recombinant GGTase-I using stoichiometric amounts of both proteins and purified the resulting complex by size exclusion chromatography. The proteins coeluted as a stoichiometric complex at a position corresponding to a molecular mass of 160 kDa (Fig. 4B). MALDI-MS analysis demonstrated that the majority of RhoA in the complex was geranylgeranylated (Fig. 4C). The availability of solubilized RhoA-GG enabled us to measure the affinity of the RhoGDI interaction with RhoA-GG using RhoA-GNBD as a fluorescent reporter. In this experiment, we titrated RhoGDI to a mixture of 50 nm RhoA-GNBD with 100, 200, or 400 nm of the RhoA-GG·GGTase-I complex. As depicted in Fig. 4D, the titration resulted in an initial increase of fluorescence, followed by its quenching. This can be interpreted as the initial displacement of GGTase-I from the RhoA-GG complex by RhoGDI and the association of GGTase-I with RhoA-GNBD. At higher concentrations, however, all RhoA-GNBD became sequestered by RhoGDI, leading to a signal decrease. We confirmed this by analyzing the interaction of GGTase-I with RhoA-GNBD (supplemental Fig. S4). The availability of the affinity values for the RhoGDI and GGTase-I interactions with RhoA-GNBD and the corresponding fluorescence yields prompted us to attempt to extract the affinity for the RhoA-GG interaction with RhoGDI and GGTase-I from these data. This is a challenging task because it requires a quantitative description of a 4-component system with 2 sets of competitive interactions, i.e. RhoA-GNBD + RhoGDI ⇔ RhoA-GNBD·RhoGDI and RhoA-GNBD + GGTase I ⇔ RhoA-GNBD·GGTase I, and RhoA-GG + RhoGDI ⇔ RhoA-GG·RhoGDI and RhoA-GG + GGTaseI ⇔ RhoA-GG·GGTase I, related to each other in a nonlinear fashion. Implementing a recursive least square minimization in Dynafit (Biokin Ltd.) (34) allowed us to calculate the Kd values for the interactions of native prenylated RhoA with RhoGDI and GGTase I. The fitting procedure consistently gave us differences of 3 orders of magnitude in the affinities of RhoA-GG/GGTase I and RhoA-GG/RhoGDI interactions, the values of which are 5.4 ± 2.5 nm and 5.6 ± 4.5 pm, respectively.
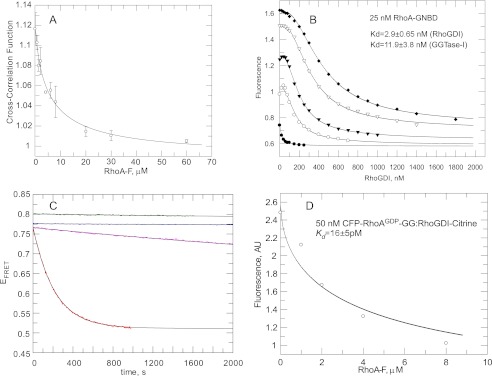
The affinity for the RhoA-GG/RhoGDI interaction was unexpectedly high, particularly in the view that RabGDI interacts with GDP-bound diprenylated Rab proteins with an affinity nearly 500 times lower (9). Thus, we sought to confirm the RhoA-GG/RhoGDI dissociation constant value by an independent and more direct method. We decided to use fluorescence cross-correlation spectroscopy (FCCS), because it allows direct monitoring of interacting molecules at very low concentrations. To this end, we genetically fused the RhoA N terminus with CFP and produced the proteins in pure recombinant form. We then took advantage of the fact that RhoGDI contains only one solution-exposed cysteine residue (Cys-79) that is located on the loop after β-strand A and that is not involved in interaction with Rho GTPases. Thus modification of this residue does not affect its association with RhoA (38). We chemically labeled RhoGDI with tetramethylrhodamine-maleimide (RhoGDI-TMR). The complex of RhoGDI-TMR with geranylgeranylated GDP-bound CFP-RhoA was formed by mixing both components and purifying the resulting complex by gel filtration (supplemental Fig. S5). As a measure of CFP-RhoA-GG·RhoGDI-TMR complex concentration in the FCCS analysis, we used the cross-correlation function (CCF) (44). We observed a decrease in the CCF value upon addition of farnesylated RhoA, which indicates competitive formation of the RhoGDI·RhoA-F complex. Fitting the competitive titration curve of the CFP-RhoA-GG·RhoGDI-TMR fluorescent complex with RhoA-F using a 3-component competitive model led to a Kd value of 20 pm (Fig. 5A). This provides direct and independent confirmation of the low picomolar affinity of RhoGDI to geranylgeranylated RhoA.
FIGURE 5.
Interaction analysis of prenylated RhoA interaction with RhoGDI. A, competitive displacement of the CFP-RhoA·GDP-GG·RhoGDI-TMR complex by farnesylated RhoA observed by FCCS. Open circles correspond to the value of the cross-correlation function at the indicated concentrations of RhoA-F. Solid line is the fit of experimental data to a competitive binding model, leading to a Kd value of 21 pm. Bars represent S.D. from two independent measurements. B, titration of RhoGDI into the 25 nm solution of RhoA-GNBD in the absence (filled circles) or presence of 100 (open circles), 200 (filled triangles), or 400 (open triangles), and 600 nm (filled diamonds) of the RhoA·GMPPNP-GG·GGTase-I complex. The data were fitted globally by numerical simulation to a competitive binding model in the program Dynafit 4.0. The Kd values obtained for the RhoA-GG/GGTase-I interaction is ∼12 nm, and for RhoA-GG/GMPPNP/RhoGDI, the affinity is ∼2.9 nm. C, kinetics of dissociation of the CFP-RhoA-GG·RhoGDI-citrine complex, measured by the changes in FRET efficiency (λex 436 nm/λem,DONOR 470 nm, and λem,ACCEPTOR 530 nm). In the experiment, 30 nm CFP-RhoA was displaced from the complex by the addition of 2 μm farnesylated RhoA. Purple curve corresponds to a EFRET change upon dissociation of CFP-RhoA·GDP-GG·RhoGDI-citrine complex, whereas red represents the change in EFRET of dissociating CFP-RhoA·GMPPNP-GG·RhoGDI-citrine. Blue and green curves are EFRET of corresponding complexes alone. Black solid lines are fits to a single exponential function with offset. D, represents titration of CFP-RhoAGDP-GG·RhoGDI-citrine complex with farnesylated RhoA. The fit of experimental data led to a Kd of the CFP-RhoAGDP-GG interaction with RhoGDI of 16 pm.
Interaction of Activated Geranylgeranylated RhoA with RhoGDI
The current model of GTPase action postulates that GTPases are activated upon membrane delivery and recycled to the cytosol following completion of the functional cycle, marked by GTP hydrolysis. However, several reports suggest that RhoGDI can also efficiently interact with GTP-loaded Rho proteins (10, 11, 15, 16) and promote their release from the intracellular membranes (45, 46). We therefore measured the affinity of RhoGDI for the activated form of prenylated RhoA. We prepared the GMPPNP-bound form of geranylgeranylated RhoA using the approach described for the production of the GDP-bound form of RhoA-GG. We then titrated RhoGDI to a mixture of 25 nm RhoA-GNBD with 100, 200, 400, or 600 nm of the RhoA·GMPPNP-GG·RhoGDI complex (Fig. 5B). Global fit of the titration data resulted in a Kd value of 3 nm for the RhoA/GMPPNP-GG/RhoGDI interaction, whereas the affinity of RhoA-GG interaction with GGTase-I was slightly lower for the GMPPNP-bound form of RhoA, with a Kd value of 12 nm. Thus, RhoGDI displays a 500-fold difference in affinity between the GDP- and GTP-bound forms of geranylgeranylated RhoA. Our data demonstrate that similar to RabGTPases, the nucleotide-bound state of RhoA dramatically affects its affinity for its GDI. Although the discrimination factor is somewhat less than in the Rab/RabGDI interaction (about 2 × 103), it is still large and likely to play an important biological role. The fact that affinities for both forms are so high provides an explanation for previous confusion in the field, because most of the biochemical methods of protein interaction analysis cannot distinguish between low nanomolar and picomolar affinities.
Analysis of Dissociation Rates of RhoGDI·RhoA-GG Complexes
The postulated biological role of RhoGDI is to sequester the Rho proteins in the cytosol and to transport them to a target membrane. Although the exact time scale of this process is unknown, it is expected to be on the order of minutes (47). This would require Rho-GG·RhoGDI complexes to have reasonably fast intrinsic or induced off-rates (10−2 to 10−3 s−1). This notion promoted us to determine the dissociation rates of the RhoA-GG·RhoGDI complex experimentally, because the determined low picomolar affinity for RhoA·GDP containing- and low-nanomolar affinity for RhoA·GMPPNP-containing complexes is most likely to reflect their slow off-rate. Unfortunately, the developed 4-component system, based on RhoA-GNBD, is not suitable for kinetic measurements due to a large number of unknown kinetic parameters. Thus, we developed an alternative sensor in which the fluorescence resonance energy transfer occurs from a CFP domain that is fused to the N terminus of RhoA to the citrine domain that is fused C-terminal to RhoGDI (supplemental Fig. S6A). ITC analysis showed that the unprenylated CFP-RhoA·RhoGDI-citrine complex has an affinity only 3 times lower than the native RhoA·RhoGDI complex (supplemental Fig. S6B). Moreover, a competitive titration of the CFP-RhoA-GG·RhoGDI-citrine complex with farnesylated RhoA resulted in a Kd value of 16 ± 5 pm (Fig. 5C, inset), indicating that it behaves similarly to the native complex. The dissociation rate was measured by mixing 30 nm of complexes containing either GDP- or GMPPNP-associated RhoA with 5 μm farnesylated RhoA and monitoring the change in FRET efficiency (Fig. 5C and supplemental Fig. S6, E and F). As can be seen in Fig. 5C, both the CFP-RhoA·GDP-GG·RhoGDI-citrine and CFP-RhoA·GMPPNP-GG·RhoGDI-citrine complexes are very stable, but the addition of excess of RhoA-F led to their dissociation, which is reflected in the decay of FRET efficiency. Remarkably, the extremely slow dissociation rate of the CFP-RhoA·GDP-containing complex (2 × 10−4 s−1) is increased by a factor of only 25 (Table 1), meaning that the half-life of the CFP-RhoA·GMPPNP-GG·RhoGDI-citrine complex is 2.5 min. This relatively modest decrease in the half-life of the complex despite almost 3 orders of magnitude change in the affinity on replacing GDP by GMPPNP means that the association rate constants for the complexes also differ by a factor of about 25 (Table 1). The rate constant for the dissociation of CFP-RhoA·GDP-containing complex (2 × 10−4 s−1) means that 50% dissociation will take about 1 h, which is probably too slow to allow the complex to operate in cellular systems that require responses on a time scale of minutes.
Crystal Structure of the RhoA·GMPPNP-GG·RhoGDI Complex
To gain insight into the molecular basis of RhoA·GTP recognition by RhoGDI, we attempted a structural analysis of the activated form of the complex. Using the in vitro prenylation approach described above, we generated preparative amounts of the RhoA·GMPPNP-GG·RhoGDI complex and subjected it to crystallization trials and diffraction testing, as described under ”Materials and Methods.“ The best diffracting crystal was used to collect the dataset to 2.8 Å using a synchrotron beam source. The structure was solved by molecular replacement using the Cdc42·GDP-GG·RhoGDI structure as a search model (48). Full data collection and refinement statistics are given in Table 2. The overall structure of the complex is shown in Fig. 6A. The structure is remarkably similar to the structure of the Cdc42·GDP-GG·RhoGDI complex (PDB 1DOA), with an overall root mean square deviation on the α-chain of 0.87 Å.
TABLE 2.
Data collection statistics for the structure of geranylgeranylated RhoA·GMPPNP-GG·RhoGDI complex
| Data collection | RhoA·GPPNHP-GG·RhoGDI |
|---|---|
| Wavelength (Å)/beamline | 0.9537/AS-MXII |
| Resolution (highest shell, Å) | 68.0–2.8 (2.9–2.8) |
| Space group | P212121 |
| Cell constants (Å; °) | a = 46.0, b = 71.6, c = 136.0; α = β = γ = 90 |
| VM | |
| Total measurements | 56,189 |
| Unique reflections | 20,741 |
| Average redundancy | 2.7 (2.7) |
| I/σ | 12.3 (3.6) |
| Completeness (%) | 97.0 (98.1) |
| Rsyma | 6.9 (26.5) |
| Wilson B-factor (Å2) | 41.7 |
| Refinement | 2.8 |
| Resolution (highest shell, Å) | 2.8 (2.87–2.8) |
| Rb | 20.6 (20.7) |
| Rfreec | 28.1 (32.5) |
| Root mean square deviation bonds (Å)/angles (°) | 0.015/1.743 |
| B-factor deviation | |
| Bonds/angles (Å2) | |
| Main chain | 0.716/1.415 |
| Side chain | 2.095/3.696 |
| Residues in ramachandran core (%)d | 92.0 |
| Protein atoms | 2997 |
| Solvent atoms | 80 |
| Ligand atoms | 54 |
| Average B-factor (Å2) | 32.7 |
| PDB accession code | |
a Rsym = ΣΣ I(h)j − 〈I(h)〉/ΣΣI(h)j, where I(h)j is the measured diffraction intensity, and the summation includes all observations.
b R is the R-factor = (Σ|Fo| − Σ|Fc|)/Σ|Fo|.
c Rfree is the R-factor calculated using 5% of the data that were excluded from the refinement.
d Ramachandran core refers to the most favored regions in the ϕ/ψ-Ramachandran plot, as defined by Laskowski et al. (61).
FIGURE 6.
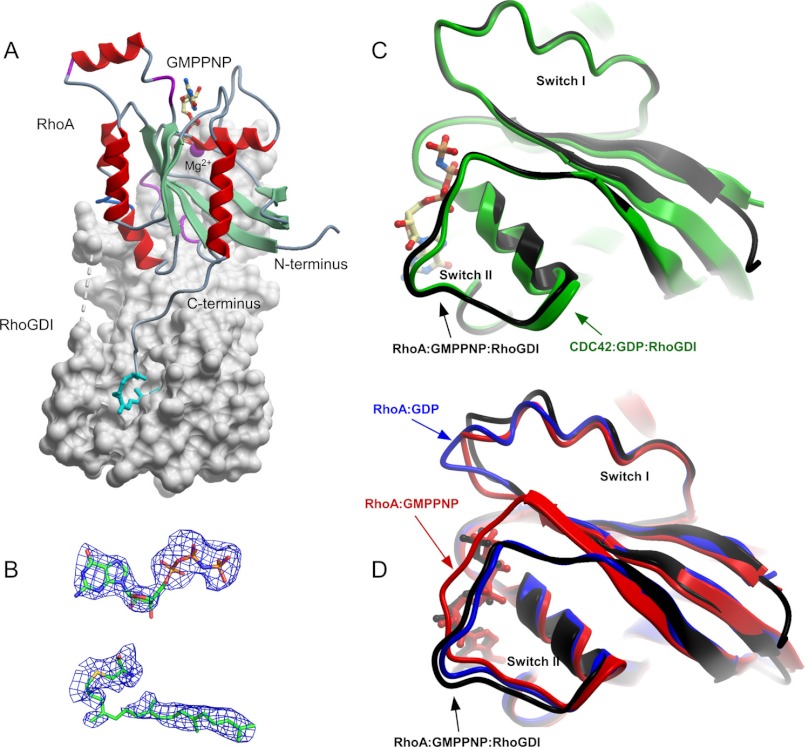
Structure of RhoA·GMPPNP-GG·RhoGDI complex (PDB 4F38). A, overall structure of the complex. RhoGDI is displayed as a gray molecular surface, whereas RhoA is displayed in ribbon representation. The nucleotide and the conjugated geranylgeranyl isoprenoid are displayed in ball-and-stick representation. The Mg2+ is displayed as a space-filling magenta ball. B, 2.5 σ (Fo − Fc) difference in electron density of the bound GMPPNP and geranylgeranylated cysteine at the RhoA C terminus before incorporation into the model. C, superimposition of the prenylated CDC42·GDP and RhoA·GPPNHP complexed with RhoGDI. D, as in C but RhoA·GMPPNP complexed with RhoGDI is superimposed with unbound GMPPNP-associated (red) and GDP-associated (blue) forms of RhoA.
Total interface area in the RhoA·GMPPNP-GG·RhoGDI complex excluding geranylgeranyl·RhoGDI hydrophobic pocket interface is somewhat smaller than in the structure of Cdc42·GDP-GG·RhoGDI complex (1340 versus 1580 Å2) with a significantly lower number of hydrogen bonds and salt bridges forming the RhoA·GMPPNP-GG·RhoGDI complex. These differences predominantly come from extensive interaction of RhoGDI with residues Arg-186 and Arg-187 of Cdc42. The C terminus of RhoA does not have a diarginine patch, instead there are 3 lysine residues that do not form hydrogen bonds with RhoGDI and are solely involved in the electrostatic interaction.
The position of geranylgeranyl moiety buried into the hydrophobic pocket on the RhoGDI is nearly identical to what is identified on the structure of Cdc42-GG·GDP·RhoGDI. The minor differences such as involvement of RhoGDILeu-170 in interaction with the RhoA geranylgeranyl group are unlikely to be functionally significant. Despite the differences in the interface of RhoGDI complexes with RhoA and Cdc42 the total free binding energy calculated by PISA (35) is similar for both complexes: 25.6 kJ/m for RhoA·RhoGDI and 27.4 kJ/m for Cdc42·RhoGDI.
The Rho-specific helix in the RhoA·GDP-GG·RhoGDI structure has different conformations than both in the free form of Cdc42 and in the Cdc42 complex with RhoGDI. In contrast to the Cdc42 the Rho insert in the structures of free RhoA and in complex with RhoGDI has an almost identical position. This might reflect differences in the signaling modes of the RhoA and Cdc42 GTPases in the cell (49).
Strikingly, the switch I and II loops of RhoA·GMPPNP adopt a nearly identical conformation to those of Cdc42·GDP in complex with RhoGDI. As can be seen in Fig. 6C, the conformations of both switch I and switch II regions in the complex differ markedly from the conformations observed in unliganded RhoA·GMPPNP. Although switch I adopts a conformation nearly identical to that of GDP-bound RhoA, the switch II region adopts a conformation distinct from both free forms, due to the extensive contacts formed with the N terminus of RhoGDI. The phosphate groups of the bound nucleotide are shifted slightly toward the exterior of the protein, possibly reflecting the fit induced by the complex formation. Overall, the guanine nucleotide in the structure of RhoA·GMPPNP-GG·RhoGDI has a 15% smaller interaction interface and less number of hydrogen bonds than in the structure of RhoAG14V·GTPγS alone (50). For instance, there is no interaction between phosphate oxygen atoms and Ala-15 from the P-loop, switch I residues Tyr-34 and Thr-37 are no longer involved in nucleotide γ-phosphate positioning. Interestingly, important for GTP hydrolysis, the Gln-63 residue in the RhoA·GMPPNP-GG·RhoGDI structure is located closer than in RhoAG14V·GTPγS, which results in hydrogen bond formation between its amide group and the oxygen of γ-phosphate with an overall distance of 3.84 Å.
The observed structure demonstrates that even in the GTP-bound form, the loop regions of RhoGTPases are sufficiently flexible to adopt an inactive conformation upon binding to RhoGDI. The resulting conformational strain can be compensated by the very high binding energy of the complex formation and results in a complex that is structurally identical to the inactive conformation but displays a faster dissociation rate.
DISCUSSION
In the present study, we have performed a comprehensive analysis of the interaction of RhoGDI with prenylated and unprenylated forms of RhoA using kinetic and thermodynamic measurements. We have focused on the role of prenylation and the nucleotide-bound state on complex formation and its implications for recycling RhoGTPases.
We constructed a sensor of the RhoGDI/RhoGTPase interaction by in vitro prenylation of RhoA with the NBD-derivatized fluorescent isoprenoid. The developed enzymatic route is significantly simpler than the previously described semisynthesis-based approach for construction of fluorescent lipidated proteins (51) and can be easily extended to other GTPases. Using the developed sensor, we were able to determine the affinities of geranylgeranylated and farnesylated RhoA for RhoGDI. The affinities for RhoA-GG/RhoGDI interactions were much higher than previously reported (16), with RhoGDI binding to GDP-bound geranylgeranylated RhoA with a Kd value of 5 pm. We demonstrated that the length of the isoprenoid chain has a dramatic impact on the RhoA interaction with RhoGDI. The affinity difference of nearly 3 orders of magnitude between farnesylated and geranylgeranylated complexes indicates that farnesylated and geranylgeranylated Rho proteins will be regulated quite differently by RhoGDI in vivo. Clearly, in the absence of a large pool of free RhoGDI, farnesylated Rhos will be outcompeted by the geranylgeranylated form and therefore are likely to distribute through alternative mechanisms. We also demonstrate that activation of RhoA results in a 500-fold decrease in the affinity of the complex. The high affinity of RhoGDI complexes with both GDP- and GMPPNP-bound prenylated RhoA molecules implies their slow dissociation rates, which we found to be 10−4 and 10−2 s−1, respectively. These results have revealed that the RhoA·GDP-GG·RhoGDI complex is very tight and has a half-life of ∼60 min. In contrast, the half-life of the GMPPNP-loaded RhoA-GG·RhoGDI complex is only 2.5 min, which fits better into the physiological context (47) and suggests the presence of two types of RhoGDI·RhoA complexes in vivo with dramatically different half-lives. These findings further support the idea that RhoGDI is actively involved in the recycling and distribution of activated RhoGTPases in the cell (52, 53). To gain insight into the mechanism underlying the ability of RhoGDI to stably associate with the activated form of RhoA, we solved the structure of the activated form of the complex and demonstrated that complex formation induces adoption of the inactive conformation without nucleotide hydrolysis. Formation of an extensive set of contacts between RhoGDI and switch II partially through hydrophobic interaction with Leu-67 and Leu-70 of RhoA stabilizes the loop and, importantly, secures the essential GTP-hydrolysis Gln-63 residue, which results in inhibition of GTPase hydrolytic activity (11). The nucleotide independence of the conformation of switch II, which forms the second largest interface in the RhoA·GMPPNP-GG·RhoGDI complex, most likely promotes association of the proteins, which involves energetically unfavorable changes in the switch I region. Notably, the role of switch II in the recognition of both nucleotide-bound forms of Rho GTPases by RhoGDI was suggested earlier (48). The structural transition of the switch I loop occurs at the cost of interaction the free energy, which is reflected by a difference of almost 3 orders of magnitude in affinities for RhoA-GG·RhoGDI complexes (Table 1). The association of RhoGDI with activated RhoA in solution is 25 times slower than with inactive RhoA, because it requires a structural transformation of the latter molecule. Additionally, the dissociation rate of such a complex is faster by almost the same factor, due to the strain in switches I and II. The ITC data, furthermore, demonstrate that the binding enthalpy of RhoGDI to unprenylated RhoA·GMPPNP is 2-fold lower than the enthalpy of the RhoGDI interaction with the GDP-bound RhoA protein. Comparison of the structures of the Cdc42·GDP-GG·RhoGDI and RhoA·GMPPNP-GG·RhoGDI complexes indicates the lack of major changes in the interaction surface area. Thus, change in the binding enthalpy most likely reflects the free energy cost of the conformational change in the switch I loop that is required for RhoA·GMPPNP·RhoGDI complex formation.
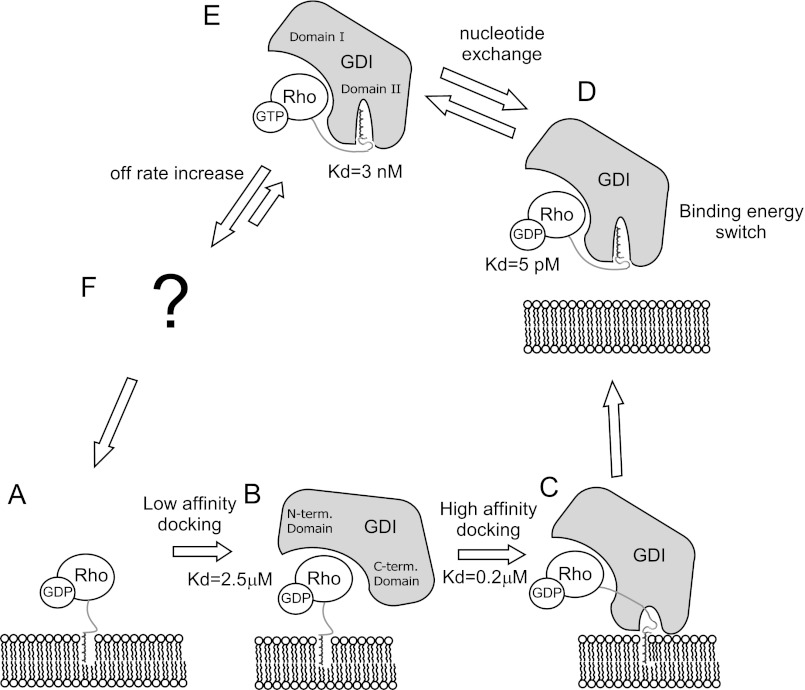
The results obtained in this work, in combination with data from previous biochemical and crystallographic studies, allow us to formulate a detailed mechanistic model of the RhoGDI-mediated recycling of prenylated RhoGTPases in vivo. Our analysis of the interaction of unprenylated RhoA with RhoGDI provides insight into the initial stages of RhoA extraction from the membranes, because the isoprenoid moiety is embedded into the membrane and unlikely to contribute to the interaction at this stage. It is likely that RhoGDI initially forms a micromolar-affinity complex with RhoA by recognizing the switch II and I regions but not the C terminus. Next, the positively charged C terminus of RhoA binds to the negatively charged residues in the C-terminal RhoGDI domain. This increases the RhoA·RhoGDI complex residence time on the membrane and allows progression to the next step in extraction, in which the isoprenoid group spontaneously flips out of the membrane and is captured by the lipid-binding pocket of RhoGDI. This eventually triggers release of the complex from the membrane (Scheme 1). In general, RhoGDI and RabGDI appear to operate in a similar way in extracting the respective GTPases from membranes, using sequential formation of progressively tighter complexes that function as a thermodynamic trap. However, the quantitative data suggest that Rho and RabGDI operate differently in the sense that the former distributes both nucleotide-bound forms between membranes and cytosol, whereas the latter acts only on inactive conformations, albeit with quantitative differences. Given the exceptionally high affinities of RhoGDI complexes with prenylated RhoA of both nucleotide-bound forms, the local concentration of RhoGDI in the cell will govern the extraction process of GDP- and GTP-bound Rho proteins. Low and limiting RhoGDI concentrations will mostly lead to the extraction of GDP-associated Rho proteins that will outcompete short-lived and weak RhoA·GTP·RhoGDI complexes. In contrast, a local abundance of RhoGDI will result in extraction of both inactive and activated RhoGTPases. The transient RhoGDI association with GTP-bound Rho molecules will favor its local distribution. The presence of slow and fast cycling pools of the RhoGTPase cycle in the cell was proposed earlier (27, 53, 54). The mechanism of dissociation of these complexes is unclear at present and, according to our data, cannot be based solely on nucleotide exchange. Therefore, unlike the case of RabGTPases, where GEFs have been suggested to be sufficient to drive targeted membrane insertion of Rabs, it seems likely that dissociation of RhoGDI·RhoGTPase complexes must be further accelerated by other factors. Recently demonstrated Arl-GTP-mediated displacement of farnesylated Rheb from its GDI-like chaperon PDEδ provides an example of a possible mechanism (60). Alternative factors may include some types of lipids and phosphorylation of both Rho and RhoGDI. Therefore, further research is needed in this area to fully understand the role of RhoGDI in the membrane targeting and recycling of RhoGTPases in the cell.
Scheme 1.
Mechanism of RhoA extraction by RhoGDI.
This work was supported in part by Australian Research Council DP Grant DP1094080, ARC FF Grant FT0991611, and National Health Research Council project Grant 569652 (to K. A.).

This article contains supplemental Figs. S1–S6.
The atomic coordinates and structure factors (code 4F38) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- GEF
- guanine nucleotide exchange factor
- GDI
- GDP dissociation inhibitor
- GMPPNP
- guanosine 5′-[β,γ-imido] triphosphate
- ITC
- isothermal titration calorimetry
- FCCS
- fluorescence cross-correlation spectroscopy
- TMR
- tetramethylrhodamine
- CCF
- cross-correlation function
- GGPP
- geranylgeranyl pyrophosphate
- FPP
- farnesyl pyrophosphate.
REFERENCES
- 1. Ridley A. J., Paterson H. F., Johnston C. L., Diekmann D., Hall A. (1992) The small GTP-binding protein Rac regulates growth factor-induced membrane ruffling. Cell 70, 401–410 [DOI] [PubMed] [Google Scholar]
- 2. Yamana N., Arakawa Y., Nishino T., Kurokawa K., Tanji M., Itoh R. E., Monypenny J., Ishizaki T., Bito H., Nozaki K., Hashimoto N., Matsuda M., Narumiya S. (2006) The Rho-mDia1 pathway regulates cell polarity and focal adhesion turnover in migrating cells through mobilizing Apc and c-Src. Mol. Cell. Biol. 26, 6844–6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hill C. S., Wynne J., Treisman R. (1995) The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell 81, 1159–1170 [DOI] [PubMed] [Google Scholar]
- 4. Saci A., Cantley L. C., Carpenter C. L. (2011) Rac1 regulates the activity of mTORC1 and mTORC2 and controls cellular size. Mol. Cell 42, 50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schmidt A., Hall A. (2002) Guanine nucleotide exchange factors for Rho GTPases. Turning on the switch. Genes Dev. 16, 1587–1609 [DOI] [PubMed] [Google Scholar]
- 6. Bernards A. (2003) GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim. Biophys. Acta 1603, 47–82 [DOI] [PubMed] [Google Scholar]
- 7. Roberts P. J., Mitin N., Keller P. J., Chenette E. J., Madigan J. P., Currin R. O., Cox A. D., Wilson O., Kirschmeier P., Der C. J. (2008) Rho family GTPase modification and dependence on CAAX motif-signaled posttranslational modification. J. Biol. Chem. 283, 25150–25163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goldberg J. (1998) Structural basis for activation of ARF GTPase. Mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell 95, 237–248 [DOI] [PubMed] [Google Scholar]
- 9. Wu Y. W., Oesterlin L. K., Tan K. T., Waldmann H., Alexandrov K., Goody R. S. (2010) Membrane targeting mechanism of Rab GTPases elucidated by semisynthetic protein probes. Nat. Chem. Biol. 6, 534–540 [DOI] [PubMed] [Google Scholar]
- 10. Hart M. J., Maru Y., Leonard D., Witte O. N., Evans T., Cerione R. A. (1992) A GDP dissociation inhibitor that serves as a GTPase inhibitor for the Ras-like protein Cdc42Hs. Science 258, 812–815 [DOI] [PubMed] [Google Scholar]
- 11. Hancock J. F., Hall A. (1993) A novel role for RhoGDI as an inhibitor of GAP proteins. EMBO J. 12, 1915–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chuang T. H., Xu X., Knaus U. G., Hart M. J., Bokoch G. M. (1993) GDP dissociation inhibitor prevents intrinsic and GTPase activating protein-stimulated GTP hydrolysis by the Rac GTP-binding protein. J. Biol. Chem. 268, 775–778 [PubMed] [Google Scholar]
- 13. Tiedje C., Sakwa I., Just U., Höfken T. (2008) The Rho GDI Rdi1 regulates Rho GTPases by distinct mechanisms. Mol. Biol. Cell 19, 2885–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sasaki T., Kato M., Takai Y. (1993) Consequences of weak interaction of Rho GDI with the GTP-bound forms of Rho p21 and Rac p21. J. Biol. Chem. 268, 23959–23963 [PubMed] [Google Scholar]
- 15. Nomanbhoy T. K., Cerione R. (1996) Characterization of the interaction between RhoGDI and Cdc42Hs using fluorescence spectroscopy. J. Biol. Chem. 271, 10004–10009 [DOI] [PubMed] [Google Scholar]
- 16. Johnson J. L., Erickson J. W., Cerione R. A. (2009) New insights into how the Rho guanine nucleotide dissociation inhibitor regulates the interaction of Cdc42 with membranes. J. Biol. Chem. 284, 23860–23871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yamashita T., Tohyama M. (2003) The p75 receptor acts as a displacement factor that releases Rho from Rho-GDI. Nat. Neurosci. 6, 461–467 [DOI] [PubMed] [Google Scholar]
- 18. Del Pozo M. A., Kiosses W. B., Alderson N. B., Meller N., Hahn K. M., Schwartz M. A. (2002) Integrins regulate GTP-Rac localized effector interactions through dissociation of Rho-GDI. Nat. Cell Biol. 4, 232–239 [DOI] [PubMed] [Google Scholar]
- 19. Ivetic A., Ridley A. J. (2004) Ezrin/radixin/moesin proteins and Rho GTPase signaling in leucocytes. Immunology 112, 165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maeda M., Matsui T., Imamura M., Tsukita S. (1999) Expression level, subcellular distribution and Rho-GDI binding affinity of merlin in comparison with Ezrin/Radixin/Moesin proteins. Oncogene 18, 4788–4797 [DOI] [PubMed] [Google Scholar]
- 21. Hirao M., Sato N., Kondo T., Yonemura S., Monden M., Sasaki T., Takai Y., Tsukita S. (1996) Regulation mechanism of ERM (ezrin/radixin/moesin) protein/plasma membrane association. Possible involvement of phosphatidylinositol turnover and Rho-dependent signaling pathway. J. Cell Biol. 135, 37–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ugolev Y., Berdichevsky Y., Weinbaum C., Pick E. (2008) Dissociation of Rac1(GDP)·RhoGDI complexes by the cooperative action of anionic liposomes containing phosphatidylinositol 3,4,5-trisphosphate, Rac guanine nucleotide exchange factor, and GTP. J. Biol. Chem. 283, 22257–22271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lang P., Gesbert F., Delespine-Carmagnat M., Stancou R., Pouchelet M., Bertoglio J. (1996) Protein kinase A phosphorylation of RhoA mediates the morphological and functional effects of cyclic AMP in cytotoxic lymphocytes. EMBO J. 15, 510–519 [PMC free article] [PubMed] [Google Scholar]
- 24. Forget M. A., Desrosiers R. R., Gingras D., Béliveau R. (2002) Phosphorylation states of Cdc42 and RhoA regulate their interactions with Rho GDP dissociation inhibitor and their extraction from biological membranes. Biochem. J. 361, 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. DerMardirossian C., Rocklin G., Seo J. Y., Bokoch G. M. (2006) Phosphorylation of RhoGDI by Src regulates Rho GTPase binding and cytosol-membrane cycling. Mol. Biol. Cell 17, 4760–4768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu Y., Moissoglu K., Wang H., Wang X., Frierson H. F., Schwartz M. A., Theodorescu D. (2009) Src phosphorylation of RhoGDI2 regulates its metastasis suppressor function. Proc. Natl. Acad. Sci. U.S.A. 106, 5807–5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tkachenko E., Sabouri-Ghomi M., Pertz O., Kim C., Gutierrez E., Machacek M., Groisman A., Danuser G., Ginsberg M. H. (2011) Protein kinase A governs a RhoA-RhoGDI protrusion-retraction pacemaker in migrating cells. Nat. Cell. Biol. 13, 660–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dursina B., Reents R., Delon C., Wu Y., Kulharia M., Thutewohl M., Veligodsky A., Kalinin A., Evstifeev V., Ciobanu D., Szedlacsek S. E., Waldmann H., Goody R. S., Alexandrov K. (2006) Identification and specificity profiling of protein prenyltransferase inhibitors using new fluorescent phosphoisoprenoids. J. Am. Chem. Soc. 128, 2822–2835 [DOI] [PubMed] [Google Scholar]
- 29. Berrow N. S., Alderton D., Sainsbury S., Nettleship J., Assenberg R., Rahman N., Stuart D. I., Owens R. J. (2007) A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 35, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rak A., Niculae A., Kalinin A., Thomä N. H., Sidorovitch V., Goody R. S., Alexandrov K. (2002) In vitro assembly, purification, and crystallization of the Rab geranylgeranyl transferase-substrate complex. Protein Expr. Purif. 25, 23–30 [DOI] [PubMed] [Google Scholar]
- 31. Simon I., Zerial M., Goody R. S. (1996) Kinetics of interaction of Rab5 and Rab7 with nucleotides and magnesium ions. J. Biol. Chem. 271, 20470–20478 [DOI] [PubMed] [Google Scholar]
- 32. Lakowicz J. R. (2006) Principles of Fluorescence Spectroscopy, Springer, New York [Google Scholar]
- 33. Rigler R., Mets U. (1993) in Diffusion of Single Molecules through a Gaussian Laser Beam (Korppi-Tommola J. E., ed) 1 Ed., pp. 235–248 SPIE, Jyvaskyla, Finland [Google Scholar]
- 34. Kuzmic P. (1996) Program DYNAFIT for the analysis of enzyme kinetic data. Application to HIV proteinase. Anal. Biochem. 237, 260–273 [DOI] [PubMed] [Google Scholar]
- 35. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 36. Ignatev A., Kravchenko S., Rak A., Goody R. S., Pylypenko O. (2008) A structural model of the GDP dissociation inhibitor Rab membrane extraction mechanism. J. Biol. Chem. 283, 18377–18384 [DOI] [PubMed] [Google Scholar]
- 37. Ugolev Y., Molshanski-Mor S., Weinbaum C., Pick E. (2006) Liposomes comprising anionic but not neutral phospholipids cause dissociation of Rac(1 or 2) × RhoGDI complexes and support amphiphile-independent NADPH oxidase activation by such complexes. J. Biol. Chem. 281, 19204–19219 [DOI] [PubMed] [Google Scholar]
- 38. Nomanbhoy T. K., Erickson J. W., Cerione R. A. (1999) Kinetics of Cdc42 membrane extraction by Rho-GDI monitored by real-time fluorescence resonance energy transfer. Biochemistry 38, 1744–1750 [DOI] [PubMed] [Google Scholar]
- 39. Armstrong S. A., Hannah V. C., Goldstein J. L., Brown M. S. (1995) CAAX geranylgeranyl transferase transfers farnesyl as efficiently as geranylgeranyl to RhoB. J. Biol. Chem. 270, 7864–7868 [DOI] [PubMed] [Google Scholar]
- 40. Takeda N., Kondo M., Ito S., Ito Y., Shimokata K., Kume H. (2006) Role of RhoA inactivation in reduced cell proliferation of human airway smooth muscle by simvastatin. Am. J. Respir. Cell Mol. Biol. 35, 722–729 [DOI] [PubMed] [Google Scholar]
- 41. Chiba Y., Sato S., Hanazaki M., Sakai H., Misawa M. (2009) Inhibition of geranylgeranyltransferase inhibits bronchial smooth muscle hyperresponsiveness in mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 297, L984–L991 [DOI] [PubMed] [Google Scholar]
- 42. Alexandrov K., Scheidig A. J., Goody R. S. (2001) Fluorescence methods for monitoring interactions of Rab proteins with nucleotides, Rab escort protein, and geranylgeranyltransferase. Methods Enzymol. 329, 14–31 [DOI] [PubMed] [Google Scholar]
- 43. Tschantz W. R., Furfine E. S., Casey P. J. (1997) Substrate binding is required for release of product from mammalian protein farnesyltransferase. J. Biol. Chem. 272, 9989–9993 [DOI] [PubMed] [Google Scholar]
- 44. Oyama R., Takashima H., Yonezawa M., Doi N., Miyamoto-Sato E., Kinjo M., Yanagawa H. (2006) Protein-protein interaction analysis by C-terminal specific fluorescence labeling and fluorescence cross-correlation spectroscopy. Nucleic Acids Res. 34, e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Golovanov A. P., Chuang T. H., DerMardirossian C., Barsukov I., Hawkins D., Badii R., Bokoch G. M., Lian L. Y., Roberts G. C. (2001) Structure-activity relationships in flexible protein domains. Regulation of Rho GTPases by RhoGDI and D4 GDI. J. Mol. Biol. 305, 121–135 [DOI] [PubMed] [Google Scholar]
- 46. Dovas A., Couchman J. R. (2005) RhoGDI. Multiple functions in the regulation of Rho family GTPase activities. Biochem. J. 390, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pertz O., Hodgson L., Klemke R. L., Hahn K. M. (2006) Spatiotemporal dynamics of RhoA activity in migrating cells. Nature 440, 1069–1072 [DOI] [PubMed] [Google Scholar]
- 48. Hoffman G. R., Nassar N., Cerione R. A. (2000) Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell 100, 345–356 [DOI] [PubMed] [Google Scholar]
- 49. Abramovitz A., Gutman M., Nachliel E. (2012) Structural coupling between the Rho-insert domain of Cdc42 and the geranylgeranyl binding site of RhoGDI. Biochemistry 51, 715–723 [DOI] [PubMed] [Google Scholar]
- 50. Ihara K., Muraguchi S., Kato M., Shimizu T., Shirakawa M., Kuroda S., Kaibuchi K., Hakoshima T. (1998) Crystal structure of human RhoA in a dominantly active form complexed with a GTP analogue. J. Biol. Chem. 273, 9656–9666 [DOI] [PubMed] [Google Scholar]
- 51. Wu Y. W., Tan K. T., Waldmann H., Goody R. S., Alexandrov K. (2007) Interaction analysis of prenylated Rab GTPase with Rab escort protein and GDP dissociation inhibitor explains the need for both regulators. Proc. Natl. Acad. Sci. U.S.A. 104, 12294–12299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wedlich-Soldner R., Altschuler S., Wu L., Li R. (2003) Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science 299, 1231–1235 [DOI] [PubMed] [Google Scholar]
- 53. Slaughter B. D., Das A., Schwartz J. W., Rubinstein B., Li R. (2009) Dual modes of Cdc42 recycling fine-tune polarized morphogenesis. Dev. Cell 17, 823–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dransart E., Olofsson B., Cherfils J. (2005) RhoGDIs revisited. Novel roles in Rho regulation. Traffic 6, 957–966 [DOI] [PubMed] [Google Scholar]
- 55. Kabsch W. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 56. McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr. D. Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 57. Murshudov G. N., Vagin A. A., Lebedev A., Wilson K. S., Dodson E. J. (1999) Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. D Biol. Crystallogr. 55, 247–255 [DOI] [PubMed] [Google Scholar]
- 58. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 59. Schuttelkopf A. W., van Aalten D. M. (2004) PRODRG. A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 60. Ismail S. A., Chen Y. X., Rusinova A., Chandra A., Bierbaum M., Gremer L., Triola G., Waldmann H., Bastiaens P. I., Wittinghofer A. (2011) Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat. Chem. Biol. 7, 942–949 [DOI] [PubMed] [Google Scholar]
- 61. Laskowski R. A., Moss D. S., Thornton J. M. (1993) Main chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231, 1049–1067 [DOI] [PubMed] [Google Scholar]